
|
||||||
|
Translation Factor: GRB7 (NCBI Gene ID:2886) |
|
 Gene Summary Gene Summary |
| Gene Information | Gene Name: GRB7 | Gene ID: 2886 | Gene Symbol | GRB7 | Gene ID | 2886 |
| Gene Name | growth factor receptor bound protein 7 | |
| Synonyms | - | |
| Cytomap | 17q12 | |
| Type of Gene | protein-coding | |
| Description | growth factor receptor-bound protein 7B47GRB7 adapter proteinepidermal growth factor receptor GRB-7 | |
| Modification date | 20200313 | |
| UniProtAcc | Q14451 | |
 Child GO biological process term(s) under GO:0006412 Child GO biological process term(s) under GO:0006412 |
| GO ID | GO term |
| GO:0017148 | Negative regulation of translation |
| GO:0006417 | Regulation of translation |
| GO:0006412 | Translation |
 Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GRB7 | GO:0030335 | positive regulation of cell migration | 10893408|12021278 |
 Inferred gene age of translation factor. Inferred gene age of translation factor. |
| Gene | Inferred gene age group among (0 - 67.6], (67.6 - 355.7], (355.7 - 733], (733 - 1119.25], >1119.25 |
| GRB7 | (733 - 1119.25] |
Top |
|
 We searched PubMed using 'GRB7[title] AND translation [title] AND human.' We searched PubMed using 'GRB7[title] AND translation [title] AND human.' |
| Gene | Title | PMID |
| GRB7 | . | . |
Top |
|
 Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. For more annotations, please visit our ExonSkipDB. |
 |
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ENST | Exon skip start (DNA) | Exon Skip end (DNA) | ORF |
| ENST00000309156 | 37899150 | 37899307 | Frame-shift |
| ENST00000394209 | 37899150 | 37899307 | Frame-shift |
| ENST00000394211 | 37899150 | 37899307 | Frame-shift |
| ENST00000309156 | 37899645 | 37899723 | In-frame |
| ENST00000394209 | 37899645 | 37899723 | In-frame |
| ENST00000394211 | 37899645 | 37899723 | In-frame |
| ENST00000309156 | 37900805 | 37900916 | In-frame |
| ENST00000394209 | 37900805 | 37900916 | In-frame |
| ENST00000394211 | 37900805 | 37900916 | In-frame |
| ENST00000309156 | 37901495 | 37901576 | In-frame |
| ENST00000394209 | 37901495 | 37901576 | In-frame |
| ENST00000394211 | 37901495 | 37901576 | In-frame |
| ENST00000309156 | 37901674 | 37901791 | In-frame |
| ENST00000394209 | 37901674 | 37901791 | In-frame |
| ENST00000394211 | 37901674 | 37901791 | In-frame |
| ENST00000309156 | 37902165 | 37902253 | Frame-shift |
| ENST00000394209 | 37902165 | 37902253 | Frame-shift |
| ENST00000394211 | 37902165 | 37902253 | Frame-shift |
 Exon skipping position in the amino acid sequence. Exon skipping position in the amino acid sequence. |
| ENST | Exon skip start (DNA) | Exon Skip end (DNA) | Len(transcript seq) | Exon skip start (mRNA) | Exon Skip end (mRNA) | Len(amino acid seq) | Exon skip start (AA) | Exon Skip end (AA) |
| ENST00000394209 | 37899645 | 37899723 | 1972 | 851 | 928 | 532 | 195 | 221 |
| ENST00000394211 | 37899645 | 37899723 | 2110 | 692 | 769 | 532 | 195 | 221 |
| ENST00000309156 | 37899645 | 37899723 | 2260 | 843 | 920 | 532 | 195 | 221 |
| ENST00000394209 | 37900805 | 37900916 | 1972 | 1067 | 1177 | 532 | 267 | 304 |
| ENST00000394211 | 37900805 | 37900916 | 2110 | 908 | 1018 | 532 | 267 | 304 |
| ENST00000309156 | 37900805 | 37900916 | 2260 | 1059 | 1169 | 532 | 267 | 304 |
| ENST00000394209 | 37901495 | 37901576 | 1972 | 1277 | 1357 | 532 | 337 | 364 |
| ENST00000394211 | 37901495 | 37901576 | 2110 | 1118 | 1198 | 532 | 337 | 364 |
| ENST00000309156 | 37901495 | 37901576 | 2260 | 1269 | 1349 | 532 | 337 | 364 |
| ENST00000394209 | 37901674 | 37901791 | 1972 | 1358 | 1474 | 532 | 364 | 403 |
| ENST00000394211 | 37901674 | 37901791 | 2110 | 1199 | 1315 | 532 | 364 | 403 |
| ENST00000309156 | 37901674 | 37901791 | 2260 | 1350 | 1466 | 532 | 364 | 403 |
 Potentially (partially) lost protein functional features of UniProt. Potentially (partially) lost protein functional features of UniProt. |
| UniProtAcc | Exon skip start (AA) | Exon Skip end (AA) | Function feature start (AA) | Function feature end (AA) | Functional feature type | Functional feature desc. |
Top |
|
 Gene expression level across TCGA pancancer Gene expression level across TCGA pancancer |
 |
 Gene expression level across GTEx pantissue Gene expression level across GTEx pantissue |
 |
 Expression level of gene isoforms across TCGA pancancer Expression level of gene isoforms across TCGA pancancer |
 |
 Expression level of gene isoforms across GTEx pantissue Expression level of gene isoforms across GTEx pantissue |
 |
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution Cancer(tissue) type-specific expression level of Translation factor using z-score distriution |
 |
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples) Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples) |
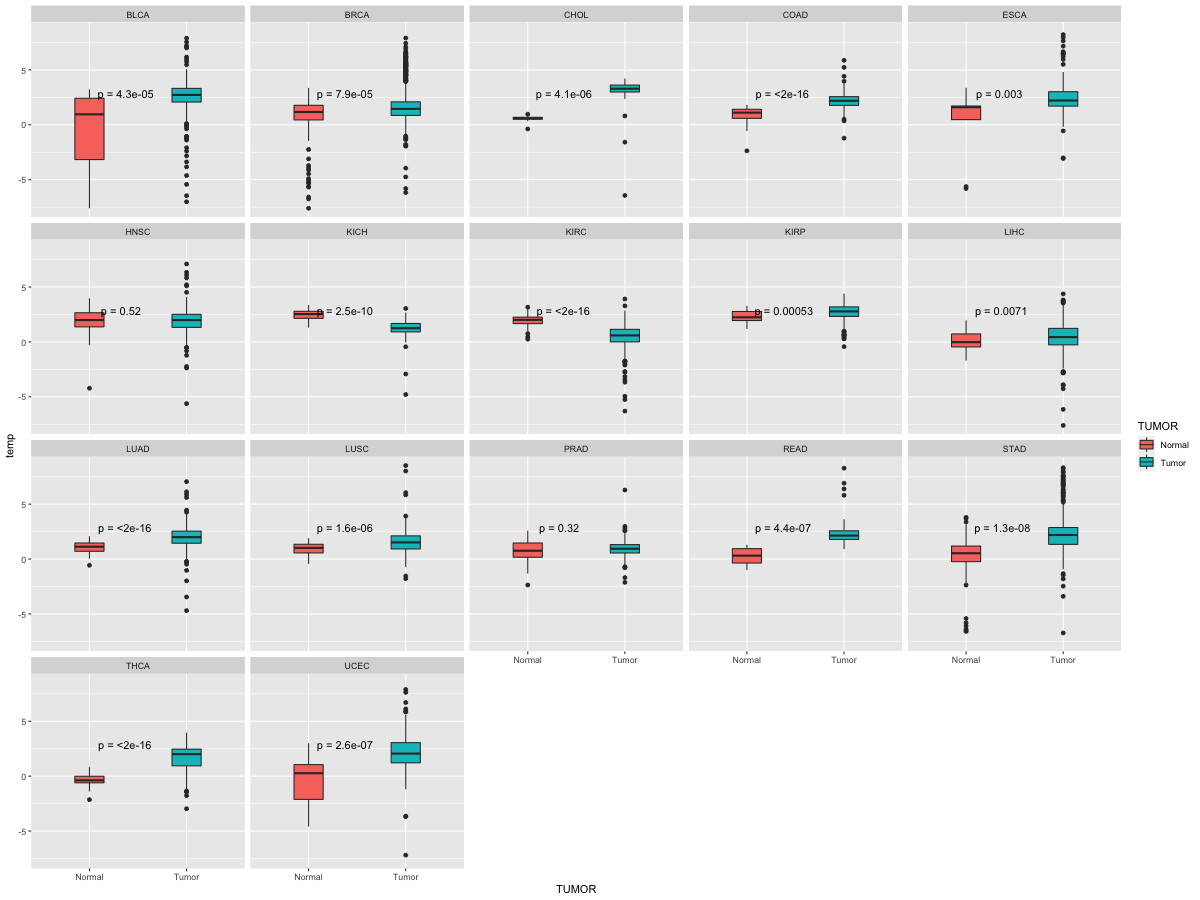 |
| Cancer type | Translation factor | FC | adj.pval |
| CHOL | GRB7 | 2.57684563734882 | 0.00390625 |
| READ | GRB7 | 4.00121793802205 | 0.03125 |
| KIRC | GRB7 | -2.08557276759602 | 1.28432097821042e-12 |
| KICH | GRB7 | -1.35886098374196 | 1.78813934326172e-07 |
| COAD | GRB7 | 1.93627228443447 | 2.98023223876954e-08 |
Top |
|
 Translation factor expression regulation through miRNA binding Translation factor expression regulation through miRNA binding |
| Cancer type | Gene | miRNA | TargetScan binding score (Context++ score percentile) | Coefficient | Pvalue |
 Translation factor expression regulation through methylation in the promoter of Translation factor Translation factor expression regulation through methylation in the promoter of Translation factor |
 |
| Cancer type | Gene | methyl group b | methyl group a | DEG pval | avg methyl in b | avg methyl in a | avg exp in b | avg exp in a |
| BLCA | GRB7 | 2 | 1 | 0.0447308207807389 | 0.293980657430046 | 0.181878666666667 | 0.71526999236041 | 0.010066227079264 |
| LGG | GRB7 | 2 | 3 | 0.0216413026325226 | 0.482977227798054 | 0.654515903054449 | -4.56975424737264 | -4.1956847290954 |
| SKCM | GRB7 | 2 | 3 | 0.0478104805630755 | 0.465116238079019 | 0.669439351851852 | -0.804220094446802 | -0.349431180328222 |
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation) Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation) |
 |
| Cancer type | Gene | methyl group b | methyl group a | DEG pval | avg methyl in b | avg methyl in a | avg exp in b | avg exp in a |
| TGCT | GRB7 | 2 | 1 | 0.0400601136430614 | 0.329289012426423 | 0.159543315508021 | 0.928562609856006 | 1.18228249982576 |
 Translation factor expression regulation through copy number variation of Translation factor Translation factor expression regulation through copy number variation of Translation factor |
 |
| Cancer type | Gene | Coefficient | Pvalue |
Top |
|
 Strongly correlated genes belong to cellular important gene groups with GRB7 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green) Strongly correlated genes belong to cellular important gene groups with GRB7 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green) |
 |
| Cancer type | Gene group | Translation factor | Correlated gene | Coefficient | Pvalue |
| CHOL | Cell metabolism gene | GRB7 | AGRN | 0.830724849 | 1.66E-12 |
| CHOL | CGC | GRB7 | WNK2 | 0.808646845 | 1.83E-11 |
| CHOL | CGC | GRB7 | ELF3 | 0.831811519 | 1.46E-12 |
| CHOL | CGC | GRB7 | ERBB3 | 0.892733231 | 1.75E-16 |
| CHOL | IUPHAR | GRB7 | WNK2 | 0.808646845 | 1.83E-11 |
| CHOL | IUPHAR | GRB7 | CD276 | 0.827004604 | 2.55E-12 |
| CHOL | IUPHAR | GRB7 | ERBB3 | 0.892733231 | 1.75E-16 |
| CHOL | Kinase | GRB7 | WNK2 | 0.808646845 | 1.83E-11 |
| CHOL | Kinase | GRB7 | ERBB3 | 0.892733231 | 1.75E-16 |
| CHOL | TF | GRB7 | ELF3 | 0.831811519 | 1.46E-12 |
| CHOL | TF | GRB7 | HNF1B | 0.893984352 | 1.37E-16 |
| CHOL | TSG | GRB7 | WNK2 | 0.808646845 | 1.83E-11 |
| CHOL | TSG | GRB7 | CTDSPL | 0.808866821 | 1.79E-11 |
| CHOL | TSG | GRB7 | KRT19 | 0.817024135 | 7.65E-12 |
| CHOL | TSG | GRB7 | RAB25 | 0.82372329 | 3.68E-12 |
| CHOL | TSG | GRB7 | DCDC2 | 0.837589741 | 7.33E-13 |
| TGCT | Cell metabolism gene | GRB7 | HCCS | 0.803982389 | 1.37E-36 |
| TGCT | Cell metabolism gene | GRB7 | DNMT3B | 0.810642014 | 1.26E-37 |
| TGCT | Cell metabolism gene | GRB7 | PSMD11 | 0.820803498 | 2.74E-39 |
| TGCT | Cell metabolism gene | GRB7 | PSMD3 | 0.837037873 | 3.56E-42 |
| TGCT | Epifactor | GRB7 | DNMT3B | 0.810642014 | 1.26E-37 |
| TGCT | TSG | GRB7 | DNMT3B | 0.810642014 | 1.26E-37 |
| TGCT | TSG | GRB7 | PHB | 0.826256098 | 3.18E-40 |
| THCA | Cell metabolism gene | GRB7 | ALDH3B1 | 0.802407429 | 6.79E-130 |
| THCA | Cell metabolism gene | GRB7 | PROS1 | 0.81771528 | 7.75E-139 |
| THCA | Cell metabolism gene | GRB7 | PLCD3 | 0.842216906 | 4.65E-155 |
| THCA | Cell metabolism gene | GRB7 | SDC4 | 0.845955799 | 8.87E-158 |
| THCA | CGC | GRB7 | FSTL3 | 0.814312727 | 8.88E-137 |
| THCA | CGC | GRB7 | ELF3 | 0.81952752 | 5.96E-140 |
| THCA | CGC | GRB7 | SLC34A2 | 0.82065141 | 1.20E-140 |
| THCA | CGC | GRB7 | ERBB3 | 0.821724089 | 2.56E-141 |
| THCA | CGC | GRB7 | SDC4 | 0.845955799 | 8.87E-158 |
| THCA | IUPHAR | GRB7 | GABRB2 | 0.809704793 | 4.68E-134 |
| THCA | IUPHAR | GRB7 | PTPRE | 0.809970791 | 3.27E-134 |
| THCA | IUPHAR | GRB7 | SLC34A2 | 0.82065141 | 1.20E-140 |
| THCA | IUPHAR | GRB7 | RARG | 0.820780957 | 9.93E-141 |
| THCA | IUPHAR | GRB7 | KCNQ3 | 0.821012565 | 7.12E-141 |
| THCA | IUPHAR | GRB7 | ERBB3 | 0.821724089 | 2.56E-141 |
| THCA | IUPHAR | GRB7 | GJB3 | 0.823580528 | 1.73E-142 |
| THCA | IUPHAR | GRB7 | TACSTD2 | 0.836759342 | 3.25E-151 |
| THCA | IUPHAR | GRB7 | EPHB3 | 0.839957115 | 1.89E-153 |
| THCA | IUPHAR | GRB7 | PLCD3 | 0.842216906 | 4.65E-155 |
| THCA | IUPHAR | GRB7 | ITGA3 | 0.848239812 | 1.78E-159 |
| THCA | IUPHAR | GRB7 | NOD1 | 0.84921455 | 3.30E-160 |
| THCA | IUPHAR | GRB7 | TMPRSS6 | 0.865940877 | 1.18E-173 |
| THCA | Kinase | GRB7 | ERBB3 | 0.821724089 | 2.56E-141 |
| THCA | Kinase | GRB7 | EPHB3 | 0.839957115 | 1.89E-153 |
| THCA | TF | GRB7 | ELF3 | 0.81952752 | 5.96E-140 |
| THCA | TF | GRB7 | RARG | 0.820780957 | 9.93E-141 |
| THCA | TSG | GRB7 | CAMK2N1 | 0.807872447 | 5.39E-133 |
| THCA | TSG | GRB7 | PPP1R1B | 0.807945542 | 4.90E-133 |
| THCA | TSG | GRB7 | KRT19 | 0.812184666 | 1.64E-135 |
| THCA | TSG | GRB7 | PDLIM4 | 0.828915512 | 6.24E-146 |
| THCA | TSG | GRB7 | EPHB3 | 0.839957115 | 1.89E-153 |
| THCA | TSG | GRB7 | TMPRSS6 | 0.865940877 | 1.18E-173 |
| THYM | TSG | GRB7 | JUP | 0.806265149 | 3.91E-29 |
| UCS | TSG | GRB7 | JUP | 0.806265149 | 3.91E-29 |
Top |
|
 Protein 3D structure Protein 3D structureVisit iCn3D. |
Top |
|
 Protein-protein interaction networks Protein-protein interaction networks * Overlap between up-regulated DEGs (log2FC<-1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P)) |
 |
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P)) Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P)) |
 |
 * Edge colors based on TCGA cancer types. |
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P)) * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P)) |
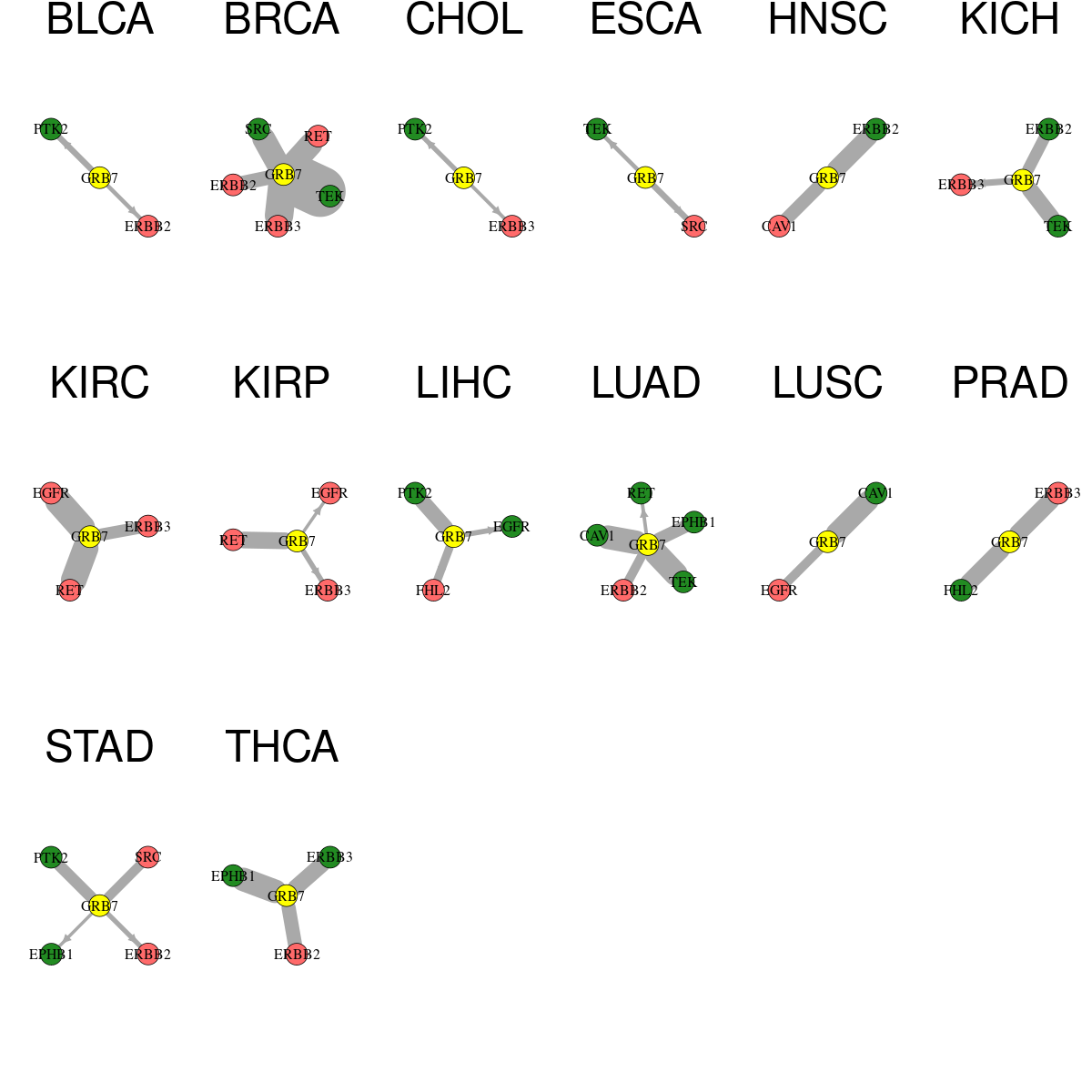 |
| Cancer type | Translation factor | Interacting protein coding gene | FC | adj.pval |
| LUSC | GRB7 | EGFR | 1.32757138391417 | 0.000102202753672845 |
| LIHC | GRB7 | FHL2 | 1.6251338358169 | 0.000209845686767642 |
| KICH | GRB7 | ERBB3 | 3.30539124394538 | 0.0014527440071106 |
| ESCA | GRB7 | SRC | 1.0077300524118 | 0.001953125 |
| BLCA | GRB7 | PTK2 | -1.24244852434456 | 0.00239944458007812 |
| LIHC | GRB7 | EGFR | -1.2230538094017 | 0.00496795806084514 |
| KIRP | GRB7 | ERBB3 | 1.05673920266283 | 0.00571200484409928 |
| STAD | GRB7 | ERBB2 | 2.08308782318447 | 0.00733334058895707 |
| CHOL | GRB7 | PTK2 | -2.09285679336748 | 0.0078125 |
| ESCA | GRB7 | TEK | -4.62660842127501 | 0.013671875 |
| BLCA | GRB7 | ERBB2 | 1.70986293820428 | 0.014068603515625 |
| CHOL | GRB7 | ERBB3 | 1.57010298428629 | 0.02734375 |
| LUAD | GRB7 | RET | -2.17383920637724 | 0.0278902380223951 |
| KIRP | GRB7 | EGFR | 1.14056399927375 | 0.0394268441013992 |
| STAD | GRB7 | EPHB1 | -5.21269873384179 | 0.0394268441013992 |
| HNSC | GRB7 | CAV1 | 1.96516854722805 | 1.49520951708837e-06 |
| PRAD | GRB7 | ERBB3 | 1.1594821213178 | 1.5966396741243e-08 |
| LIHC | GRB7 | PTK2 | -1.87535325648584 | 2.06383403084721e-07 |
| LUAD | GRB7 | EPHB1 | -2.93922759093794 | 2.0741790728873e-06 |
| PRAD | GRB7 | FHL2 | -1.82979561467933 | 2.19100533620049e-08 |
| KIRC | GRB7 | EGFR | 1.55836383937799 | 2.25998111659284e-12 |
| BRCA | GRB7 | ERBB3 | 2.28105620192688 | 2.73353544828495e-13 |
| LUAD | GRB7 | CAV1 | -3.06294584300431 | 3.59849856594825e-11 |
| LUAD | GRB7 | TEK | -3.06350124532268 | 3.59910232594763e-11 |
| KIRC | GRB7 | ERBB3 | 1.53916110157728 | 3.91927479910437e-06 |
| THCA | GRB7 | EPHB1 | -2.55178321527736 | 4.09207132288577e-11 |
| KIRP | GRB7 | RET | 2.48155495633 | 4.09781932830811e-08 |
| BRCA | GRB7 | TEK | -1.6632249483129 | 4.10483912789294e-23 |
| BRCA | GRB7 | ERBB2 | 2.10379074903324 | 4.64802044953281e-07 |
| KIRC | GRB7 | RET | 4.56498206164407 | 4.81926317698848e-12 |
| THCA | GRB7 | ERBB2 | 1.02793927266444 | 5.71717197842929e-07 |
| KICH | GRB7 | TEK | -2.30291883829546 | 5.96046447753906e-08 |
| STAD | GRB7 | SRC | 1.29268590267835 | 6.0301274061203e-05 |
| BRCA | GRB7 | RET | 1.29262215816003 | 6.3625505327664e-11 |
| KICH | GRB7 | ERBB2 | -2.13618034410539 | 6.55651092529297e-06 |
| BRCA | GRB7 | SRC | -1.01389608845799 | 7.35163663752073e-11 |
| STAD | GRB7 | PTK2 | -3.29131709886752 | 8.74791294336319e-06 |
| THCA | GRB7 | ERBB3 | -1.82448909246289 | 8.9058794211314e-08 |
| LUAD | GRB7 | ERBB2 | 1.65046120667728 | 8.94245335020896e-05 |
| HNSC | GRB7 | ERBB2 | -3.64938375773393 | 9.1349647846073e-09 |
| LUSC | GRB7 | CAV1 | -2.61693164564285 | 9.58407745365481e-10 |
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Protein-protein interactors with this translation factor (BIOGRID-3.4.160) |
| PPI interactors with GRB7 |
| SFN, LY6G6F, ERBB3, KIT, RND1, RET, ERBB2, PTK2, EPHB1, INSR, NPM1, RPS2, ZBTB16, A1BG, ATP5B, CPNE6, PKM, DYNC1I1, AGAP1, GNB2, TMSB4X, PDHB, FLAD1, DOCK7, MSH2, HADHB, TLE1, KMT2B, FGB, PHAX, SETDB1, CALM1, LAX1, TRIM36, KCTD6, OLIG1, ZNHIT1, GIGYF1, GIGYF2, PDGFRB, SHC1, USP2, PICK1, TGM5, FAM124B, FCHO1, TCF12, TRIM43, CREBRF, STAC3, RSL1D1, MRPS7, MRPS34, MRPS14, RPL6, RPL39P5, RPL39, RPL12, NOC2L, MRPS2, C19orf53, RPF2, MRPS26, RIOK2, C1orf35, PURB, MRPS24, MRPS27, BRIX1, RPP25L, MCAT, MINA, RPL19, MRPS21, MRPS5, MRPS35, MRPS10, RPL32, RPL31, POLR2L, RPL23A, RPL35, NEMF, RIOK3, RPL18, RPL37A, RPL27, RPL15, RPL14, RPL13A, RPL4, RPL10L, RPL10, RPL35A, GRB7, AR, MET, EGFR, ERBB4, FARSA, HDAC2, RBM10, LTV1, PPIL4, STAU1, PTBP1, G3BP2, PSMD3, LUC7L3, CCT7, RPN1, HNRNPM, RBM14-RBM4, RBM14, EIF4B, SDHA, PUF60, HSPA8, CAPRIN1, |
Top |
|
 Clinically associated variants from ClinVar. Clinically associated variants from ClinVar. |
| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| GRB7 | chr17 | 37899147 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GRB7 | chr17 | 37899452 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| GRB7 | chr17 | 37900387 | G | A | single_nucleotide_variant | Uncertain_significance | High_myopia | SO:0001583|missense_variant | SO:0001583|missense_variant |
| GRB7 | chr17 | 37900864 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| GRB7 | chr17 | 37901569 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| GRB7 | chr17 | 37901977 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GRB7 | chr17 | 37902027 | C | A | single_nucleotide_variant | Benign | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
 nsSNVs with sample frequency (size of circle) from TCGA 33 cancers. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers. |
 SNVs and Indels SNVs and Indels |
| Gene | Cancer type | Chromosome | Start | End | RefSeeq | MutSeq | Mutation type | AAchange | # samples |
| GRB7 | SKCM | chr17 | 37898852 | 37898852 | C | T | Silent | p.S63S | 8 |
| GRB7 | SKCM | chr17 | 37898880 | 37898880 | G | A | Missense_Mutation | p.E73K | 3 |
| GRB7 | HNSC | chr17 | 37899470 | 37899470 | G | T | Silent | p.V167V | 3 |
| GRB7 | STAD | chr17 | 37901173 | 37901173 | G | A | Missense_Mutation | p.R316Q | 3 |
| GRB7 | BRCA | chr17 | 37898827 | 37898827 | G | A | Missense_Mutation | p.R55Q | 3 |
| GRB7 | BRCA | chr17 | 37899236 | 37899236 | T | A | Missense_Mutation | p.L131Q | 3 |
| GRB7 | BRCA | chr17 | 37899253 | 37899253 | G | A | Missense_Mutation | p.A137T | 3 |
| GRB7 | SKCM | chr17 | 37898851 | 37898851 | C | T | Missense_Mutation | p.S63F | 3 |
| GRB7 | UCEC | chr17 | 37899223 | 37899223 | G | A | Missense_Mutation | p.V127M | 3 |
| GRB7 | ESCA | chr17 | 37898911 | 37898911 | T | A | Missense_Mutation | 2 | |
| GRB7 | SARC | chr17 | 37899243 | 37899243 | G | T | Missense_Mutation | 2 | |
| GRB7 | BLCA | chr17 | 37901554 | 37901554 | C | G | Missense_Mutation | p.S357C | 2 |
| GRB7 | UCEC | chr17 | 37900339 | 37900339 | G | A | Missense_Mutation | p.G227D | 2 |
| GRB7 | KIRP | chr17 | 37898588 | 37898588 | A | G | Missense_Mutation | p.S12G | 2 |
| GRB7 | SKCM | chr17 | 37902191 | 37902191 | C | T | Silent | p.F432F | 2 |
| GRB7 | ESCA | chr17 | 37898928 | 37898928 | A | C | Missense_Mutation | 2 | |
| GRB7 | UCEC | chr17 | 37901530 | 37901531 | CA | - | Frame_Shift_Del | p.Q350fs | 2 |
| GRB7 | SKCM | chr17 | 37901701 | 37901701 | G | A | Silent | p.V373V | 2 |
| GRB7 | BRCA | chr17 | 37898674 | 37898674 | G | C | Missense_Mutation | p.K40N | 2 |
| GRB7 | HNSC | chr17 | 37898909 | 37898909 | A | C | Silent | 2 | |
| GRB7 | UCEC | chr17 | 37901539 | 37901539 | G | A | Missense_Mutation | p.R352H | 2 |
| GRB7 | CHOL | chr17 | 37899545 | 37899545 | G | T | Missense_Mutation | 2 | |
| GRB7 | SKCM | chr17 | 37898596 | 37898596 | G | A | Silent | p.P14P | 2 |
| GRB7 | PAAD | chr17 | 37901227 | 37901227 | G | A | Missense_Mutation | p.R334H | 2 |
| GRB7 | SKCM | chr17 | 37899207 | 37899207 | T | A | Silent | p.G121G | 2 |
| GRB7 | BRCA | chr17 | 37899229 | 37899229 | G | C | Missense_Mutation | p.E129Q | 2 |
| GRB7 | SKCM | chr17 | 37902204 | 37902204 | T | C | Missense_Mutation | p.S437P | 2 |
| GRB7 | PAAD | chr17 | 37899533 | 37899533 | C | T | Silent | p.Y188Y | 2 |
| GRB7 | SKCM | chr17 | 37899515 | 37899515 | G | A | Silent | p.R182R | 2 |
| GRB7 | HNSC | chr17 | 37900456 | 37900456 | C | A | Missense_Mutation | p.S266Y | 2 |
| GRB7 | SKCM | chr17 | 37900842 | 37900842 | G | A | Missense_Mutation | p.E280K | 2 |
| GRB7 | UCS | chr17 | 37899524 | 37899524 | C | T | Silent | p.F185F | 2 |
| GRB7 | SKCM | chr17 | 37901727 | 37901727 | G | A | Missense_Mutation | p.G382E | 2 |
| GRB7 | PAAD | chr17 | 37900913 | 37900913 | C | A | Silent | 2 | |
| GRB7 | ESCA | chr17 | 37898911 | 37898911 | T | A | Missense_Mutation | p.I106N | 2 |
| GRB7 | LIHC | chr17 | 37902437 | 37902437 | G | T | Missense_Mutation | 2 | |
| GRB7 | SKCM | chr17 | 37899506 | 37899506 | C | T | Silent | p.F179F | 2 |
| GRB7 | CESC | chr17 | 37899650 | 37899650 | C | A | Missense_Mutation | 2 | |
| GRB7 | KICH | chr17 | 37900401 | 37900401 | C | T | Missense_Mutation | p.R248C | 2 |
| GRB7 | SKCM | chr17 | 37899717 | 37899717 | C | T | Silent | p.L219L | 2 |
| GRB7 | ESCA | chr17 | 37898928 | 37898928 | A | C | Missense_Mutation | p.S112R | 2 |
| GRB7 | LIHC | chr17 | 37902252 | 37902252 | G | T | Missense_Mutation | 2 | |
| GRB7 | STAD | chr17 | 37901173 | 37901173 | G | A | Missense_Mutation | 2 | |
| GRB7 | CESC | chr17 | 37899685 | 37899685 | G | A | Missense_Mutation | 2 | |
| GRB7 | SKCM | chr17 | 37898921 | 37898921 | C | T | Silent | p.G86G | 2 |
| GRB7 | THCA | chr17 | 37901566 | 37901566 | C | T | Missense_Mutation | p.S361F | 2 |
| GRB7 | PRAD | chr17 | 37898891 | 37898891 | T | G | Missense_Mutation | p.S76R | 2 |
| GRB7 | BLCA | chr17 | 37899524 | 37899524 | C | A | Missense_Mutation | 2 | |
| GRB7 | CESC | chr17 | 37901738 | 37901738 | G | A | Missense_Mutation | 2 | |
| GRB7 | KIRP | chr17 | 37898588 | 37898588 | A | G | Missense_Mutation | p.S35G | 2 |
| GRB7 | PAAD | chr17 | 37899533 | 37899533 | C | T | Silent | 2 | |
| GRB7 | BLCA | chr17 | 37901234 | 37901234 | C | T | Silent | p.F336F | 2 |
| GRB7 | SKCM | chr17 | 37901571 | 37901571 | C | T | Missense_Mutation | p.P363S | 1 |
| GRB7 | LIHC | chr17 | 37902437 | 37902437 | G | T | Missense_Mutation | p.K478N | 1 |
| GRB7 | PAAD | chr17 | 37901227 | 37901227 | G | A | Missense_Mutation | 1 | |
| GRB7 | STAD | chr17 | 37900873 | 37900873 | G | A | Missense_Mutation | p.R313H | 1 |
| GRB7 | STAD | chr17 | 37900454 | 37900454 | C | A | Silent | p.T265T | 1 |
| GRB7 | HNSC | chr17 | 37901747 | 37901747 | C | T | Missense_Mutation | 1 | |
| GRB7 | CESC | chr17 | 37899685 | 37899685 | G | A | Missense_Mutation | p.D232N | 1 |
| GRB7 | LUAD | chr17 | 37902407 | 37902407 | C | T | Missense_Mutation | p.S439F | 1 |
| GRB7 | PAAD | chr17 | 37899533 | 37899533 | C | T | Silent | p.Y211Y | 1 |
| GRB7 | SKCM | chr17 | 37898819 | 37898819 | G | A | Splice_Site | p.R52_splice | 1 |
| GRB7 | LIHC | chr17 | 37900452 | 37900452 | A | - | Frame_Shift_Del | p.T265fs | 1 |
| GRB7 | SARC | chr17 | 37903135 | 37903135 | G | T | Silent | 1 | |
| GRB7 | STAD | chr17 | 37898611 | 37898611 | A | G | Silent | p.P19P | 1 |
| GRB7 | BLCA | chr17 | 37898867 | 37898867 | C | T | Silent | p.P68P | 1 |
| GRB7 | HNSC | chr17 | 37899470 | 37899470 | G | T | Silent | 1 | |
| GRB7 | CESC | chr17 | 37901738 | 37901738 | G | A | Missense_Mutation | p.E409K | 1 |
| GRB7 | KIRP | chr17 | 37898588 | 37898588 | A | G | Missense_Mutation | 1 | |
| GRB7 | LUAD | chr17 | 37903087 | 37903087 | C | T | Silent | p.H512H | 1 |
| GRB7 | PAAD | chr17 | 37901227 | 37901227 | G | A | Missense_Mutation | p.R357H | 1 |
| GRB7 | SKCM | chr17 | 37902393 | 37902393 | C | T | Missense_Mutation | p.P464S | 1 |
| GRB7 | STAD | chr17 | 37900454 | 37900454 | C | A | Silent | p.T288T | 1 |
| GRB7 | ESCA | chr17 | 37898911 | 37898911 | T | A | Missense_Mutation | p.I83N | 1 |
| GRB7 | LIHC | chr17 | 37899662 | 37899662 | A | - | Frame_Shift_Del | p.E201fs | 1 |
| GRB7 | STAD | chr17 | 37899172 | 37899172 | G | A | Missense_Mutation | p.D110N | 1 |
| GRB7 | LGG | chr17 | 37902408 | 37902408 | C | T | Missense_Mutation | p.L469F | 1 |
| GRB7 | LUAD | chr17 | 37902034 | 37902034 | C | T | Silent | p.L421L | 1 |
| GRB7 | SKCM | chr17 | 37898848 | 37898848 | C | T | Missense_Mutation | p.T62I | 1 |
| GRB7 | STAD | chr17 | 37898828 | 37898828 | A | T | Silent | p.R78R | 1 |
| GRB7 | ESCA | chr17 | 37898928 | 37898928 | A | C | Missense_Mutation | p.S89R | 1 |
| GRB7 | LIHC | chr17 | 37901499 | 37901499 | G | - | Frame_Shift_Del | p.G339fs | 1 |
| GRB7 | LIHC | chr17 | 37899516 | 37899516 | A | - | Frame_Shift_Del | p.K183fs | 1 |
| GRB7 | TGCT | chr17 | 37901196 | 37901196 | C | T | Nonsense_Mutation | p.Q347X | 1 |
| GRB7 | UCS | chr17 | 37899147 | 37899147 | G | A | Silent | 1 | |
| GRB7 | CHOL | chr17 | 37899545 | 37899545 | G | T | Missense_Mutation | p.K215N | 1 |
| GRB7 | LGG | chr17 | 37901696 | 37901696 | C | A | Missense_Mutation | p.L372M | 1 |
| GRB7 | LUAD | chr17 | 37901538 | 37901538 | C | T | Missense_Mutation | p.R352C | 1 |
| GRB7 | SKCM | chr17 | 37899512 | 37899512 | C | T | Silent | p.F181F | 1 |
| GRB7 | ESCA | chr17 | 37902406 | 37902407 | - | C | Frame_Shift_Ins | p.A468fs | 1 |
| GRB7 | LIHC | chr17 | 37898604 | 37898604 | T | - | Frame_Shift_Del | p.L17fs | 1 |
| GRB7 | TGCT | chr17 | 37901196 | 37901196 | C | T | Nonsense_Mutation | p.Q324* | 1 |
| GRB7 | UCS | chr17 | 37899524 | 37899524 | C | T | Silent | p.F208F | 1 |
| GRB7 | COAD | chr17 | 37898577 | 37898577 | C | T | Missense_Mutation | p.P8L | 1 |
| GRB7 | LGG | chr17 | 37899152 | 37899152 | T | C | Splice_Site | p.V103_splice | 1 |
| GRB7 | LUAD | chr17 | 37898958 | 37898958 | A | T | Missense_Mutation | p.S99C | 1 |
| GRB7 | SKCM | chr17 | 37898943 | 37898943 | C | T | Missense_Mutation | p.L94F | 1 |
| GRB7 | PAAD | chr17 | 37899218 | 37899218 | G | A | Missense_Mutation | p.R125H | 1 |
| GRB7 | SKCM | chr17 | 37899513 | 37899513 | C | T | Missense_Mutation | p.R182W | 1 |
| GRB7 | STAD | chr17 | 37900873 | 37900873 | G | A | Missense_Mutation | p.R290H | 1 |
| GRB7 | GBM | chr17 | 37902194 | 37902194 | C | A | Missense_Mutation | p.H433Q | 1 |
| GRB7 | LIHC | chr17 | 37899714 | 37899714 | C | - | Frame_Shift_Del | p.D218fs | 1 |
| GRB7 | TGCT | chr17 | 37901196 | 37901196 | C | T | Nonsense_Mutation | 1 | |
| GRB7 | HNSC | chr17 | 37901747 | 37901747 | C | T | Missense_Mutation | p.R389W | 1 |
| GRB7 | COAD | chr17 | 37900874 | 37900874 | C | T | Silent | p.R290R | 1 |
| GRB7 | LGG | chr17 | 37902408 | 37902408 | C | T | Missense_Mutation | 1 | |
| GRB7 | LUAD | chr17 | 37900375 | 37900375 | G | T | Missense_Mutation | p.R239L | 1 |
| GRB7 | SKCM | chr17 | 37898600 | 37898600 | G | A | Missense_Mutation | p.D16N | 1 |
| GRB7 | STAD | chr17 | 37900851 | 37900851 | G | A | Missense_Mutation | p.V283M | 1 |
| GRB7 | BLCA | chr17 | 37901554 | 37901554 | C | G | Missense_Mutation | 1 | |
| GRB7 | GBM | chr17 | 37901165 | 37901165 | G | C | Missense_Mutation | p.K313N | 1 |
| GRB7 | LIHC | chr17 | 37902178 | 37902178 | C | - | Frame_Shift_Del | p.T428fs | 1 |
| GRB7 | SKCM | chr17 | 37898819 | 37898819 | G | A | Silent | p.R52R | 1 |
| GRB7 | THCA | chr17 | 37898639 | 37898639 | C | A | Silent | 1 | |
| GRB7 | KICH | chr17 | 37900401 | 37900401 | C | T | Missense_Mutation | 1 | |
| GRB7 | LUAD | chr17 | 37900435 | 37900435 | A | G | Missense_Mutation | p.Y259C | 1 |
| GRB7 | PRAD | chr17 | 37898891 | 37898891 | T | G | Missense_Mutation | p.S99R | 1 |
| GRB7 | SKCM | chr17 | 37903012 | 37903012 | G | A | Silent | p.E487E | 1 |
| GRB7 | STAD | chr17 | 37902012 | 37902012 | T | C | Missense_Mutation | p.M414T | 1 |
| GRB7 | BLCA | chr17 | 37901234 | 37901234 | C | T | Silent | 1 | |
| GRB7 | GBM | chr17 | 37903076 | 37903076 | G | A | Missense_Mutation | 1 | |
| GRB7 | LUAD | chr17 | 37903138 | 37903138 | G | A | Silent | p.R529R | 1 |
| GRB7 | THCA | chr17 | 37901566 | 37901566 | C | T | Missense_Mutation | 1 | |
| GRB7 | OV | chr17 | 37903121 | 37903121 | C | T | Missense_Mutation | p.R524C | 1 |
| GRB7 | SKCM | chr17 | 37899245 | 37899245 | G | A | Missense_Mutation | p.R134Q | 1 |
| GRB7 | PRAD | chr17 | 37903005 | 37903005 | G | A | Splice_Site | p.S485_splice | 1 |
| GRB7 | STAD | chr17 | 37902041 | 37902041 | G | A | Missense_Mutation | p.A424T | 1 |
| GRB7 | BLCA | chr17 | 37900352 | 37900352 | G | C | Missense_Mutation | 1 | |
| GRB7 | GBM | chr17 | 37901165 | 37901165 | G | C | Missense_Mutation | 1 | |
| GRB7 | KIRC | chr17 | 37899552 | 37899552 | C | T | Missense_Mutation | p.P218S | 1 |
| GRB7 | LUAD | chr17 | 37900365 | 37900365 | C | G | Missense_Mutation | p.L236V | 1 |
| GRB7 | ESCA | chr17 | 37902406 | 37902407 | - | C | Frame_Shift_Ins | p.L492fs | 1 |
| GRB7 | LIHC | chr17 | 37901708 | 37901708 | G | A | Missense_Mutation | p.D399N | 1 |
| GRB7 | PAAD | chr17 | 37899218 | 37899218 | G | A | Missense_Mutation | 1 | |
| GRB7 | SKCM | chr17 | 37898526 | 37898526 | C | T | Missense_Mutation | p.S14F | 1 |
| GRB7 | STAD | chr17 | 37901173 | 37901173 | G | A | Missense_Mutation | p.R339Q | 1 |
| GRB7 | STAD | chr17 | 37898828 | 37898828 | A | T | Silent | p.R55R | 1 |
| GRB7 | GBM | chr17 | 37902175 | 37902175 | G | A | Missense_Mutation | 1 | |
| GRB7 | LUAD | chr17 | 37898639 | 37898639 | C | T | Missense_Mutation | p.R29W | 1 |
| GRB7 | ESCA | chr17 | 37900347 | 37900347 | C | T | Missense_Mutation | 1 | |
| GRB7 | LIHC | chr17 | 37901211 | 37901211 | T | C | Missense_Mutation | p.W352R | 1 |
| GRB7 | SKCM | chr17 | 37903036 | 37903036 | G | A | Missense_Mutation | p.M495I | 1 |
| GRB7 | PRAD | chr17 | 37903005 | 37903005 | G | A | Splice_Site | p.S485N | 1 |
| GRB7 | STAD | chr17 | 37898611 | 37898611 | A | G | Silent | p.P42P | 1 |
| GRB7 | STAD | chr17 | 37901697 | 37901697 | T | C | Missense_Mutation | p.L372P | 1 |
| GRB7 | HNSC | chr17 | 37900456 | 37900456 | C | A | Missense_Mutation | 1 | |
| GRB7 | CESC | chr17 | 37899650 | 37899650 | C | A | Missense_Mutation | p.S220Y | 1 |
| GRB7 | KIRP | chr17 | 37903002 | 37903002 | A | C | Splice_Site | . | 1 |
| GRB7 | LUAD | chr17 | 37898907 | 37898907 | C | T | Missense_Mutation | p.P82S | 1 |
| GRB7 | UCEC | chr17 | 37901530 | 37901531 | CA | - | Frame_Shift_Del | p.A349fs | 1 |
 Copy number variation (CNV) of GRB7 Copy number variation (CNV) of GRB7 * Click on the image to open the original image in a new window. |
 |
 Fusion gene breakpoints (product of the structural variants (SVs)) across GRB7 Fusion gene breakpoints (product of the structural variants (SVs)) across GRB7 * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion genes with this translation factor from FusionGDB2.0. Fusion genes with this translation factor from FusionGDB2.0. |
| FusionGDB2 ID | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 99269 | LUSC | TCGA-60-2715 | FBXL20 | chr17 | 37557613 | - | GRB7 | chr17 | 37898504 | + |
| 99269 | LUSC | TCGA-60-2715-01A | FBXL20 | chr17 | 37558426 | - | GRB7 | chr17 | 37898505 | + |
| 99269 | READ | TCGA-AF-A56L | FBXL20 | chr17 | 37557614 | - | GRB7 | chr17 | 37898505 | + |
| 101356 | BLCA | TCGA-HQ-A2OF-01A | GRB7 | chr17 | 37902455 | + | ACACA | chr17 | 35567404 | - |
| 79881 | BRCA | TCGA-BH-A1EN-01A | GRB7 | chr17 | 37902455 | + | ACTN3 | chr11 | 66318710 | + |
| 97709 | BRCA | TCGA-AR-A0TQ-01A | GRB7 | chr17 | 37902041 | + | BRIP1 | chr17 | 59934592 | - |
| 34681 | BLCA | TCGA-XF-A9SK-01A | GRB7 | chr17 | 37901237 | + | CD300E | chr17 | 72610172 | - |
| 99264 | OV | TCGA-61-1736-01B | GRB7 | chr17 | 37894386 | + | CDK12 | chr17 | 37721013 | + |
| 99264 | READ | TCGA-AF-A56L-01A | GRB7 | chr17 | 37902455 | + | CDK12 | chr17 | 37700502 | + |
| 99264 | STAD | TCGA-CG-4449 | GRB7 | chr17 | 37902455 | + | CDK12 | chr17 | 37680926 | + |
| 99264 | STAD | TCGA-CG-4449-01A | GRB7 | chr17 | 37902455 | + | CDK12 | chr17 | 37680927 | + |
| 101608 | BLCA | TCGA-GU-AATP | GRB7 | chr17 | 37894386 | + | ERBB2 | chr17 | 37844949 | + |
| 101608 | BRCA | TCGA-C8-A1HK-01A | GRB7 | chr17 | 37902253 | + | ERBB2 | chr17 | 37883942 | + |
| 101608 | CESC | TCGA-C5-A1M9-01A | GRB7 | chr17 | 37902455 | + | ERBB2 | chr17 | 37849514 | + |
| 101608 | CESC | TCGA-LP-A5U3-01A | GRB7 | chr17 | 37897160 | + | ERBB2 | chr17 | 37879791 | + |
| 101608 | CESC | TCGA-LP-A5U3-01A | GRB7 | chr17 | 37898709 | + | ERBB2 | chr17 | 37855790 | + |
| 101608 | CESC | TCGA-LP-A5U3-01A | GRB7 | chr17 | 37898709 | + | ERBB2 | chr17 | 37863243 | + |
| 101608 | CESC | TCGA-VS-A9V5 | GRB7 | chr17 | 37894386 | + | ERBB2 | chr17 | 37886276 | + |
| 101608 | CESC | TCGA-VS-A9V5 | GRB7 | chr17 | 37894645 | + | ERBB2 | chr17 | 37886276 | + |
| 100675 | N/A | DL050887 | GRB7 | chr17 | 37903538 | + | FBXL17 | chr5 | 107440884 | - |
| 103189 | READ | TCGA-EI-6509 | GRB7 | chr17 | 37894645 | + | IKZF3 | chr17 | 37988404 | - |
| 103189 | STAD | TCGA-D7-8573 | GRB7 | chr17 | 37894386 | + | IKZF3 | chr17 | 37949186 | - |
| 103189 | STAD | TCGA-FP-7735-01A | GRB7 | chr17 | 37898986 | + | IKZF3 | chr17 | 37921231 | - |
| 97711 | BRCA | TCGA-A2-A0CX-01A | GRB7 | chr17 | 37900460 | + | ITGB3 | chr17 | 45351785 | + |
| 98049 | STAD | TCGA-CG-4466 | GRB7 | chr17 | 37894645 | + | KRT222 | chr17 | 38816459 | - |
| 98049 | STAD | TCGA-CG-4466-01A | GRB7 | chr17 | 37894386 | + | KRT222 | chr17 | 38816459 | - |
| 34681 | STAD | TCGA-D7-8573 | GRB7 | chr17 | 37900916 | + | KRT25 | chr17 | 38906849 | - |
| 41073 | STAD | TCGA-D7-8573 | GRB7 | chr17 | 37900916 | + | KRT27 | chr17 | 38933984 | - |
| 41073 | STAD | TCGA-D7-8573-01A | GRB7 | chr17 | 37900916 | + | KRT27 | chr17 | 38950298 | - |
| 34681 | BRCA | TCGA-AO-A12C-01A | GRB7 | chr17 | 37894645 | + | KRT35 | chr17 | 39637007 | - |
| 97197 | ESCA | TCGA-2H-A9GF | GRB7 | chr17 | 37898709 | + | MIEN1 | chr17 | 37886544 | - |
| 97197 | ESCA | TCGA-M9-A5M8 | GRB7 | chr17 | 37894386 | + | MIEN1 | chr17 | 37886544 | - |
| 97197 | ESCA | TCGA-M9-A5M8 | GRB7 | chr17 | 37894645 | + | MIEN1 | chr17 | 37886544 | - |
| 97197 | STAD | TCGA-EQ-8122-01A | GRB7 | chr17 | 37898541 | + | MIEN1 | chr17 | 37886516 | - |
| 99077 | BRCA | TCGA-A8-A09I | GRB7 | chr17 | 37894645 | + | PGAP3 | chr17 | 37830932 | - |
| 99077 | BRCA | TCGA-A8-A09I-01A | GRB7 | chr17 | 37894386 | + | PGAP3 | chr17 | 37830932 | - |
| 99077 | BRCA | TCGA-BH-A202 | GRB7 | chr17 | 37898709 | + | PGAP3 | chr17 | 37842272 | - |
| 99077 | BRCA | TCGA-BH-A202 | GRB7 | chr17 | 37898969 | + | PGAP3 | chr17 | 37842272 | - |
| 99077 | BRCA | TCGA-BH-A202 | GRB7 | chr17 | 37899307 | + | PGAP3 | chr17 | 37842272 | - |
| 99077 | BRCA | TCGA-BH-A202-01A | GRB7 | chr17 | 37898708 | + | PGAP3 | chr17 | 37842271 | - |
| 99077 | BRCA | TCGA-BH-A202-01A | GRB7 | chr17 | 37898968 | + | PGAP3 | chr17 | 37842271 | - |
| 99077 | BRCA | TCGA-BH-A202-01A | GRB7 | chr17 | 37899306 | + | PGAP3 | chr17 | 37842271 | - |
| 99077 | ESCA | TCGA-R6-A8W5 | GRB7 | chr17 | 37902455 | + | PGAP3 | chr17 | 37830932 | - |
| 99665 | ESCA | TCGA-L5-A88V | GRB7 | chr17 | 37894386 | + | PIP4K2B | chr17 | 36927525 | - |
| 99665 | ESCA | TCGA-L5-A88V | GRB7 | chr17 | 37894645 | + | PIP4K2B | chr17 | 36927525 | - |
| 99665 | ESCA | TCGA-L5-A88V | GRB7 | chr17 | 37898709 | + | PIP4K2B | chr17 | 36927525 | - |
| 99665 | ESCA | TCGA-L5-A88V | GRB7 | chr17 | 37899307 | + | PIP4K2B | chr17 | 36927525 | - |
| 99665 | ESCA | TCGA-L5-A88V-01A | GRB7 | chr17 | 37898969 | + | PIP4K2B | chr17 | 36927525 | - |
| 88698 | BLCA | TCGA-CU-A0YR-01A | GRB7 | chr17 | 37900460 | + | PLXDC1 | chr17 | 37224212 | - |
| 99282 | STAD | TCGA-EQ-8122-01A | GRB7 | chr17 | 37902455 | + | PSMD3 | chr17 | 38144936 | + |
| 96982 | BRCA | TCGA-AR-A24U-01A | GRB7 | chr17 | 37899723 | + | SAMHD1 | chr20 | 35547922 | - |
| 99287 | CESC | TCGA-Q1-A73R | GRB7 | chr17 | 37894645 | + | STARD3 | chr17 | 37813260 | + |
| 99287 | CESC | TCGA-Q1-A73R-01A | GRB7 | chr17 | 37894645 | + | STARD3 | chr17 | 37813261 | + |
| 99287 | STAD | TCGA-BR-7901 | GRB7 | chr17 | 37894645 | + | STARD3 | chr17 | 37809733 | + |
| 76256 | STAD | TCGA-BR-A4PE-01A | GRB7 | chr17 | 37895013 | + | TCAP | chr17 | 37822070 | + |
| 93090 | STAD | TCGA-BR-A4PE-01A | GRB7 | chr17 | 37894386 | + | TOP2A | chr17 | 38548989 | - |
| 99295 | STAD | TCGA-BR-8381 | GRB7 | chr17 | 37894645 | + | WIPF2 | chr17 | 38430041 | + |
| 99295 | STAD | TCGA-BR-8483 | GRB7 | chr17 | 37894386 | + | WIPF2 | chr17 | 38430041 | + |
| 99295 | STAD | TCGA-BR-8483 | GRB7 | chr17 | 37894386 | + | WIPF2 | chr17 | 38433334 | + |
| 99295 | STAD | TCGA-BR-8483-01A | GRB7 | chr17 | 37894386 | + | WIPF2 | chr17 | 38430042 | + |
| 99295 | STAD | TCGA-BR-8483-01A | GRB7 | chr17 | 37894645 | + | WIPF2 | chr17 | 38430042 | + |
| 99295 | STAD | TCGA-EQ-8122 | GRB7 | chr17 | 37902455 | + | WIPF2 | chr17 | 38420741 | + |
| 99295 | STAD | TCGA-EQ-8122-01A | GRB7 | chr17 | 37902455 | + | WIPF2 | chr17 | 38420742 | + |
| 99269 | STAD | TCGA-CG-4466-01A | KAT6A | chr8 | 41905896 | - | GRB7 | chr17 | 37898505 | + |
| 99269 | OV | TCGA-09-1659 | LAMA5 | chr20 | 60907427 | - | GRB7 | chr17 | 37903003 | + |
| 99269 | STAD | TCGA-BR-8486-01A | MED1 | chr17 | 37607291 | - | GRB7 | chr17 | 37898505 | + |
| 99269 | STAD | TCGA-BR-A4PE | MIEN1 | chr17 | 37885937 | - | GRB7 | chr17 | 37898504 | + |
| 99269 | STAD | TCGA-BR-A4PE-01A | MIEN1 | chr17 | 37885938 | - | GRB7 | chr17 | 37898505 | + |
| 99269 | UCEC | TCGA-D1-A16S-01A | MSL1 | chr17 | 38282659 | + | GRB7 | chr17 | 37898505 | + |
| 99269 | LUSC | TCGA-56-A4BX | PGAP3 | chr17 | 37844087 | - | GRB7 | chr17 | 37898505 | + |
| 99269 | UCEC | TCGA-EY-A3QX-01A | PPP1R1B | chr17 | 37791979 | + | GRB7 | chr17 | 37898505 | + |
| 99269 | STAD | TCGA-HU-8604 | RPL41 | chr12 | 56510583 | + | GRB7 | chr17 | 37898504 | + |
| 99269 | BRCA | TCGA-E2-A1LE | STARD3 | chr17 | 37793484 | + | GRB7 | chr17 | 37898504 | + |
| 99269 | BRCA | TCGA-E2-A1LE-01A | STARD3 | chr17 | 37793484 | + | GRB7 | chr17 | 37898505 | + |
| 99269 | UCEC | TCGA-EY-A3QX-01A | STARD3 | chr17 | 37793483 | + | GRB7 | chr17 | 37898504 | + |
| 99298 | BRCA | TCGA-C8-A132-01A | WIPF2 | chr17 | 38375744 | + | GRB7 | chr17 | 37901139 | + |
Top |
|
 Kaplan-Meier plots with logrank tests of overall survival (OS) Kaplan-Meier plots with logrank tests of overall survival (OS) |
 |
| Cancer type | Translation factor | Coefficent | Hazard ratio | Wald test pval | Likelihool ratio pval | Logrank test pval | # samples |
Top |
|
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05) Differential gene expression between female and male. (Wilcoxon test, pval<0.05) |
 |
| Cancer type | Translation factor | pval | adj.p |
| HNSC | GRB7 | 0.00141313607782347 | 0.04 |
| STAD | GRB7 | 0.00237962254345471 | 0.064 |
| LIHC | GRB7 | 0.00849012759528567 | 0.22 |
| SARC | GRB7 | 0.0303487188204106 | 0.76 |
| LGG | GRB7 | 0.0330076288435039 | 0.79 |
| COAD | GRB7 | 0.0426390573469971 | 0.98 |
Top |
|
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05) Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05) |
 |
| Cancer type | Translation factor | pval | adj.p |
| LGG | GRB7 | 0.00141630467103485 | 0.047 |
| LAML | GRB7 | 0.0479848772150928 | 1 |
| PRAD | GRB7 | 0.013459670869531 | 0.42 |
| UCS | GRB7 | 0.0241055224698056 | 0.72 |
| THYM | GRB7 | 0.00424965323036572 | 0.14 |
| READ | GRB7 | 0.0399026804483136 | 1 |
Top |
|
 Drugs targeting genes involved in this translation factor. Drugs targeting genes involved in this translation factor. (DrugBank Version 5.1.8 2021-05-08) |
| UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
|
 Diseases associated with this translation factor. Diseases associated with this translation factor. (DisGeNet 4.0) |
| Disease ID | Disease Name | # PubMeds | Disease source |
| C0006142 | Malignant neoplasm of breast | 2 | CTD_human |
| C0678222 | Breast Carcinoma | 2 | CTD_human |
| C1257931 | Mammary Neoplasms, Human | 2 | CTD_human |
| C4704874 | Mammary Carcinoma, Human | 2 | CTD_human |
| C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |

