|
Home |
Download |
Statistics |
Landscape |
Examples |
Help |
Contact |
Navigation
1. Translation factor gene information.2. Understanding of TranslFac's Annotation Category.
- Search Page, example: EIF5A (translation initiation factor).
- Gene Search Result Page.
- Gene Annotation Result Page.
-- 1) Translation Factor Gene Summary.
-- 2) Translation Studies in PubMed.
-- 3) Exon Skippping Events in Translaction Factor gene.
-- 4) Expression of Translation Factor.
-- 5) Expression Regulation of Translation Factor.
-- 6) Associated Genes.
-- 7) Translation Factor Protein's 3D Structure.
-- 8) Protein-Protein Interaction Category.
-- 9) Mutation in Translation Factor Gene .
-- 10) Prognostic Analysis of Translation Factor.
-- 11) Translation Factor Expression in Different Genders.
-- 12) Translation Factor Expression in Different Age Group.
-- 13) Related Drugs.
-- 14) Related Diseases.
3. Download Data and Contact Us.
1. Translation factor gene information
Defining the human translation factor genes is very critical to performing our study. After many trials, we noticed to use the genes belonging to the biological process of the ‘translation’ is a reasonable way. The GO term (GO:0006412) includes 617 genes. Following the definition of this translation factor process from the GO like ‘The cellular metabolic process in which a protein is formed, using the sequence of a mature mRNA molecule to specify the sequence of amino acids in a polypeptide chain. The translation is mediated by the ribosome and begins with the formation of a ternary complex between aminoacylated initiator methionine tRNA, GTP, and initiation factor 2, which subsequently associates with the small subunit of the ribosome and an mRNA. Translation ends with the release of a polypeptide chain from the ribosome.’, we aimed to study the dynamic molecular profiles, features, and aberrations in cancer of these translation factors. The alternative synonyms for this biological process are GO:0006416, GO:0006453, and GO:0043037. The translation factor process has these synonyms including protein anabolism, protein biosynthesis, protein biosynthetic process, protein formation, protein synthesis, and protein translation. As shown in Table 1, there are 14 child nodes with specific functions. The number of genes was counted after taking the UniProt protein-coding genes.2. Understanding TranslFac's Annotation Categories
Search page, example: EIF5A (translation initiation factor)
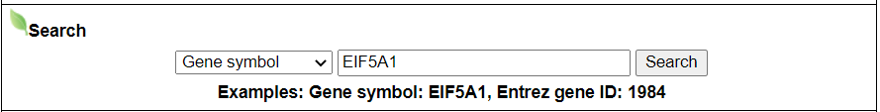
Input query - Official HUGO gene symbol or Entrez gene ID.
Gene Search Result Page

Select your translation factor gene from the gene list.
Gene Annotation Result Page

These are GeneDB's annotation categories for your query with links to their corresponding annotation parts.
1) Gene Summary.
This category shows the information of the translation factor gene. Firstly, it shows each partner gene's overall information from basic information such as symbol, alias, and locations and ENST accessions. It also shows the GO terms under the translation process (GO:0006412), which include this gene.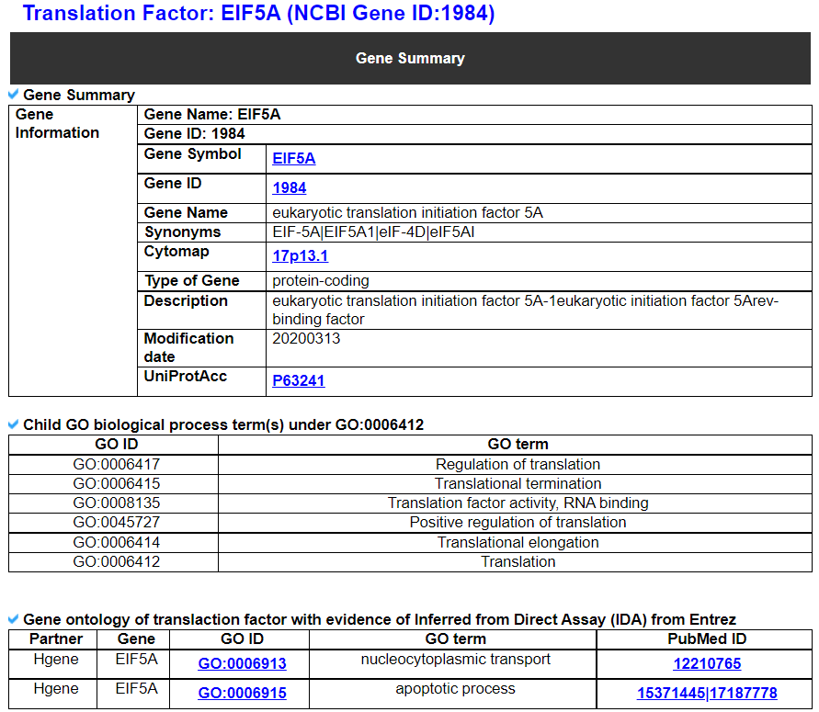
2) Translation Studies in PubMed.
For all TLFs, literature query of PubMed was performed in April 2022 using the search expression that applied to each TLF gene. Using EIF5A as an example, it is ‘((EIF5A [Title/Abstract]) AND translation [Title/Abstract])’.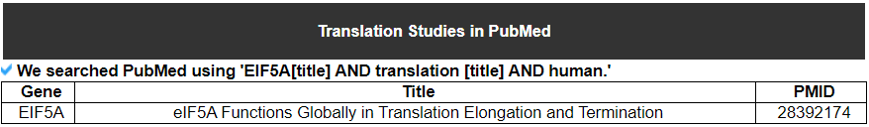
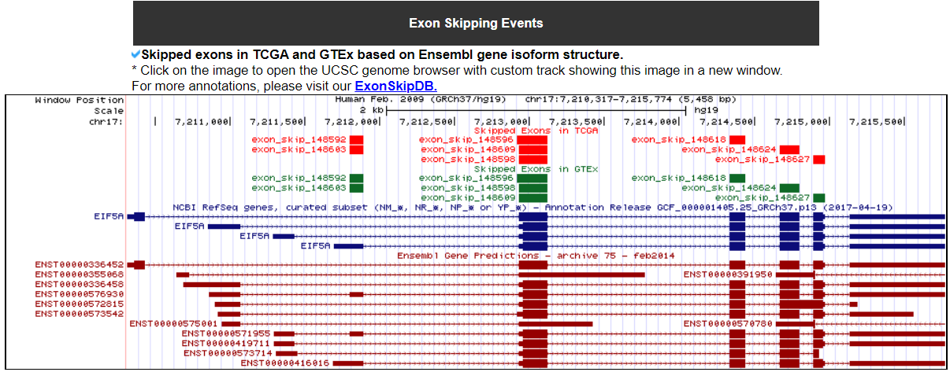
3) Exon Skippping Events in Translaction Factor gene.
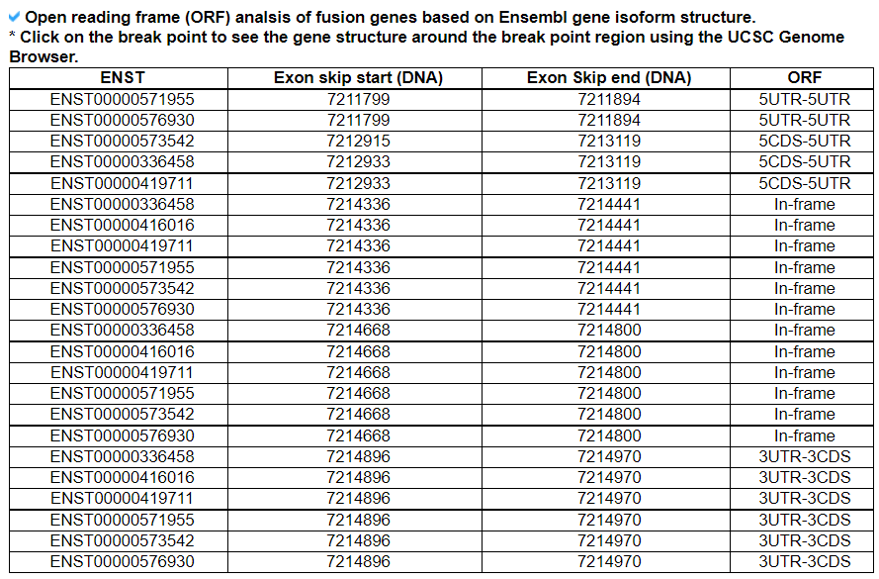
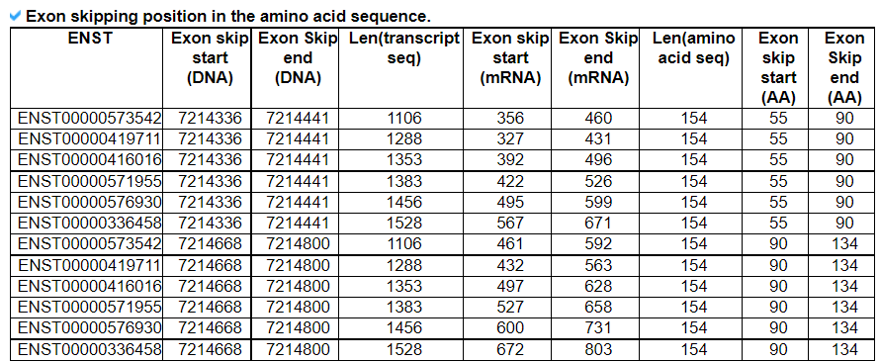
We downloaded the exon skippping event information of 8,705 patients across 33 cancer types from TCGA and of over 3,000 normal samples across 31 different tissues from GTEx(Kahles A et al. 2018). We only used the exon skipping events when they have conserved loci among the six splice sites of three exons, which involved in exon skipping such as upstream exon-skipped exon-downstream exon. Of these six sites, the four sites are the upstream exon’s donor site, skipped exons' acceptor and donor sitex, and the acceptor site of downstream exon. Based on the GENCODE v19 annotation reference genome, we have obtained 90,616 and 89,845 annotated gene structure-based exon skipping events from TCGA and GTEx, respectively.We examined the open reading frames of major isoforms' transcript sequences when there is the specific exon skipping event. When both of the nucleotide start and end positions of exon skipping are located inside of the coding region (CDS) and the number of transcript sequences after exclusion of the skipped exon sequence is a multiple of three, then we reported such exon skipped gene isoform as ‘in-frame’. If there are one or two nucleotide insertions, then we reported such transcript as ‘frame-shift’.
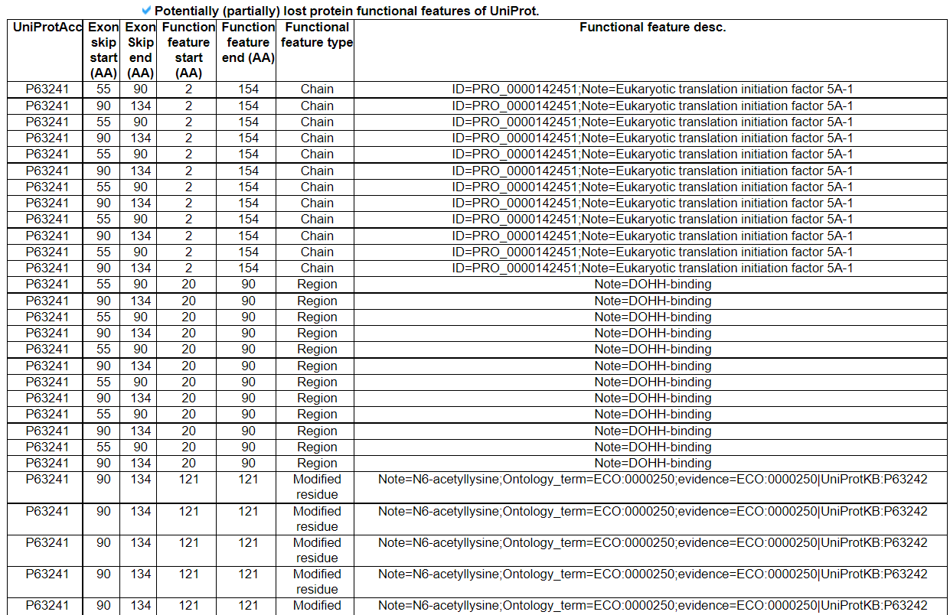
We searched the retention of 39 protein features of UniProt (6 molecule processing features, 13 region features, the 4 site features, 6 amino acid modification features, 2 natural variation features, 5 experimental info features, and 3 secondary structure features) at the canonical amino acid sequence level with skipped exon loci information. Detailed information about all of the protein features is in UniProt.
4) Expression of Translation Factor.
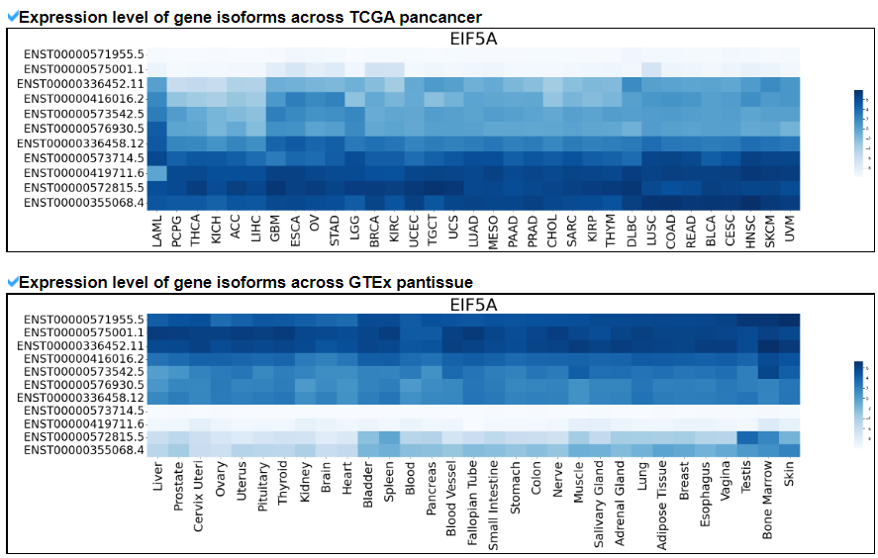
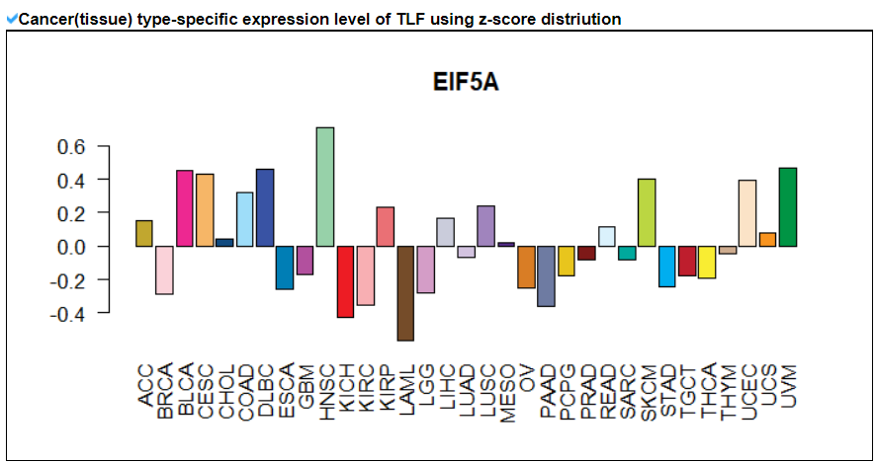
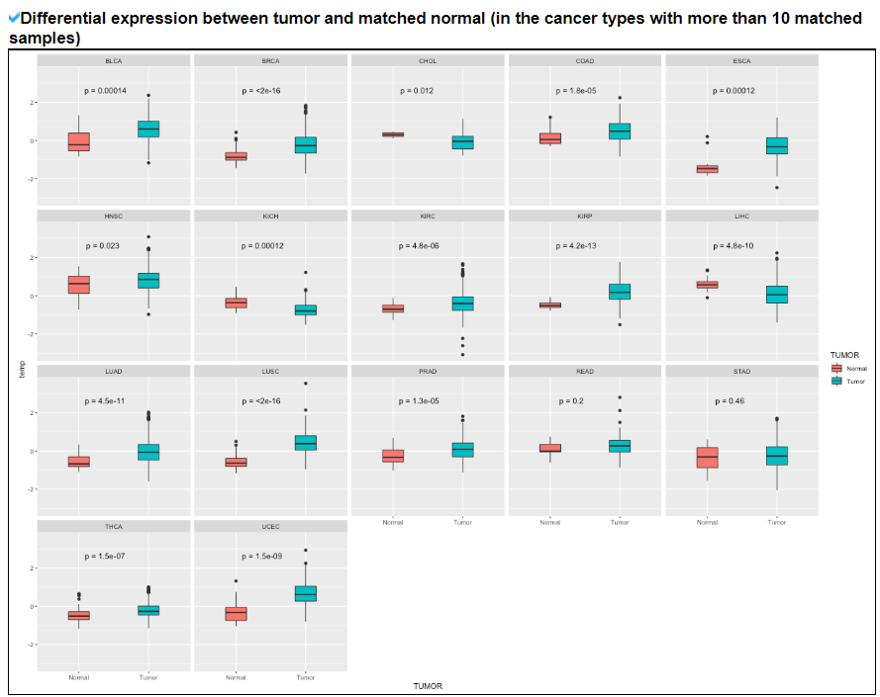
This category provides the overall landscape of gene expression with diverse aspects including gene/isoform expressoin in cancer (TCGA) and normal (GTEx), tissue specific expression patterns using z-scores, and differential expressions between tumor and matched normal samples.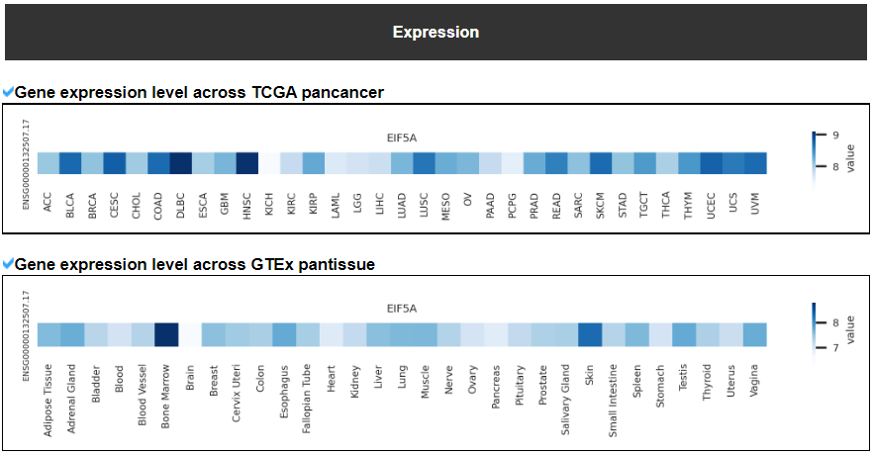
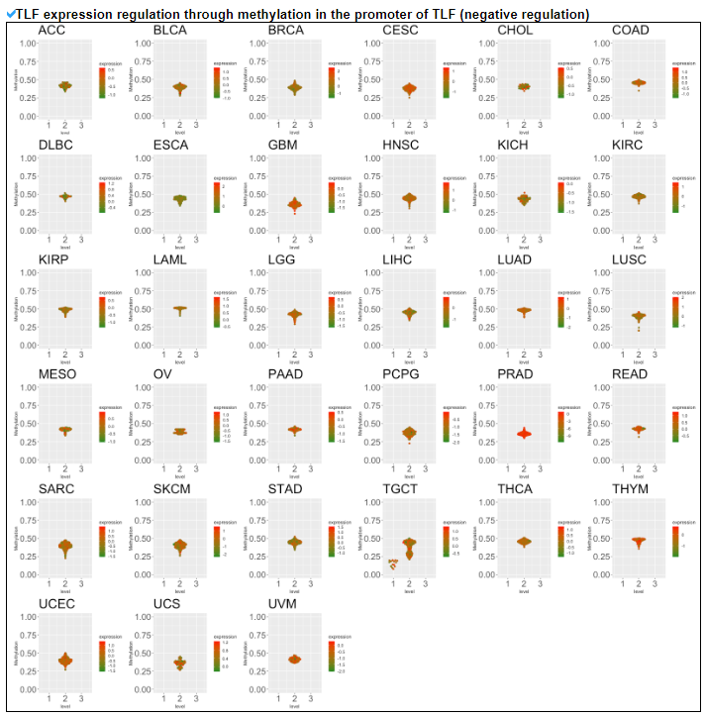
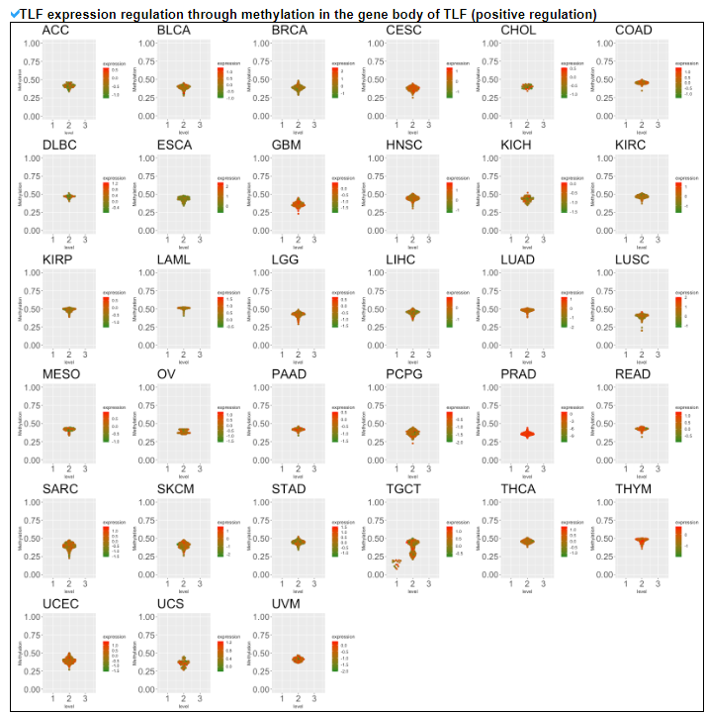
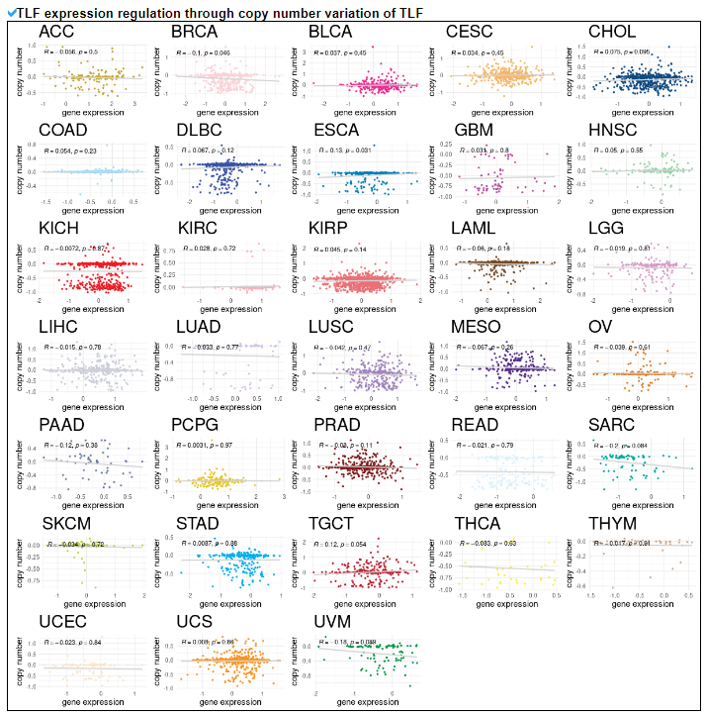
5) Expression Regulation of Translation Factor.
This category provides the potentially associated epigenetic gene expression regulatories including miRNA, methylation, and CNV on individual translation factors in 33 cancers.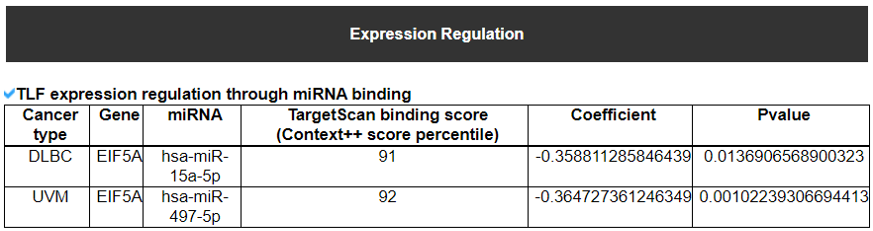
6) Associated Genes
This category provides the potentially associated important cellular gene group genes with individual human translation factors such as essential genes, cell metabolism genes, IUPHAR drug target genes, human kinase genes, cancer gene census genes, tumor suppressors, epigenetic factors, and transcription factors. We filtered with the stringent criteria (Spearman correlation coefficient>0.8 and p-value<0.05).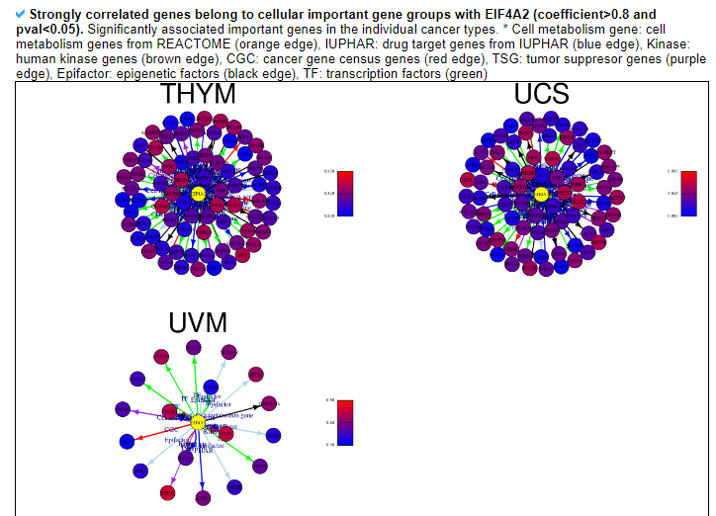
7) Translation Factor Protein's 3D Structure
This category provides the 3D structure of the translation factor protein.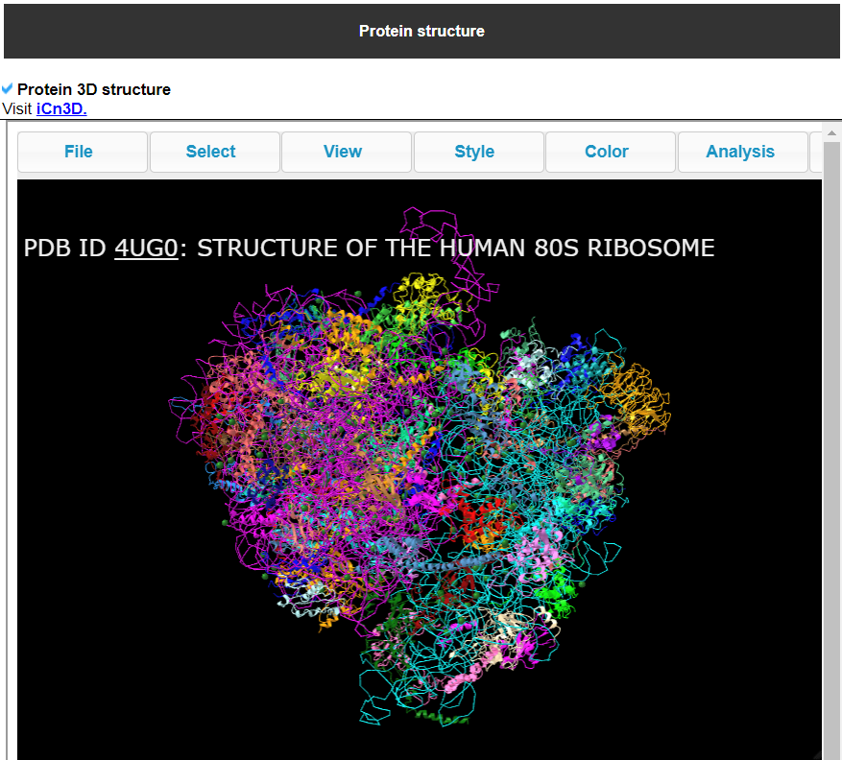
8) Protein-Protein Interaction Category
This category provides the protein-protein interaction information of individual human translation factors per cancer type. Based on the backbone of the PPI element from STRING network of each translation factor gene, we have checked the DEGs in each cancer and added these information into the network node colors. We also made up/down-regulated PPI network in pancancer.
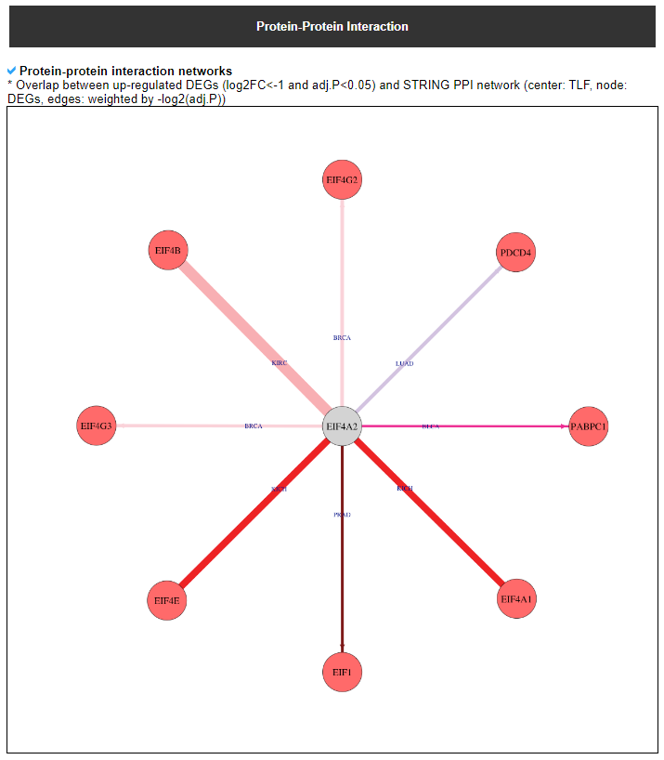
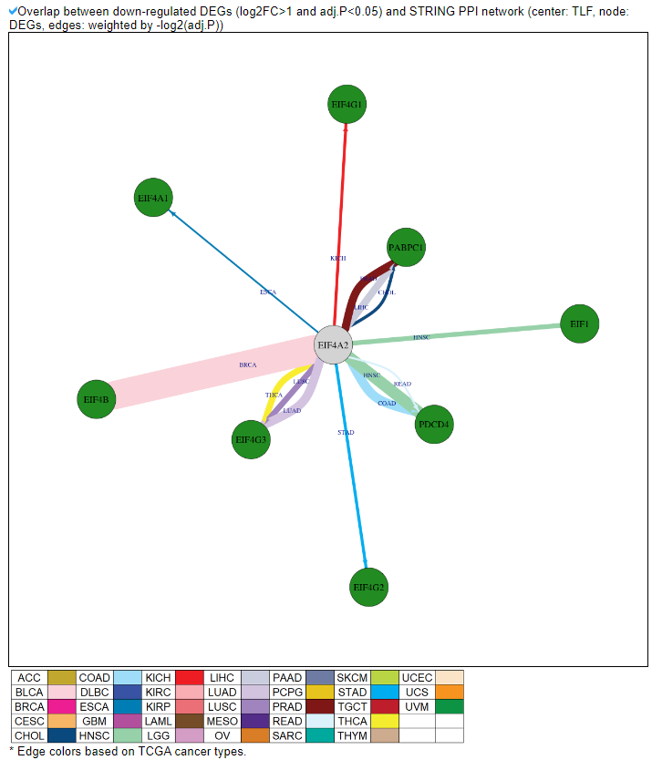
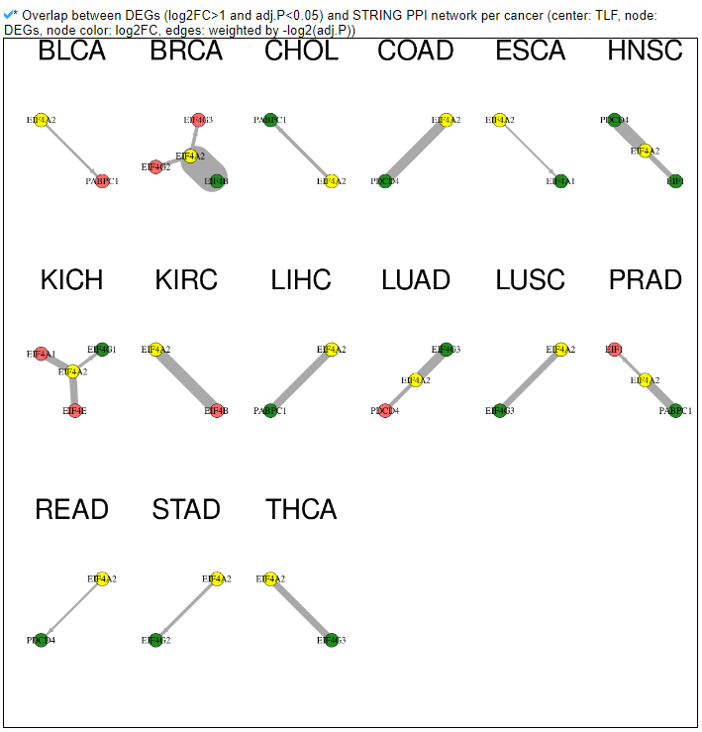
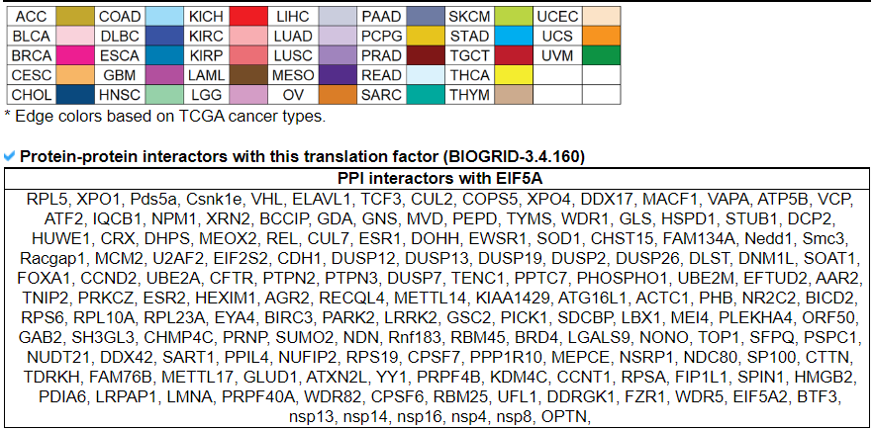
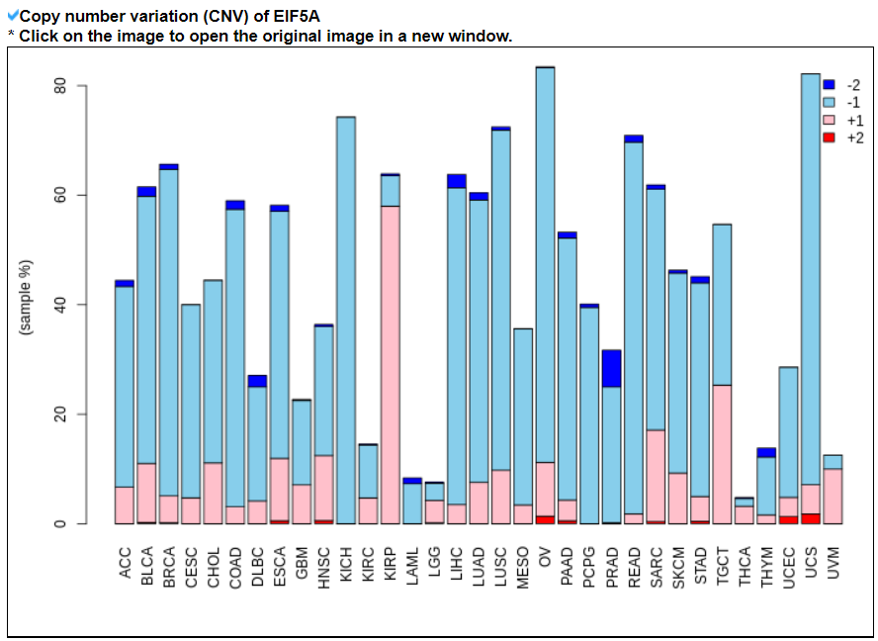
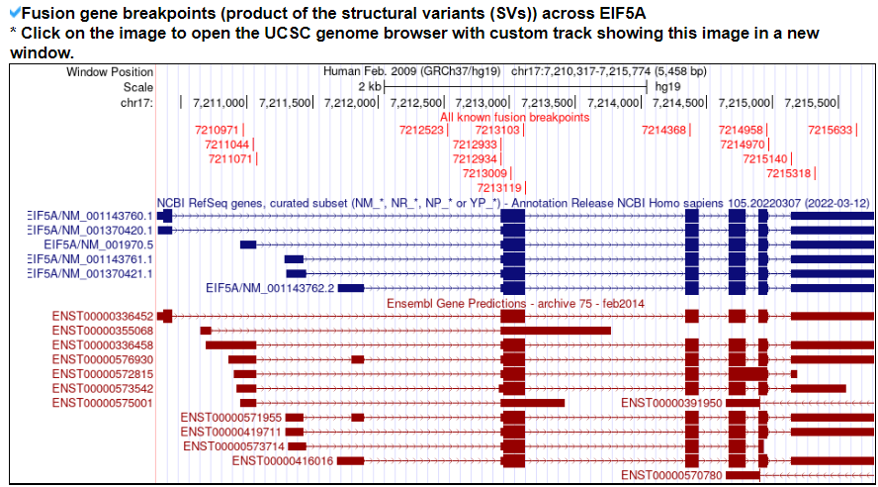
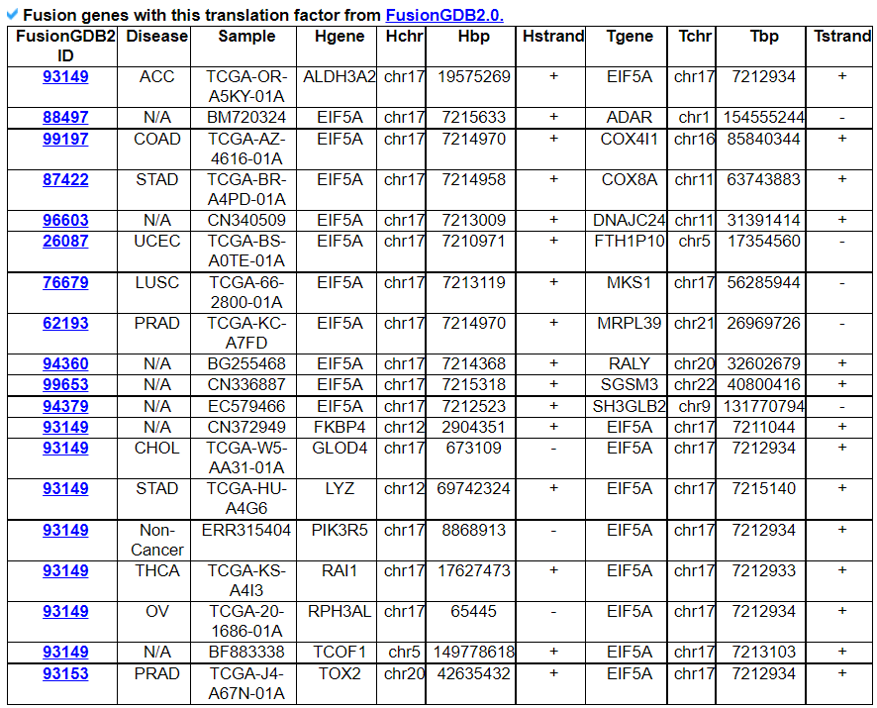
9) Mutation in Translation Factor Gene
This category provides the all available mutations on the translation factors including SNVs, Indels, CNVs, and fusion genes from TCGA. It also provides the ClinVar information of individual translation factors.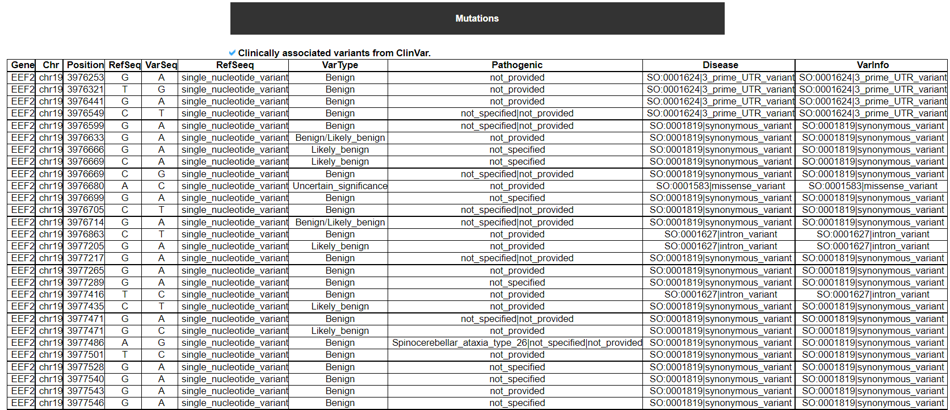
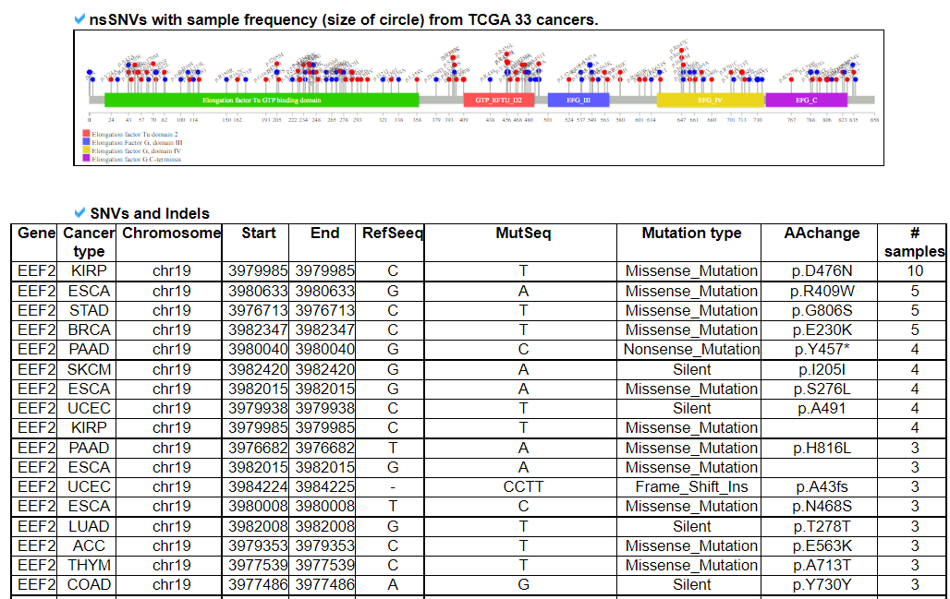
10) Prognostic Analysis of Translation Factor.
This category provides the potency of individual translation factors as the potential prognostic marker per cancer based on its expression groups (high vs low based on the median value).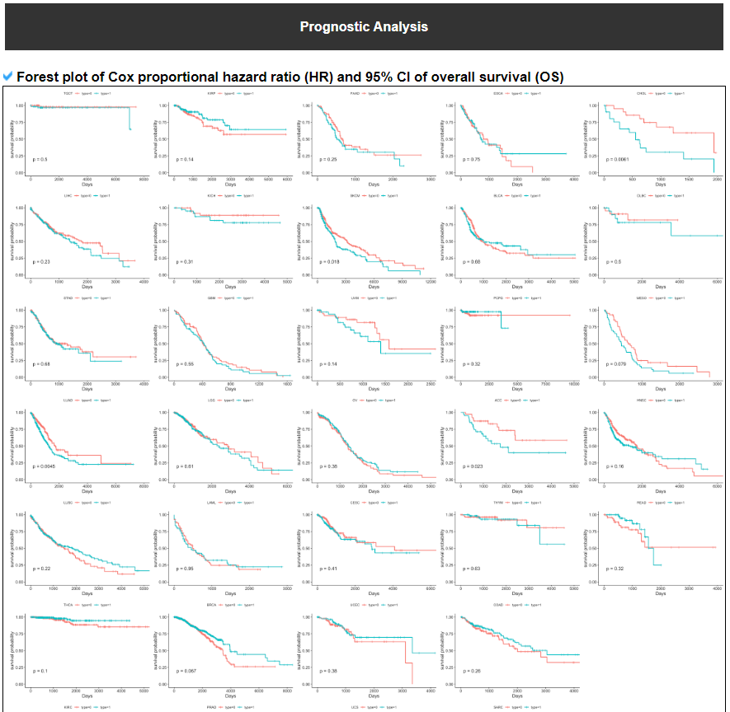
11) Translation Factor Expression in Different Genders
This category provides the analysis result of DEG between different genders (male vs female).
12) Translation Factor Expression in Different Age Group
This category provides the analysis result of DEG between different age groups (old vs young based on the median age).
13) Translation Factor Related Drugs
This table provides the DrugBank information related to each translation factor gene.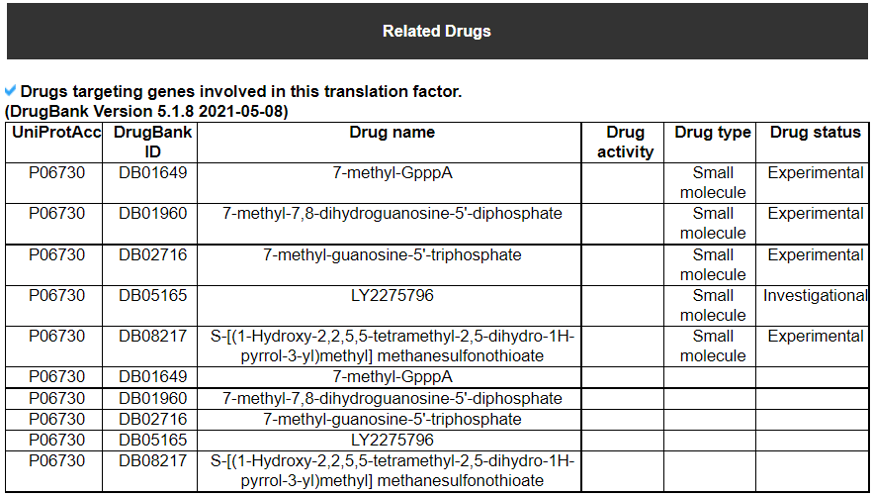
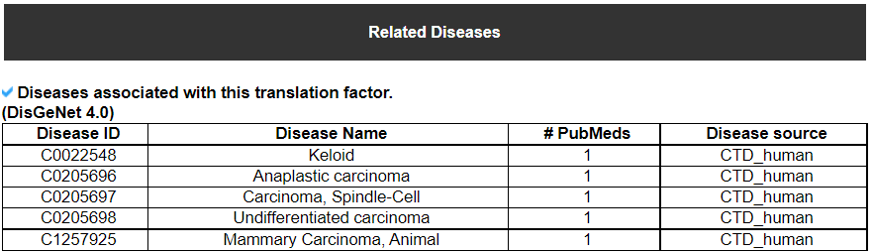
14) Translation Factor Related Diseases
This table provides disease information related to each translation factor gene.