| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| HARS1 | chr5 | 140053561 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HARS1 | chr5 | 140053598 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HARS1 | chr5 | 140053846 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140053853 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140053880 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140053888 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140053893 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140053903 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140053904 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001587|nonsense | SO:0001587|nonsense |
| HARS1 | chr5 | 140053910 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140053911 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140053935 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140053956 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054253 | A | G | single_nucleotide_variant | Uncertain_significance | Pes_cavus | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054257 | C | T | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054276 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054277 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Retinal_dystrophy|Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054277 | G | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054280 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054283 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054289 | A | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054302 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054312 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054315 | C | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054320 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054323 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054324 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054328 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054329 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054329 | T | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Urinary_urgency|Microcephaly|Intellectual_disability|Cerebellar_ataxia|Dysarthria|Motor_delay|Joint_laxity|Pes_planus|Hammertoe|Distal_muscle_weakness|Spastic_ataxia|Scoliosis|Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054350 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|not_specified|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054353 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054361 | T | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|Inborn_genetic_diseases|not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054378 | T | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054387 | GTTC | G | Microsatellite | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| HARS1 | chr5 | 140054396 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054399 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054407 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054418 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054485 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054598 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054652 | G | C | single_nucleotide_variant | Benign/Likely_benign | Usher_syndrome,_type_3B|not_specified|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054655 | GCTT | G | Microsatellite | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| HARS1 | chr5 | 140054657 | TT | AG | Indel | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054664 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001587|nonsense | SO:0001587|nonsense |
| HARS1 | chr5 | 140054685 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001587|nonsense | SO:0001587|nonsense |
| HARS1 | chr5 | 140054692 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054693 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054696 | G | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054699 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054700 | G | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140054712 | C | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054712 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054713 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054717 | G | A | single_nucleotide_variant | Benign/Likely_benign | Usher_syndrome,_type_3B|not_specified|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140054968 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054993 | C | CT | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140054993 | CT | C | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140055015 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056146 | GC | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056219 | A | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056235 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056256 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056257 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056271 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056273 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056274 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056277 | C | T | single_nucleotide_variant | Uncertain_significance | Retinal_dystrophy | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056282 | A | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056300 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056306 | T | C | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056309 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056310 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|Charcot-Marie-Tooth_disease,_axonal,_type_2w|Usher_Syndrome,_Type_III | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056322 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056323 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056343 | C | A | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056347 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056349 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056364 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056366 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056379 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056380 | G | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056381 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056386 | T | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056388 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056402 | G | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056405 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056424 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001587|nonsense | SO:0001587|nonsense |
| HARS1 | chr5 | 140056424 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056433 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056444 | T | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056452 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056456 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056457 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| HARS1 | chr5 | 140056457 | G | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056470 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056473 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056477 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056484 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056485 | T | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056504 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056555 | C | T | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056556 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056566 | T | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056584 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056601 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056602 | TACTCAAAGAGCA | T | Deletion | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| HARS1 | chr5 | 140056612 | G | GCAA | Duplication | Pathogenic | Urinary_urgency|Microcephaly|Intellectual_disability|Cerebellar_ataxia|Dysarthria|Motor_delay|Joint_laxity|Pes_planus|Hammertoe|Distal_muscle_weakness|Spastic_ataxia|Scoliosis | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| HARS1 | chr5 | 140056622 | G | A | single_nucleotide_variant | Benign/Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056628 | C | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056640 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056643 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056645 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056673 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056696 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056699 | C | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056714 | A | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056775 | A | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056874 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056879 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056906 | C | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140056924 | C | G | single_nucleotide_variant | Uncertain_significance | not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056924 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056938 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056938 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056940 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056946 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140056951 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056953 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056983 | T | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056990 | C | A | single_nucleotide_variant | Uncertain_significance | not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140056992 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057004 | AC | A | Deletion | Pathogenic | Oculomotor_apraxia|Nystagmus|Intellectual_disability|Dysarthria|Choreoathetosis|Cerebellar_atrophy|Tremor|Dysmetria|Spastic_ataxia|Peripheral_neuropathy | | |
| HARS1 | chr5 | 140057015 | C | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057022 | C | T | single_nucleotide_variant | Benign/Likely_benign | Usher_syndrome,_type_3B|none_provided|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057027 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057216 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057237 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057241 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057243 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057246 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057256 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057274 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057275 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057279 | C | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057280 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057290 | A | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|not_specified|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057292 | A | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057295 | CCA | C | Microsatellite | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HARS1 | chr5 | 140057300 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057314 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057316 | T | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057319 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057330 | A | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057332 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057386 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057481 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057485 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057488 | CTGACCT | C | Deletion | Uncertain_significance | Usher_syndrome | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| HARS1 | chr5 | 140057501 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057507 | C | A | single_nucleotide_variant | Pathogenic | Oculomotor_apraxia|Nystagmus|Intellectual_disability|Dysarthria|Choreoathetosis|Cerebellar_atrophy|Tremor|Dysmetria|Spastic_ataxia|Peripheral_neuropathy | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057508 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057509 | C | T | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_specified|none_provided|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057511 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057521 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001587|nonsense | SO:0001587|nonsense |
| HARS1 | chr5 | 140057534 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057535 | G | A | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057542 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057561 | C | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057562 | A | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140057578 | A | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057591 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057596 | A | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057598 | A | C | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140057623 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140057838 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140058494 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140058579 | A | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140058590 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058608 | T | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058615 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058632 | A | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058636 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058644 | T | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058645 | A | C | single_nucleotide_variant | Pathogenic | Usher_syndrome,_type_3B|Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001575|splice_donor_variant,SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001575|splice_donor_variant,SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058652 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058664 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058678 | A | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058699 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|Charcot-Marie-Tooth_disease,_axonal,_type_2w|not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058700 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058702 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058708 | G | T | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058712 | C | A | single_nucleotide_variant | Likely_pathogenic | Charcot-Marie-Tooth_disease,_axonal,_type_2w | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140058720 | CAA | C | Deletion | Uncertain_significance | Retinitis_pigmentosa-deafness_syndrome | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140058893 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140059374 | G | A | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease,_axonal,_type_2w|not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059379 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059386 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059387 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059405 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059405 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059406 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059414 | C | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059420 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140059508 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140059738 | GT | G | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140062389 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140062547 | C | T | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140062596 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140062689 | A | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062698 | AC | TA | Indel | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001587|nonsense,SO:0001627|intron_variant | SO:0001587|nonsense,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062728 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062741 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062750 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062751 | G | A | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062765 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_specified | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062767 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062782 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062783 | G | A | single_nucleotide_variant | Uncertain_significance | Spastic_ataxia|Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062785 | G | A | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062786 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062797 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140062819 | C | G | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140063091 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070432 | G | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070443 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070446 | G | A | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070446 | G | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070446 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070455 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070461 | T | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070465 | T | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140070478 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140070488 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HARS1 | chr5 | 140070505 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140070516 | ACCT | A | Microsatellite | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| HARS1 | chr5 | 140070517 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140070520 | C | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HARS1 | chr5 | 140070536 | A | G | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070545 | AG | A | Deletion | Benign | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070719 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070796 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070796 | T | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070797 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HARS1 | chr5 | 140070799 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|Charcot-Marie-Tooth_disease,_axonal,_type_2w|not_provided | SO:0001575|splice_donor_variant,SO:0001627|intron_variant | SO:0001575|splice_donor_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070801 | A | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070802 | G | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070818 | C | T | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B|not_provided | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070828 | A | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070829 | G | A | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070838 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B|not_specified | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070847 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070850 | G | T | single_nucleotide_variant | Benign | Usher_syndrome,_type_3B|not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070851 | A | C | single_nucleotide_variant | Likely_benign | Usher_syndrome,_type_3B | SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070873 | G | C | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070876 | G | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Usher_syndrome,_type_3B|not_specified|none_provided|not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| HARS1 | chr5 | 140070886 | C | T | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001575|splice_donor_variant,SO:0001583|missense_variant | SO:0001575|splice_donor_variant,SO:0001583|missense_variant |
| HARS1 | chr5 | 140070888 | A | G | single_nucleotide_variant | Uncertain_significance | Usher_syndrome,_type_3B | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant |
| HARS1 | chr5 | 140070953 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'HARS1[title] AND translation [title] AND human.'
We searched PubMed using 'HARS1[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer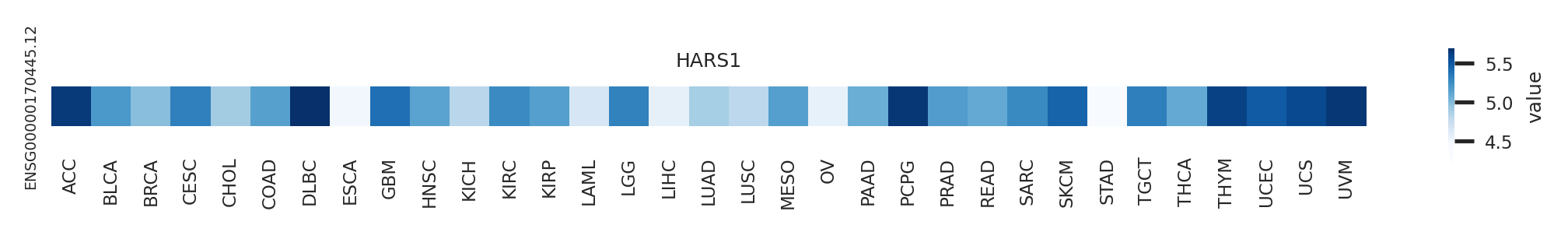
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer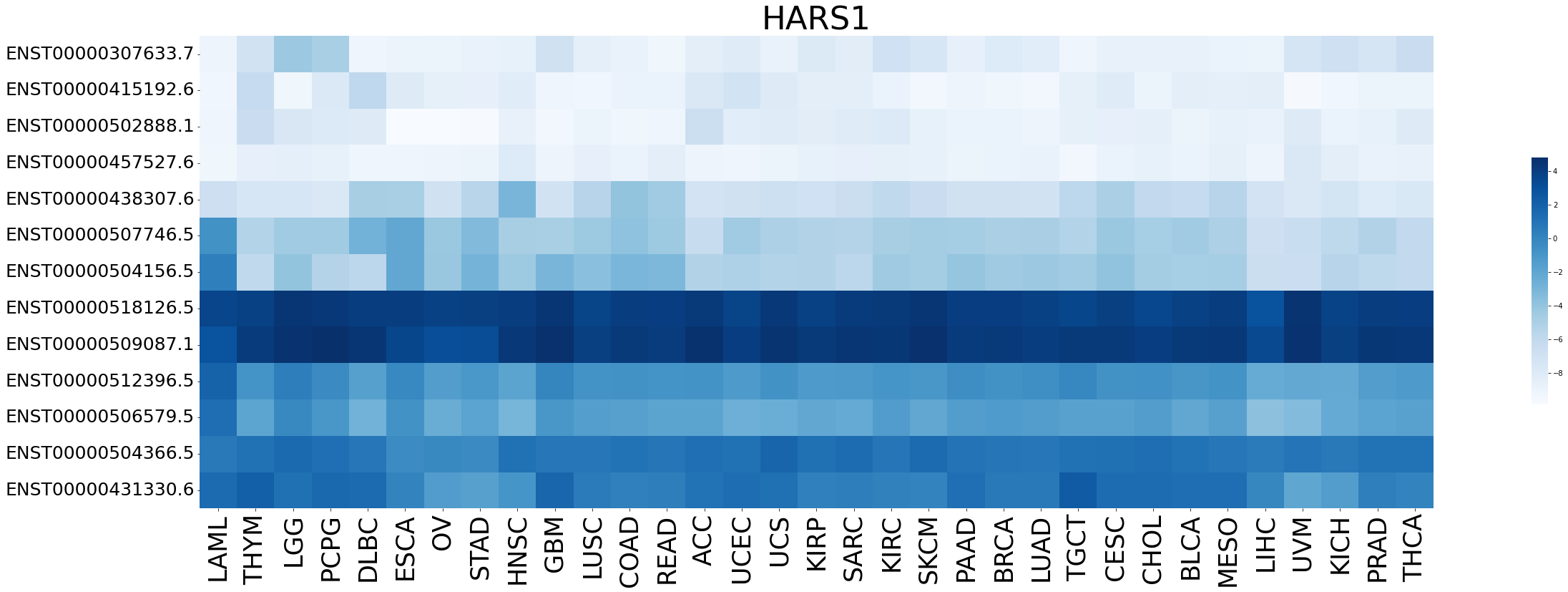
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples) Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation) Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor  Strongly correlated genes belong to cellular important gene groups with HARS1 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with HARS1 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green) Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 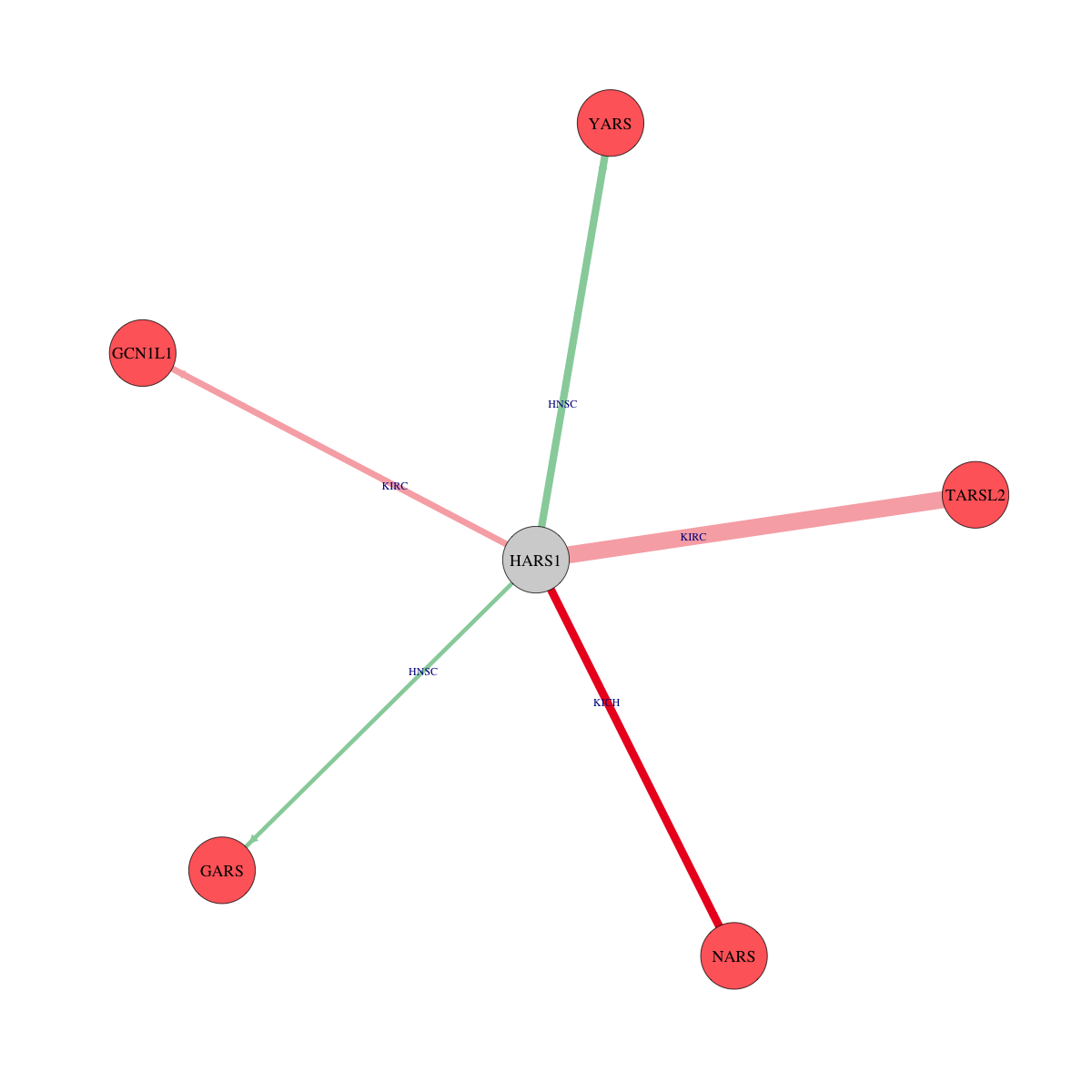
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))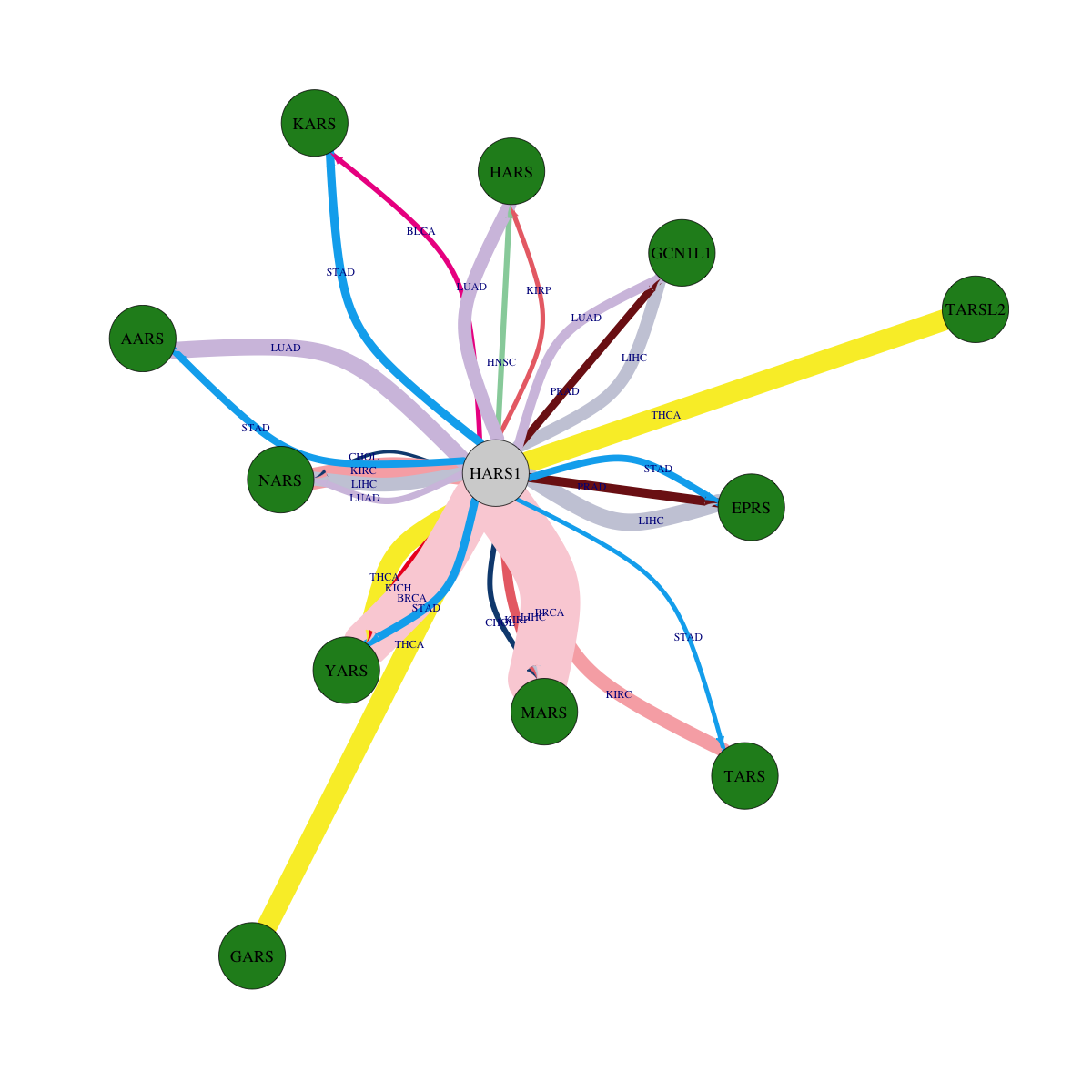
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of HARS1
Copy number variation (CNV) of HARS1 
 Fusion gene breakpoints (product of the structural variants (SVs)) across HARS1
Fusion gene breakpoints (product of the structural variants (SVs)) across HARS1 
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS)  Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05) Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05) Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
