| PPI interactors with HSPB1 |
| HSPB8, MAGED1, ILK, IGSF21, PPA1, MED31, RBM48, LRIF1, TP53, AKT1, MAPKAPK2, CRYAB, CYCS, MAPKAPK5, CRYAA, CRYBB2, HNRNPD, TGFB1I1, USP38, USP1, DMWD, SRRM2, PRKD1, UBC, MYC, G6PD, GATA1, UCHL5, YWHAQ, MME, UBE2I, HSF1, EBNA-LP, TNF, UNG, STAT2, HDAC6, CASP3, PRKCD, SNCA, CUL3, COPS5, lwr, ISG15, CRYGC, PHLDA1, SMN1, DAXX, SMURF2, PNPT1, PUM1, PALLD, NRD1, PEX19, CRK, TPM3, RNF114, CFTR, CHUK, IKBKB, VCP, FN1, VCAM1, SF3B3, EFTUD2, PRPF19, MFAP1, HSPB1, WDR83, ATF2, IQCB1, NOS2, UBL4A, ITGA4, CS, INS, SGA1, MAPKAPK3, PLG, NPM1, SOX2, LBP, PPM1A, SMARCD1, SERTAD1, ESR1, RPA1, RPA2, RPA3, WWOX, ASB4, ASB6, ASB10, ASB17, ZBTB1, MAP3K7, AARSD1, BAG3, LCE3A, IRS4, HAUS1, CEP57, HUWE1, PCSK9, EZH2, AATK, ARAF, BUB1B, IRAK1, LIMK2, SIK2, SRMS, TYK2, BTRC, HSPB6, HNRNPA1, ACBD6, ACTN1, ACTN4, ADCY9, ADPRHL2, AGAP5, AGAP4, ALPI, ANK1, ANXA1, SLC7A9, C11orf98, C15orf57, UQCC2, C6orf211, CALR, CCHCR1, CDC123, CDK5RAP3, CENPK, CEP78, AGAP7P, DFFA, EIF1AX, EIF2S1, EIF4A2, EPB41L1, FTH1, GARS, HAUS8, HECTD1, HNRNPH2, HSP90AA1, KCMF1, KCTD3, KIAA0368, KPNA3, KRT18, LSM3, MCM6, MYL12A, MRPS23, MSL1, MTIF2, NAP1L1, NEXN, NSMCE4A, NVL, PELO, PSMA3, PSMA6, PSMB2, PSMD1, PTGES3, PTPRA, RASSF9, RBM25, RBX1, RPAP3, RYBP, S100A4, SAT1, SF3A3, SNRNP200, SNRPF, SQSTM1, STUB1, TARBP2, TARS, TOMM70A, TOX4, TPT1, UFD1L, UQCRB, WWP1, XPO1, XRCC5, YWHAB, ZBTB17, ZNF197, EIF3M, EIF5, EIF4G3, FLNB, FKBP4, GSTO1, MRPL40, OBSL1, PGM2, PAF1, RAD23A, RBPJ, TJP2, THOC5, ACTN2, ADRBK2, AGAP3, AMD1, ANAPC7, ANKRD7, AP2B1, ARHGAP28, ARHGEF7, ARIH1, ARRDC5, BVES, C12orf10, SPACA7, ERICH6, CAMK1, CBX1, CCDC53, CCDC64, CCDC70, CCNC, CETN3, CLCN3, CPSF3, CRNKL1, DES, DMAP1, EHD1, ENPP2, EPB41L3, ERCC5, FAM71D, C2orf73, FXR1, GAPVD1, GLMN, GNPAT, GUCA1A, HNRNPA2B1, HNRNPU, HSPA4, IARS2, INPP5K, ISG20L2, KANSL1, KLC1, KRT72, LONRF1, LUZP1, MRPL28, MYL9, MYLK, NBAS, NAPA, NCKIPSD, NEK10, NFU1, OPTN, OSBPL9, PALM3, PHYHIPL, PPP2R3C, PRPF40A, PTPN3, RAB41, RALA, RPSA, RSPH3, SBF1, SEC13, PNISR, SMARCA2, SPATA7, SPIN1, SPTBN1, SRP72, SUPT5H, SYCE1, TBC1D1, TBC1D3B, TMCO3, TRIM24, TRIM54, TSNAXIP1, TTC3, TTC39C, TXNL1, WDR66, YWHAE, ZBTB37, ZFAT, ZNRF3, ACAP2, RBM39, ADD3, ATXN10, KNSTRN, MRGBP, CCNDBP1, CCNO, DNAJC21, EIF4G2, EPB41L2, GDI2, GOLGA8A, GRIPAP1, GTSF1, HDLBP, HNRNPF, HSP90AB1, HSPA5, IGBP1, LIG1, MPHOSPH6, MYL12B, PSMC1, SMARCA4, SNW1, VIM, YWHAH, YWHAZ, ZNF131, TCPT, AHCY, AHNAK, ARHGDIA, ATIC, CARHSP1, CDK1, CSRP1, DUT, EEF1A1, EEF1B2, GLO1, GLOD4, GMPS, GRPEL1, HMGN5, HN1, HN1L, HNRNPH1, HSPA8, HSPA9, EEF1D, EIF4B, ERP29, HNRNPH3, LASP1, NAMPT, OXCT1, PAICS, PKM, PTBP1, SARS, SDHA, SEPHS1, USP14, IPO7, ITPA, MSN, OLA1, PDLIM1, PFDN2, PITPNA, PITPNB, PRDX6, PSAT1, STIP1, STMN1, TARDBP, TES, TKT, TUBA4A, UAP1, UBA2, UBE2N, UGDH, ZYX, NTRK1, STRN, TUBA1C, ARMC6, Bmpr1a, Kctd5, MCM2, CDC5L, EGFR, CTGLF10P, CDC73, PHF12, HYPK, DUSP16, PTPN22, CYLD, DLST, PDHA1, EDEM3, TRIM25, CCND2, BRCA1, YAP1, MAPK14, E2F6, PCGF5, TRIM14, RIPK4, MAPK6, ADSS, ARNT, BCL2L1, CTNNB1, HDAC4, JUP, MAP2K1, MTCH2, POLD1, TGFB1, BRAF, CCNT1, KRAS, PRPF8, AAR2, PIH1D1, RNF4, PRKCZ, MDH2, CSNK2A1, CSNK2A2, PPT1, MB21D1, CDK9, KRT17, KIAA1429, AMBRA1, RC3H1, RC3H2, LRRK2, NR2C2, MECOM, AGRN, ATXN3, MPO, ALB, ELANE, HIST1H4A, CREBBP, LUCAT1, RARA, SCARB2, ABCD3, HAX1, CLU, TFRC, NNT, LETM1, PDE3A, CYC1, NUP93, BAG2, RUVBL2, RUVBL1, MCM7, HADHA, GCN1L1, AKAP8, PANK4, RCN2, TUBA1B, SLC25A13, PRKDC, XPOT, MSH6, MCCC1, ATP2A2, SEC16A, AIFM1, MAP1B, AKAP8L, MCCC2, TOMM40, STT3A, TIMM21, ERLIN2, PHB, PHB2, SLC25A6, GLUD1, SLC25A5, ATP5A1, CHCHD3, DNAJC11, ERLIN1, EMD, LBR, IMMT, ACADM, GEMIN4, SFXN3, KIAA1217, SLC25A12, MLF2, SLC25A3, UQCRC2, SLC25A1, PC, SUCLG2, YME1L1, PTPN1, GFPT1, MAPT, FUS, TAF15, MATR3, CLUAP1, BIRC3, LMBR1L, WWP2, ATXN1, HTT, PSEN1, PLEKHA4, FANCD2, NGB, HCVgp1, SP1, ETS1, STAT3, ZC3H18, ABI1, BAD, BLK, CAMK2A, CASP9, CAV1, CAV2, CDC42, CSK, DOK2, DUSP1, DUSP4, DUSP6, ELK1, ELK4, FGR, GAB2, GRB10, GRB2, HCK, HDAC1, LCK, MAP2K3, MAP2K5, MAPK7, MYLK2, PAK1, PEBP1, PRKAR1A, PRKCA, PRKCB, PRKCE, PRKCG, PRKCI, PTK6, PTPN12, PXN, RAB5A, RAC1, RAF1, RASA1, RHOA, RIPK1, RPS6KA3, SH2D3C, SH3BGRL, SH3GL3, SH3KBP1, SHC1, SHOC2, SOCS1, VAV1, NME1, APPL1, CIT, ANLN, AURKB, CHMP4B, ECT2, KIF14, KIF20A, KIF23, PRC1, TRIM55, TRIM63, SUMO2, BRD4, ASXL1, JADE1, LGALS7, AR, DDX39B, TRIM37, SLC26A4-AS1, NFKBIA, ABCG2, FZR1, FXR2, HSPB2, NPAS1, METTL21B, SPRTN, BTF3, F13A1, CCNF, MAP1LC3B, |
| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| HSPB1 | chr7 | 75931676 | C | T | single_nucleotide_variant | Likely_benign | not_provided | | |
| HSPB1 | chr7 | 75931729 | G | C | single_nucleotide_variant | Likely_benign | not_provided | | |
| HSPB1 | chr7 | 75931901 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | | |
| HSPB1 | chr7 | 75931908 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | | |
| HSPB1 | chr7 | 75931974 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | | |
| HSPB1 | chr7 | 75931984 | C | T | single_nucleotide_variant | Benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | | |
| HSPB1 | chr7 | 75932011 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_specified|not_provided | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| HSPB1 | chr7 | 75932015 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease|not_specified | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| HSPB1 | chr7 | 75932025 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| HSPB1 | chr7 | 75932026 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_provided | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| HSPB1 | chr7 | 75932030 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932032 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932036 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932038 | G | A | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932045 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932046 | T | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932048 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932049 | C | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932049 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932050 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932053 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932056 | G | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932058 | T | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932065 | G | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932066 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932068 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932074 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932076 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75932081 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932081 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932089 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932099 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932100 | C | CGCATAGCCGCCTCTTCGACCAG | Duplication | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001587|nonsense,SO:0001821|inframe_insertion | SO:0001587|nonsense,SO:0001821|inframe_insertion |
| HSPB1 | chr7 | 75932109 | G | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932109 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932113 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932118 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932128 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932129 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932137 | C | G | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932139 | G | C | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932145 | C | T | single_nucleotide_variant | Pathogenic | HSPB1-related_axonal_neuropathies|Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932146 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932150 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932150 | G | GAGTGGTCGC | Duplication | Uncertain_significance | not_provided | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| HSPB1 | chr7 | 75932155 | G | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75932156 | T | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932158 | G | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932163 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75932167 | A | G | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932168 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932171 | G | A | single_nucleotide_variant | Uncertain_significance | not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932178 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932182 | G | A | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75932187 | G | A | single_nucleotide_variant | Likely_pathogenic | Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932193 | T | TGCGCCCC | Duplication | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75932196 | GC | G | Deletion | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease,_dominant_intermediate_C | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75932197 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932198 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932200 | C | CGCGCCCT | Insertion | Pathogenic | Charcot-Marie-Tooth_disease | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75932200 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932203 | G | GC | Duplication | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75932204 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932207 | C | CCCG | Microsatellite | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| HSPB1 | chr7 | 75932207 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_specified|none_provided|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932213 | G | A | single_nucleotide_variant | Uncertain_significance | HSPB1-Related_Disorder|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932213 | G | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932219 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932221 | G | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932227 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932231 | G | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932233 | G | C | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932239 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932245 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932254 | C | G | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932258 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932262 | GC | TT | Indel | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932264 | C | T | single_nucleotide_variant | Benign | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932265 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932265 | G | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932269 | A | G | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932272 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932277 | GC | AT | Indel | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932279 | G | A | single_nucleotide_variant | Pathogenic/Likely_pathogenic | HSPB1-Related_Disorders|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932279 | G | C | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932279 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932280 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932282 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932284 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932286 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932293 | C | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932295 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932306 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932309 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932310 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932315 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932315 | CG | C | Deletion | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75932323 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932324 | C | A | single_nucleotide_variant | Likely_pathogenic | Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932334 | A | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932334 | A | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932347 | G | C | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932356 | G | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932360 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932362 | C | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932368 | C | G | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932369 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932378 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932380 | G | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75932385 | A | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932393 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75932399 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932400 | CCCCCTGCTCCTG | C | Deletion | Likely_benign | Charcot-Marie-Tooth_disease | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932403 | C | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932403 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932427 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932475 | C | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932701 | C | CA | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932861 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932862 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75932930 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933052 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_type_4 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933059 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933106 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933108 | T | TC | Duplication | Likely_benign | Charcot-Marie-Tooth_disease | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933108 | TC | T | Deletion | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933109 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933112 | C | G | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933112 | C | T | single_nucleotide_variant | Benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933113 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933113 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|Charcot-Marie-Tooth_disease_type_4 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933114 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933115 | A | G | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933122 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933123 | G | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933126 | C | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933126 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933126 | CGAG | C | Microsatellite | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| HSPB1 | chr7 | 75933133 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933133 | C | T | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933134 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933134 | G | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933137 | A | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933146 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933148 | G | A | single_nucleotide_variant | Uncertain_significance | not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933149 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933152 | A | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933153 | C | T | single_nucleotide_variant | Benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933154 | A | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933157 | T | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933158 | C | A | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933158 | C | G | single_nucleotide_variant | Likely_pathogenic | Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933158 | C | T | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933160 | C | T | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933161 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933161 | G | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933165 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933169 | A | G | single_nucleotide_variant | Likely_pathogenic | Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933170 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933171 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933172 | C | G | single_nucleotide_variant | Pathogenic | HSPB1-related_axonal_neuropathies|Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933175 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933179 | A | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933182 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933187 | G | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933191 | T | C | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933195 | G | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933226 | T | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933242 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933294 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001627|intron_variant | SO:0001627|intron_variant |
| HSPB1 | chr7 | 75933304 | G | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933304 | G | GC | Duplication | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933309 | C | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933310 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933321 | C | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933323 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933324 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933332 | T | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933341 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933347 | CCT | C | Deletion | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933362 | A | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933368 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933370 | G | A | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933373 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933376 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933376 | CA | C | Deletion | Pathogenic | Charcot-Marie-Tooth_disease | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933379 | GC | G | Deletion | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933380 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933382 | CA | C | Deletion | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|not_specified | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933394 | GC | CT | Indel | Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75933395 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75933404 | G | T | single_nucleotide_variant | Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001587|nonsense | SO:0001587|nonsense |
| HSPB1 | chr7 | 75933410 | A | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933411 | C | G | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933411 | C | T | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933416 | C | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933416 | C | G | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933416 | C | T | single_nucleotide_variant | Pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933417 | C | T | single_nucleotide_variant | Pathogenic | Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933423 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933424 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933426 | T | C | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933427 | C | T | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933432 | C | T | single_nucleotide_variant | Pathogenic | Distal_hereditary_motor_neuronopathy_type_2B | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933434 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933437 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933439 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933442 | G | C | single_nucleotide_variant | Likely_pathogenic | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933443 | CTTGGGGGCCCAGA | C | Deletion | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933445 | T | C | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Charcot-Marie-Tooth_disease|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933446 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933470 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933478 | TGCC | T | Microsatellite | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| HSPB1 | chr7 | 75933479 | G | GC | Duplication | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| HSPB1 | chr7 | 75933482 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933482 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933487 | G | A | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| HSPB1 | chr7 | 75933487 | G | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F | SO:0001583|missense_variant | SO:0001583|missense_variant |
| HSPB1 | chr7 | 75933501 | C | T | single_nucleotide_variant | Benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B|Charcot-Marie-Tooth_disease|not_specified|none_provided | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HSPB1 | chr7 | 75933510 | A | C | single_nucleotide_variant | Likely_benign | Charcot-Marie-Tooth_disease | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HSPB1 | chr7 | 75933512 | C | T | single_nucleotide_variant | Benign/Likely_benign | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HSPB1 | chr7 | 75933576 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HSPB1 | chr7 | 75933586 | CAAAT | C | Deletion | Uncertain_significance | Distal_hereditary_motor_neuronopathy|Charcot-Marie-Tooth_disease,_type_2 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HSPB1 | chr7 | 75933606 | C | T | single_nucleotide_variant | Uncertain_significance | Charcot-Marie-Tooth_disease_axonal_type_2F|Distal_hereditary_motor_neuronopathy_type_2B | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| HSPB1 | chr7 | 75933712 | T | G | single_nucleotide_variant | Benign | not_provided | | |
| HSPB1 | chr7 | 75933718 | C | T | single_nucleotide_variant | Likely_benign | not_provided | | |
| HSPB1 | chr7 | 75933750 | C | T | single_nucleotide_variant | Likely_benign | not_provided | | |
| HSPB1 | chr7 | 75933766 | G | C | single_nucleotide_variant | Likely_benign | not_provided | | |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'HSPB1[title] AND translation [title] AND human.'
We searched PubMed using 'HSPB1[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)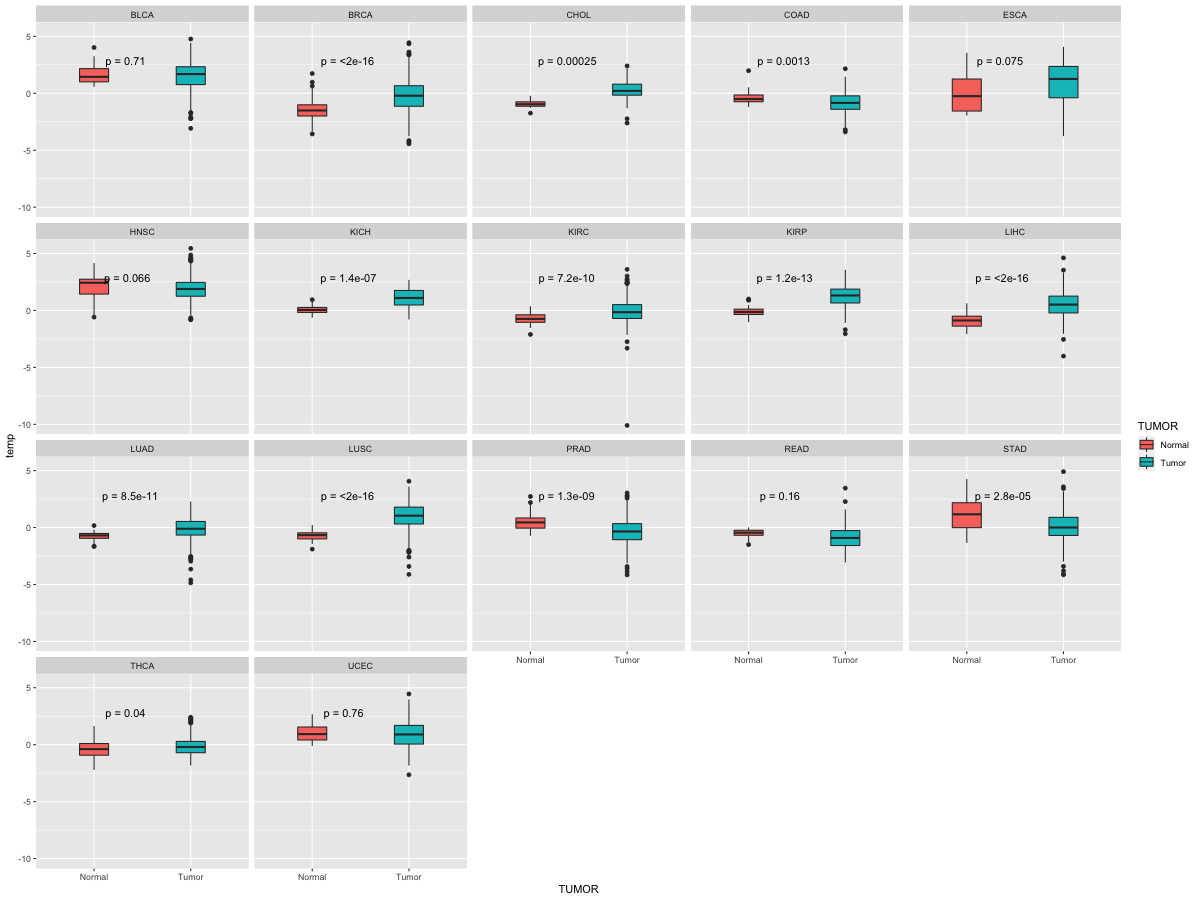
 Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
 Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor 
 Strongly correlated genes belong to cellular important gene groups with HSPB1 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with HSPB1 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)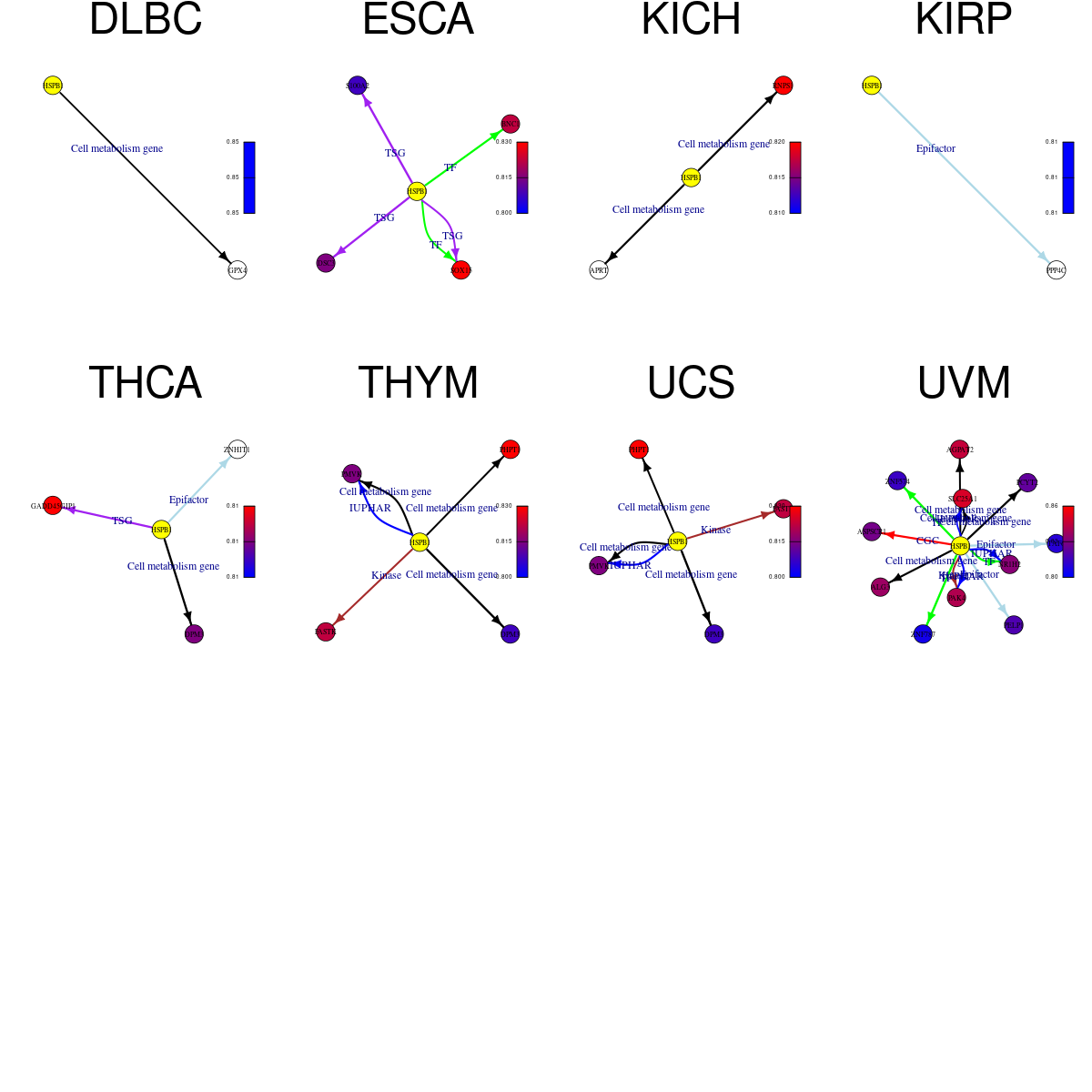
 Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 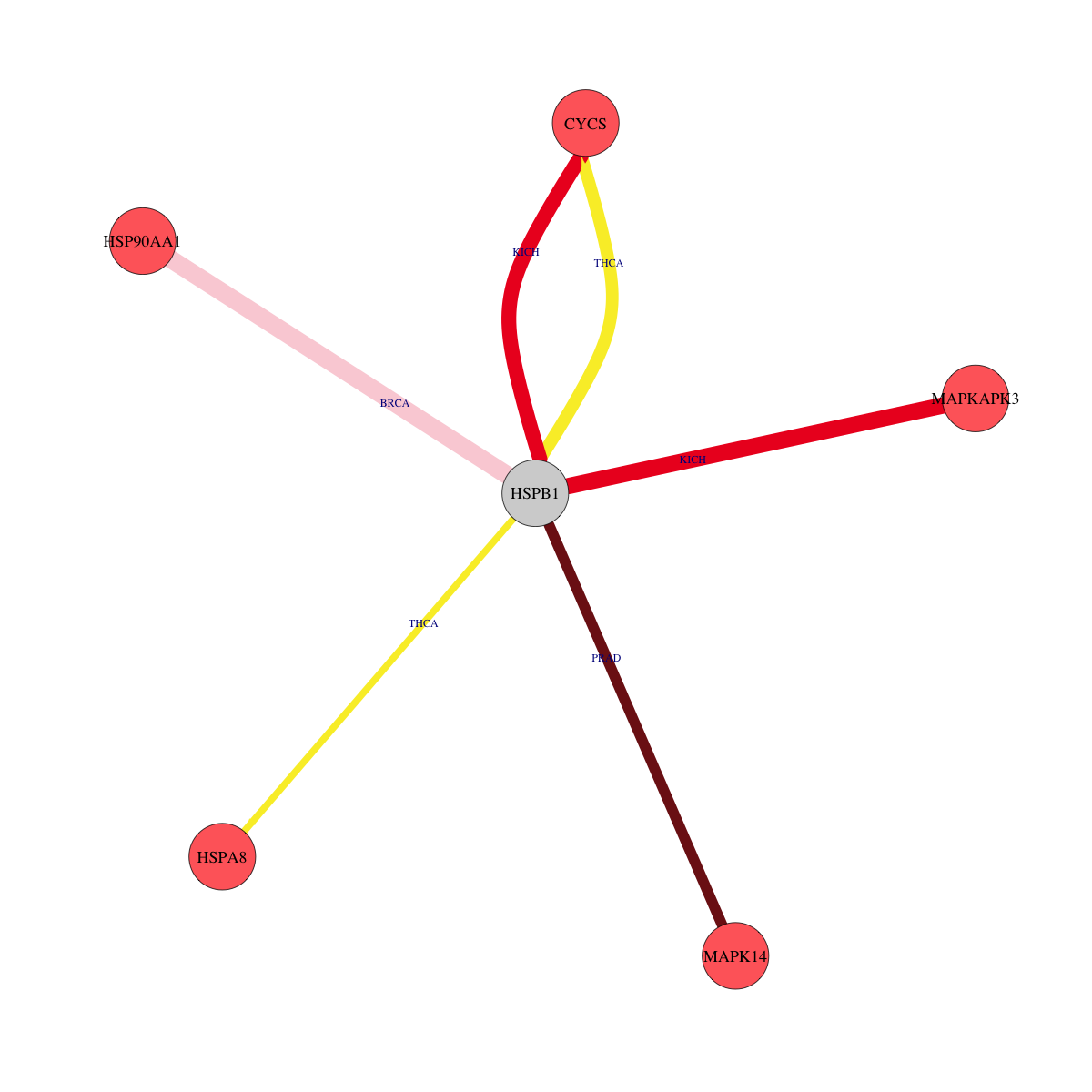
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))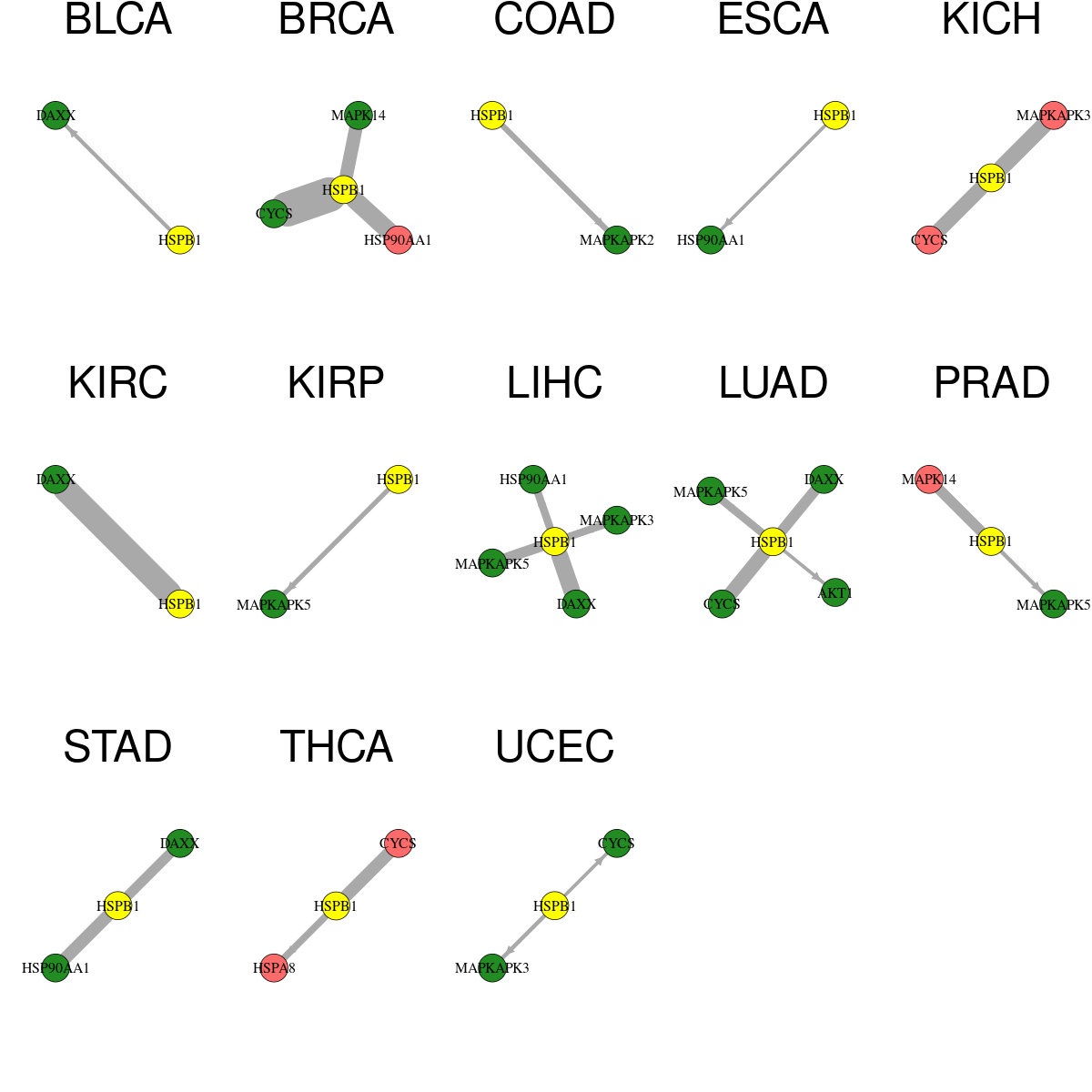
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of HSPB1
Copy number variation (CNV) of HSPB1 
 Fusion gene breakpoints (product of the structural variants (SVs)) across HSPB1
Fusion gene breakpoints (product of the structural variants (SVs)) across HSPB1 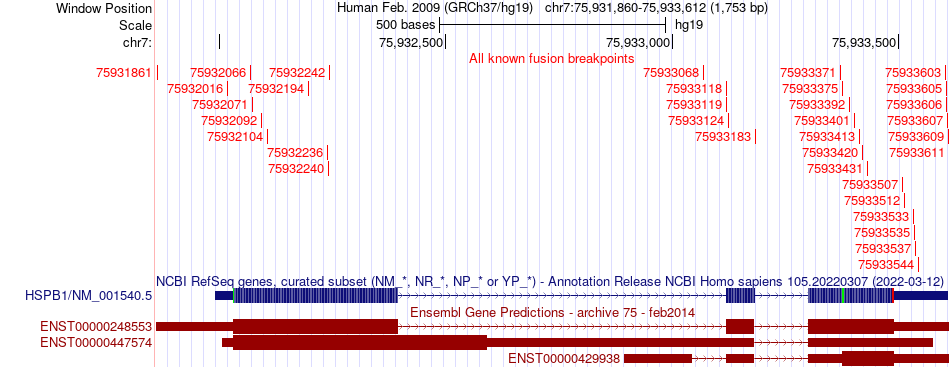
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS) 
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)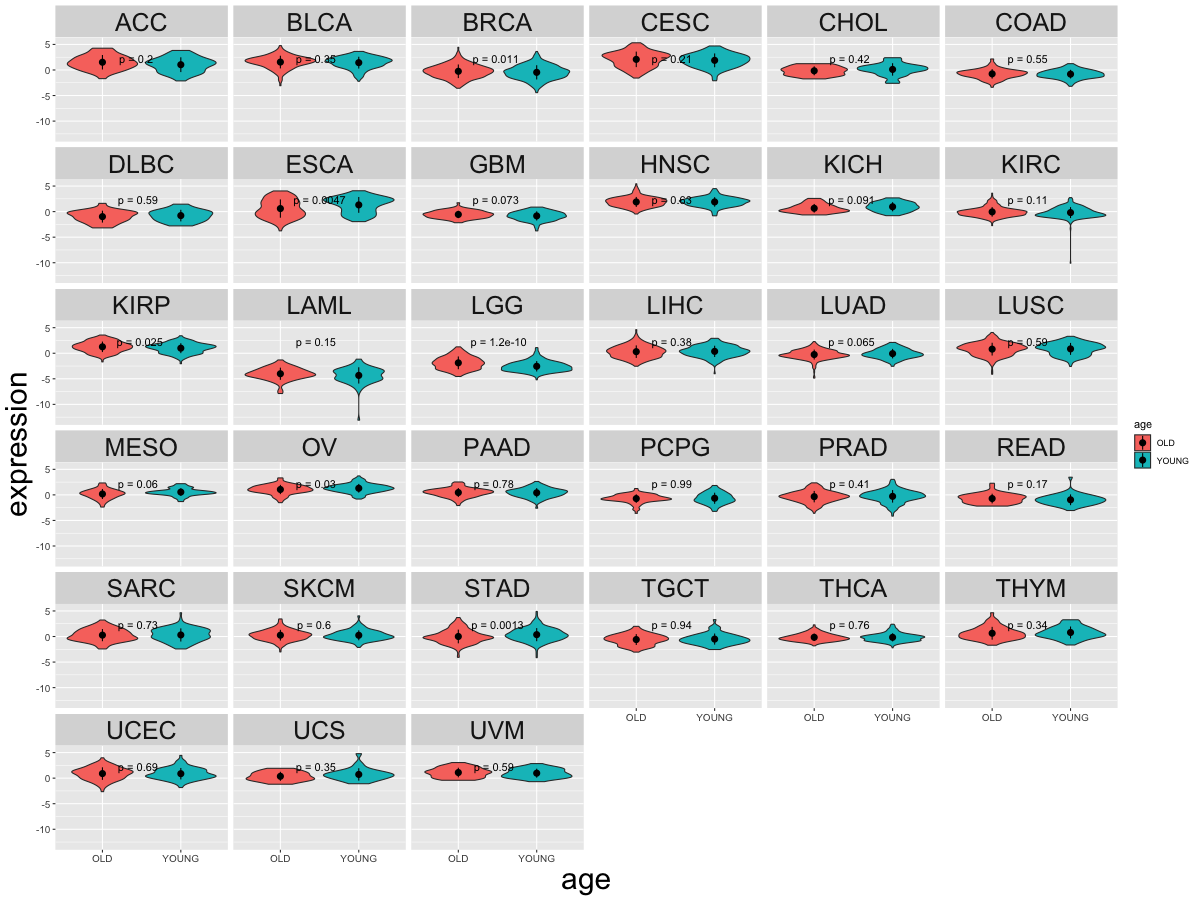
 Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
