| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| ITGA2 | chr5 | 52285077 | C | G | single_nucleotide_variant | Benign | not_provided | | |
| ITGA2 | chr5 | 52285098 | G | T | single_nucleotide_variant | Benign | not_provided | | |
| ITGA2 | chr5 | 52285117 | T | C | single_nucleotide_variant | Benign | not_provided | | |
| ITGA2 | chr5 | 52285175 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | | |
| ITGA2 | chr5 | 52285200 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| ITGA2 | chr5 | 52285210 | T | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| ITGA2 | chr5 | 52285264 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| ITGA2 | chr5 | 52285286 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| ITGA2 | chr5 | 52285309 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52285644 | G | GCA | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52285644 | G | GCACA | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52285663 | CACAG | C | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52285667 | G | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52322369 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52322598 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52322636 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52322684 | T | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52322708 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52322721 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52322982 | T | TGATA | Duplication | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52337756 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52337783 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52337908 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52337983 | G | A | single_nucleotide_variant | Benign/Likely_benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52338024 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52338083 | T | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52338227 | T | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52338241 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52338276 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52340862 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52341064 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52341110 | T | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52343901 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52344046 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52344319 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52344487 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52344543 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52344572 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52344584 | G | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52344610 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52344780 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52344837 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52344842 | CTT | C | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52347243 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52347246 | G | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52347366 | A | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52347369 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52347561 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52351182 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52351236 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52351242 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52351357 | A | T | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52351377 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52351413 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52351437 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52351459 | A | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52351746 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52351838 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52351876 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52351879 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52351989 | A | AT | Duplication | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52353613 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52353866 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52353899 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52354103 | A | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52355516 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52355566 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52355706 | T | C | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52355719 | G | A | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52355832 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52355854 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52356006 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52356692 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52356701 | T | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52356790 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52356859 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52358681 | G | A | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52358751 | A | C | single_nucleotide_variant | Benign/Likely_benign | Platelet-type_bleeding_disorder_9|Fetal_and_neonatal_alloimmune_thrombocytopenia | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52358757 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52358761 | T | TA | Duplication | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52358762 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52360777 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52360928 | C | T | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52361219 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52361665 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ITGA2 | chr5 | 52362765 | A | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52362941 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52362962 | C | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52363073 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52365954 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52366090 | G | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52366138 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52366162 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52366284 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52366336 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52366381 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52367549 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52367666 | A | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52367706 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52367715 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52367773 | C | T | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52367956 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52368366 | A | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52368472 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52368473 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52368522 | C | CT | Duplication | Uncertain_significance | See_cases | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52368525 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52368546 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52368594 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52368922 | T | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52368952 | C | A | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52369001 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52369002 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52369003 | C | G | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52369021 | A | G | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52369086 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52369104 | C | G | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52369193 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52369335 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52370174 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52370226 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52370251 | T | C | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52370432 | G | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52370732 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52370900 | A | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52370904 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52370944 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52370956 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52371047 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52371089 | A | G | single_nucleotide_variant | Benign/Likely_benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52371105 | C | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52371370 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52374590 | A | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52374690 | A | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52376119 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52376415 | C | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52376433 | T | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52376446 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52377181 | T | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52377489 | CT | C | Deletion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52378850 | GT | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52378956 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52379019 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52379179 | A | T | single_nucleotide_variant | Benign/Likely_benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52379277 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52382655 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52382748 | A | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52382847 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ITGA2 | chr5 | 52382925 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52385806 | A | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52386231 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ITGA2 | chr5 | 52386401 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ITGA2 | chr5 | 52386458 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386464 | A | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386489 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386489 | G | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386496 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386516 | A | AT | Duplication | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386637 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386660 | TG | T | Deletion | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386727 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386851 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386859 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386891 | A | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386894 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386901 | TAAC | T | Deletion | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386908 | AG | A | Deletion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52386912 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387038 | T | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387042 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387090 | T | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387111 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387136 | G | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387150 | A | C | single_nucleotide_variant | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387151 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387187 | C | CATTT | Insertion | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387232 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387274 | GTTCTC | G | Deletion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387288 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387323 | C | T | single_nucleotide_variant | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387471 | C | CAA | Duplication | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387628 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387681 | G | A | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387729 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387751 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387775 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387785 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387794 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52387878 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388064 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388070 | C | G | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388100 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388178 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388186 | GA | G | Deletion | Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388246 | C | CCTTGG | Insertion | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388254 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388326 | A | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388344 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388350 | G | T | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388376 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388386 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388409 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388438 | C | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388454 | TA | T | Deletion | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388454 | TAAA | T | Deletion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388480 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388493 | C | T | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388504 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388518 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388580 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388627 | T | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388694 | C | CT | Duplication | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388796 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388821 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388843 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388858 | C | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388879 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388933 | TCAAA | T | Microsatellite | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388950 | A | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388954 | AAGTT | A | Deletion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388999 | C | A | single_nucleotide_variant | Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52388999 | C | T | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389003 | C | G | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389037 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389077 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389082 | T | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389103 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389111 | T | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389119 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389145 | C | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389152 | G | C | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389211 | C | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389311 | G | GGTATGTTTAGC | Duplication | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389350 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389368 | C | T | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389369 | G | A | single_nucleotide_variant | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389428 | A | G | single_nucleotide_variant | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389455 | A | G | single_nucleotide_variant | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389457 | G | A | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389477 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389542 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389591 | TG | T | Deletion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389593 | G | A | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389595 | G | A | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389666 | A | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389734 | ACTC | A | Deletion | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389739 | T | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389760 | A | G | single_nucleotide_variant | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389787 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389811 | T | TAAAG | Insertion | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389827 | T | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389833 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389838 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389843 | A | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52389884 | A | G | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390023 | AACA | TTTATAAACAACTTTGTAGGACT | Indel | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390026 | A | AATTATATAAACAACTTTGTAGGACT | Insertion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390026 | A | AATTTATAAACAACTTTGTAGGACT | Insertion | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390026 | A | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390153 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390190 | TAA | T | Deletion | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390242 | G | A | single_nucleotide_variant | Uncertain_significance | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390249 | T | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390318 | A | G | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390347 | G | C | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390415 | A | C | single_nucleotide_variant | Benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390425 | G | T | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390430 | CATTTG | C | Deletion | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390457 | A | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390469 | CAGTTAGT | C | Deletion | Likely_benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390471 | G | A | single_nucleotide_variant | Benign | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390472 | TTAG | T | Microsatellite | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390512 | G | C | single_nucleotide_variant | Uncertain_significance | Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52390527 | C | G | single_nucleotide_variant | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52391850 | T | G | single_nucleotide_variant | Benign | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52392017 | G | A | single_nucleotide_variant | Benign | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52392274 | G | A | single_nucleotide_variant | Benign | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52392387 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52392400 | C | T | single_nucleotide_variant | Benign | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52392519 | C | T | single_nucleotide_variant | Benign/Likely_benign | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52392560 | G | A | single_nucleotide_variant | Benign | Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52394018 | GC | G | Deletion | Benign/Likely_benign | Combined_molybdoflavoprotein_enzyme_deficiency|Platelet-type_bleeding_disorder_9|not_provided | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52394111 | T | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_molybdoflavoprotein_enzyme_deficiency|Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52394261 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_molybdoflavoprotein_enzyme_deficiency|Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |
| ITGA2 | chr5 | 52394388 | T | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_molybdoflavoprotein_enzyme_deficiency|Molybdenum_cofactor_deficiency,_complementation_group_B|Platelet-type_bleeding_disorder_9 | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'ITGA2[title] AND translation [title] AND human.'
We searched PubMed using 'ITGA2[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 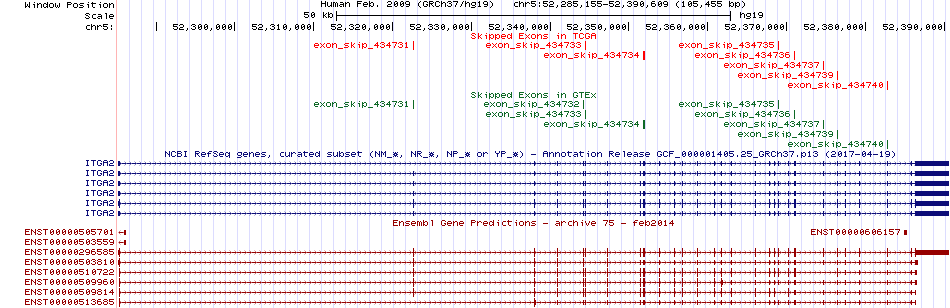
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue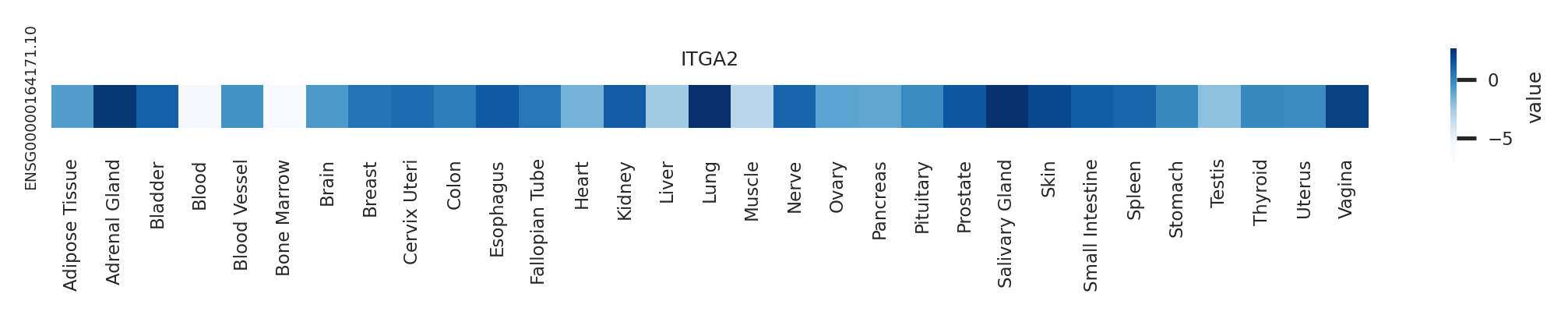
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
 Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor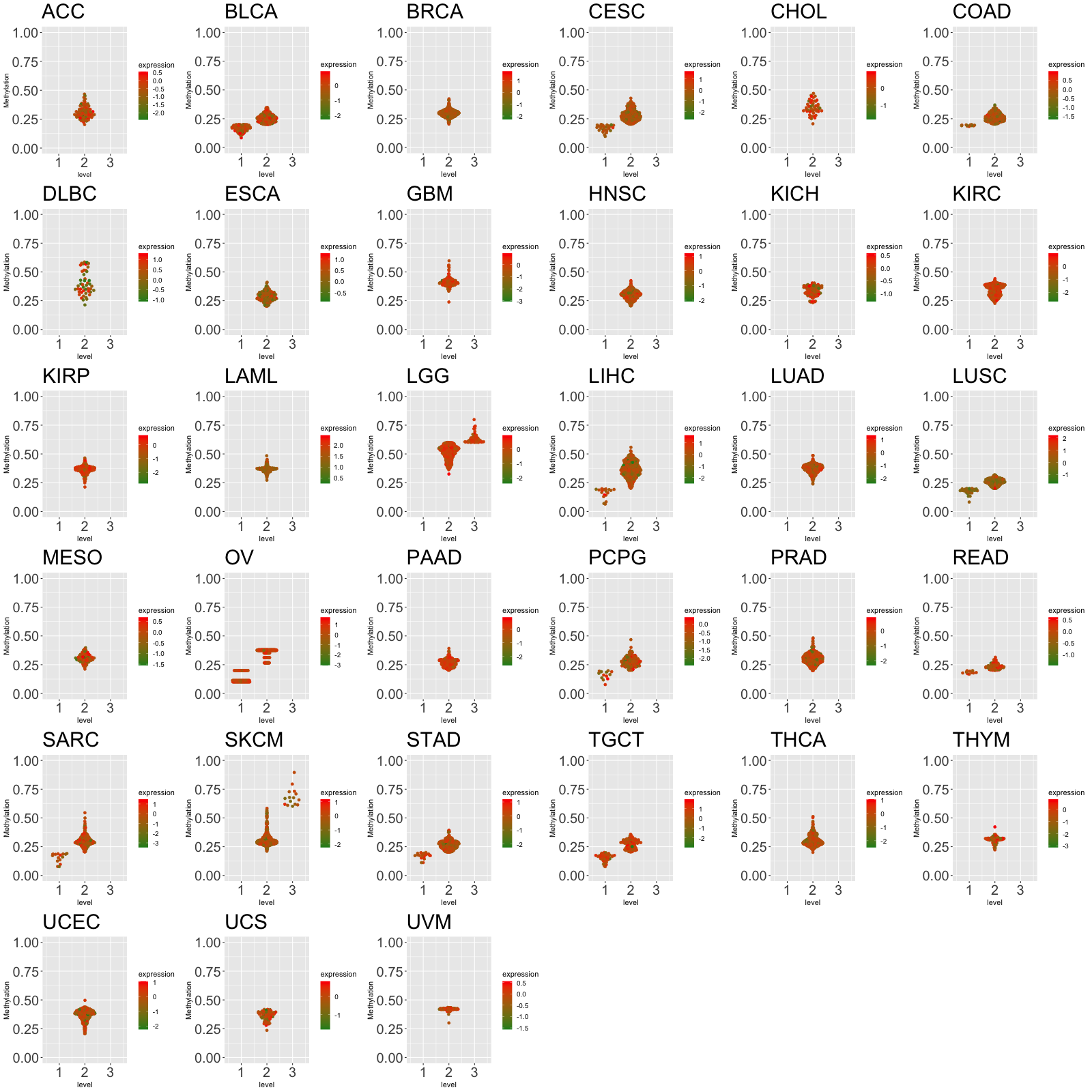
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
 Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor 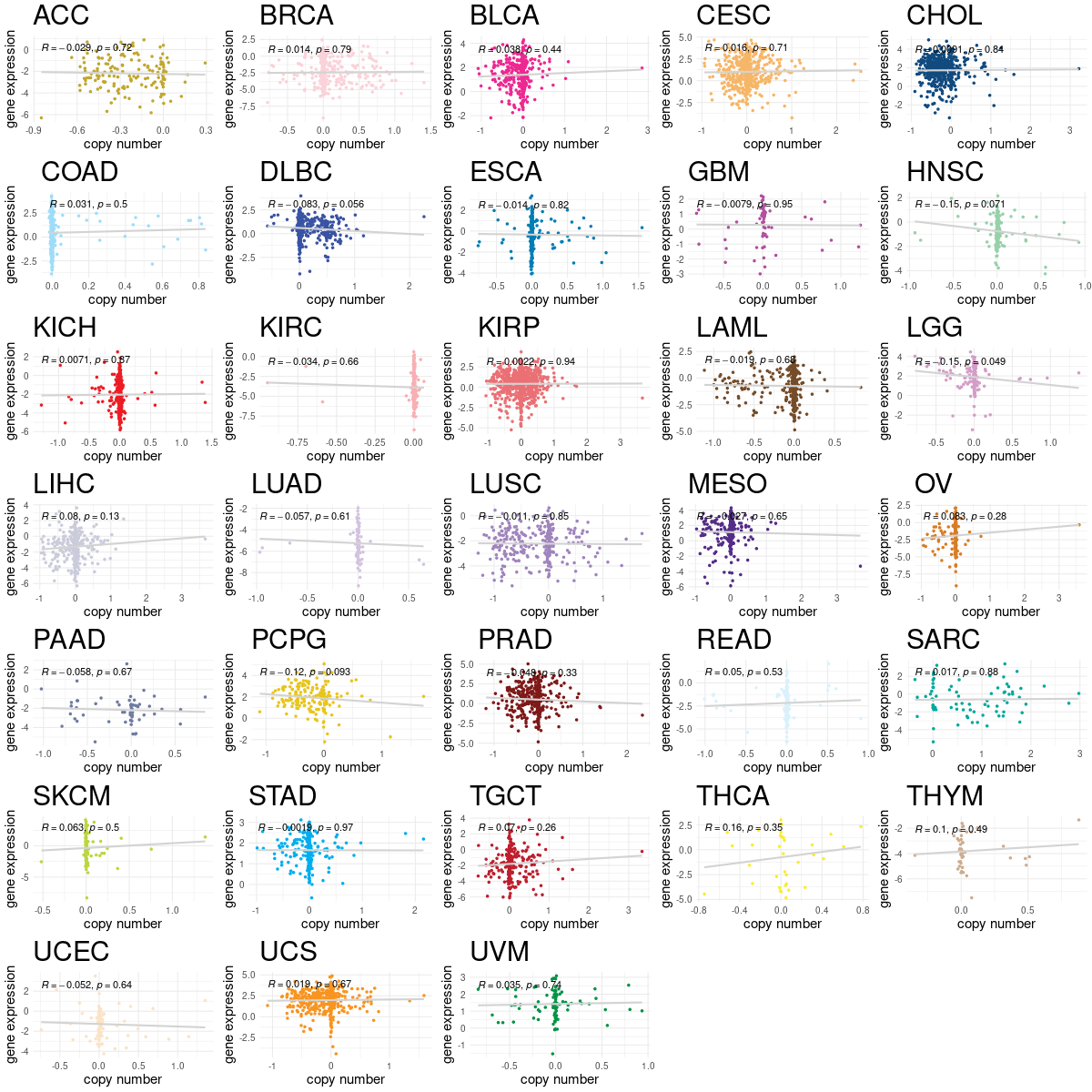
 Strongly correlated genes belong to cellular important gene groups with ITGA2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with ITGA2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
 Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of ITGA2
Copy number variation (CNV) of ITGA2 
 Fusion gene breakpoints (product of the structural variants (SVs)) across ITGA2
Fusion gene breakpoints (product of the structural variants (SVs)) across ITGA2 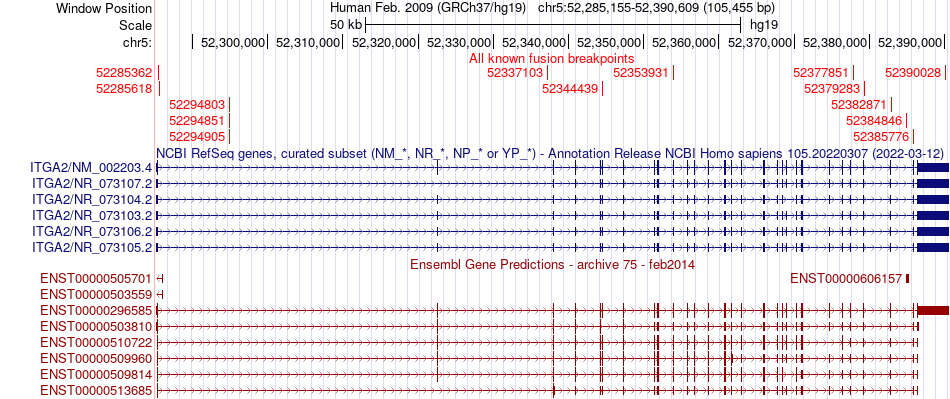
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS) 
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05)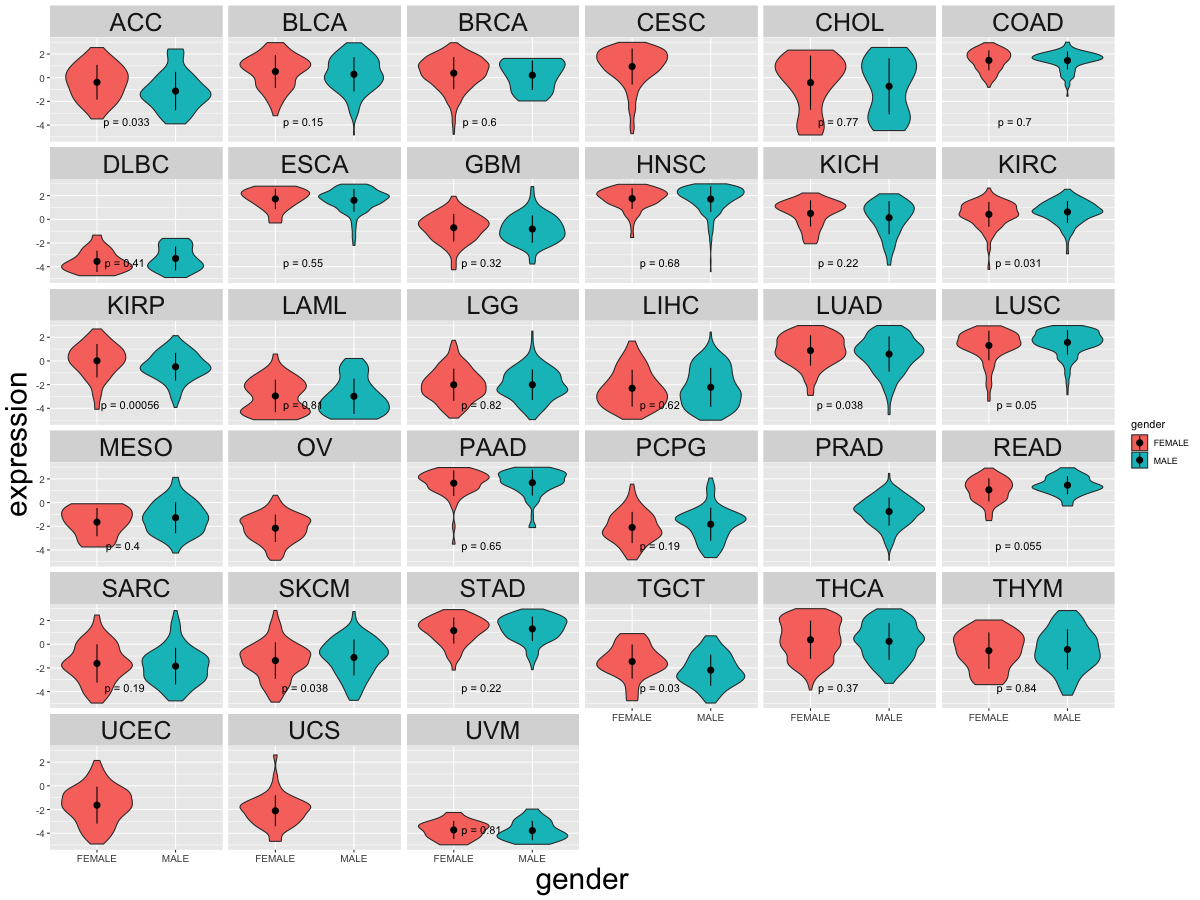
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
 Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
