| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| RARS2 | chr6 | 88223890 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| RARS2 | chr6 | 88224016 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| RARS2 | chr6 | 88224029 | C | CA | Duplication | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88224029 | CA | C | Deletion | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88224076 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88224132 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88224143 | T | TA | Duplication | Uncertain_significance | not_specified | SO:0001575|splice_donor_variant,SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001575|splice_donor_variant,SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224160 | G | A | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|Inborn_genetic_diseases | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224176 | G | C | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224189 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224190 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224191 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224193 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224214 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224225 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224226 | TAAATC | T | Deletion | Likely_benign | not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224229 | A | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224248 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224304 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224467 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224495 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224670 | C | T | single_nucleotide_variant | Likely_pathogenic | Congenital_cerebellar_hypoplasia|Pontocerebellar_hypoplasia_type_6 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224688 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224690 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224696 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224708 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224709 | A | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224712 | GT | G | Deletion | Pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88224723 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224729 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88224747 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88224748 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88226526 | T | TAA | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226529 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226532 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226543 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226546 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226547 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226555 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226555 | TG | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226565 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226566 | T | C | single_nucleotide_variant | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226589 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226591 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226592 | G | A | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226592 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88226595 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88226602 | A | G | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88226604 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88226604 | AGAT | A | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88226605 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88226641 | A | G | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88226851 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88227878 | ACAT | A | Microsatellite | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88227884 | T | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88227895 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88227906 | T | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227916 | CTG | C | Microsatellite | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227952 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88227956 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227959 | C | T | single_nucleotide_variant | Uncertain_significance | Pontoneocerebellar_hypoplasia|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227960 | C | T | single_nucleotide_variant | Uncertain_significance | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227966 | CAA | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227975 | C | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88227988 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228134 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228193 | A | AT | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228350 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228353 | G | T | single_nucleotide_variant | Benign/Likely_benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228357 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228358 | G | A | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228359 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228362 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228375 | AG | A | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228386 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228386 | T | TG | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228392 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228394 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228395 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228395 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228396 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228397 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228397 | GACTCTGGAAAACACGATCCCAGCTGA | G | Deletion | Pathogenic/Likely_pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228425 | C | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228429 | T | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228436 | A | G | single_nucleotide_variant | Likely_pathogenic | Abnormality_of_brain_morphology | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228437 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228451 | T | A | single_nucleotide_variant | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228464 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228465 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228522 | A | G | single_nucleotide_variant | Uncertain_significance | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228524 | G | A | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228541 | C | T | single_nucleotide_variant | Pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| RARS2 | chr6 | 88228544 | GAAT | G | Microsatellite | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion |
| RARS2 | chr6 | 88228551 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228560 | G | A | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228563 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228565 | C | T | single_nucleotide_variant | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228566 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228571 | T | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228584 | T | C | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88228594 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88228652 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228857 | ATT | A | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228866 | ATATAT | A | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228882 | G | GTA | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88228902 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229132 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229263 | C | T | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229264 | G | GT | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229291 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229292 | T | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229300 | C | T | single_nucleotide_variant | Pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| RARS2 | chr6 | 88229302 | C | G | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229317 | G | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229327 | A | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229368 | G | A | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229370 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229371 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229382 | G | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229383 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229386 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229394 | C | AAT | Indel | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229395 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229416 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229432 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88229902 | C | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229915 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229915 | T | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229924 | C | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229933 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229954 | CTT | C | Deletion | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229966 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88229973 | G | A | single_nucleotide_variant | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88229983 | G | A | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88230110 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88230127 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88230887 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231172 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231172 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231191 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|Pontoneocerebellar_hypoplasia|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88231193 | T | C | single_nucleotide_variant | Pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88231218 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88231226 | T | C | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88231256 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231285 | C | T | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231415 | T | TA | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231443 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88231454 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88234185 | A | G | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88234267 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88234277 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88234279 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234281 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234283 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88234284 | TAG | T | Microsatellite | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234296 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234306 | G | A | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234320 | A | AT | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234333 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234346 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88234348 | G | A | single_nucleotide_variant | Uncertain_significance | Secondary_microcephaly|Intellectual_disability|Seizures | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234350 | T | TC | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234361 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88234361 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88234362 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234364 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88234371 | C | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001574|splice_acceptor_variant,SO:0001619|non-coding_transcript_variant | SO:0001574|splice_acceptor_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88234376 | A | G | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88234380 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88234383 | A | G | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88234543 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239166 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239241 | A | C | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239243 | T | C | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239250 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239253 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239266 | T | C | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239271 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88239274 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88239290 | A | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Congenital_cerebellar_hypoplasia|Pontocerebellar_hypoplasia_type_6|Pontoneocerebellar_hypoplasia | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239299 | A | C | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239320 | C | A | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239320 | C | G | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239329 | G | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239355 | T | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88239363 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239365 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239366 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88239368 | TA | T | Deletion | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239368 | TAA | T | Deletion | Uncertain_significance | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239370 | A | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239376 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88239616 | TA | T | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240202 | A | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240492 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240495 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240501 | C | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| RARS2 | chr6 | 88240516 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240519 | A | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240534 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240538 | C | T | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240541 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240547 | T | C | single_nucleotide_variant | Benign/Likely_benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240556 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240564 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240570 | C | T | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240585 | A | G | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240588 | G | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240603 | G | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240613 | T | G | single_nucleotide_variant | Benign/Likely_benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240616 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240625 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240628 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240632 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240636 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240636 | CTTCT | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| RARS2 | chr6 | 88240652 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88240667 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240669 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240670 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88240708 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88244587 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88244648 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88244770 | T | TTCTTA | Insertion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88244797 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88251437 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88251642 | G | A | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88251654 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88251660 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88251660 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88251666 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88251668 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| RARS2 | chr6 | 88251717 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88251719 | T | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88251817 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88251827 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88251869 | T | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255203 | GA | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255205 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255274 | A | G | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255344 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255352 | C | T | single_nucleotide_variant | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255365 | TATTC | T | Deletion | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255380 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255381 | T | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255391 | CTTCT | C | Deletion | Pathogenic | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255394 | CTTT | C | Deletion | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255396 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255413 | A | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88255433 | GTA | G | Microsatellite | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255438 | T | C | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255499 | A | AG | Insertion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88255716 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258169 | C | T | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258299 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258300 | A | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258311 | A | G | single_nucleotide_variant | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258316 | G | A | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258318 | T | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258322 | AC | A | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258327 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258334 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258341 | A | C | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Pontocerebellar_hypoplasia_type_6|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258346 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258353 | A | G | single_nucleotide_variant | Uncertain_significance | Severe_intellectual_deficiency|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88258368 | C | CA | Duplication | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258373 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258377 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258469 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258607 | G | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88258652 | C | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88265020 | AAGTTTTCC | A | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88265112 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88265116 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88265150 | TG | T | Deletion | Likely_pathogenic | Pontocerebellar_hypoplasia_type_6 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265164 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265181 | A | G | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265182 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265185 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265198 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265203 | A | T | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265208 | CTTG | C | Microsatellite | Uncertain_significance | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88265227 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88265481 | TGTAG | T | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88272418 | A | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| RARS2 | chr6 | 88272418 | AC | A | Deletion | Pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| RARS2 | chr6 | 88272419 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| RARS2 | chr6 | 88272423 | T | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272431 | GCT | G | Microsatellite | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272431 | GCTCT | G | Microsatellite | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272454 | A | ACAGT | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272455 | C | T | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272472 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272482 | TCA | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272483 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272503 | G | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88272625 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273838 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273843 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273854 | T | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273860 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273865 | T | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273871 | GA | G | Deletion | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273877 | G | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273899 | ATT | A | Deletion | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273906 | T | A | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273906 | T | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273909 | T | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88273965 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273969 | A | C | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273970 | C | T | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273971 | G | A | single_nucleotide_variant | Benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88273991 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88274000 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88274068 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88274145 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88274253 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88278941 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88279225 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88279227 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88279230 | T | C | single_nucleotide_variant | Pathogenic | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88279235 | T | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279249 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279267 | T | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279276 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279282 | T | C | single_nucleotide_variant | Benign/Likely_benign | Pontocerebellar_hypoplasia_type_6|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279285 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279302 | TG | T | Deletion | Pathogenic/Likely_pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88279397 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88279611 | TA | T | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88279644 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88299357 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| RARS2 | chr6 | 88299447 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88299631 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88299632 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88299633 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88299636 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| RARS2 | chr6 | 88299639 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001575|splice_donor_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299641 | T | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299642 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299647 | G | A | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299648 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299650 | A | G | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299652 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299655 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299657 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299661 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299663 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299667 | G | A | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299667 | G | C | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299667 | G | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299669 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299670 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299670 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299670 | C | T | single_nucleotide_variant | Likely_benign | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299671 | G | A | single_nucleotide_variant | Uncertain_significance | Pontocerebellar_hypoplasia_type_6 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299673 | C | T | single_nucleotide_variant | Pathogenic | Pontoneocerebellar_hypoplasia|not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299674 | A | C | single_nucleotide_variant | Pathogenic | not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299675 | T | A | single_nucleotide_variant | Pathogenic | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299675 | T | C | single_nucleotide_variant | Pathogenic | Pontocerebellar_hypoplasia_type_6|Inborn_genetic_diseases|not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299676 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299677 | T | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Pontocerebellar_hypoplasia_type_6|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299683 | T | G | single_nucleotide_variant | Benign/Likely_benign | Pontocerebellar_hypoplasia_type_6|not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299687 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299688 | G | A | single_nucleotide_variant | Benign/Likely_benign | Pontocerebellar_hypoplasia_type_6|not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| RARS2 | chr6 | 88299709 | A | C | single_nucleotide_variant | Benign | not_specified | SO:0002153|genic_upstream_transcript_variant | SO:0002153|genic_upstream_transcript_variant |
| RARS2 | chr6 | 88299805 | T | C | single_nucleotide_variant | Benign | not_provided | | |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'RARS2[title] AND translation [title] AND human.'
We searched PubMed using 'RARS2[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue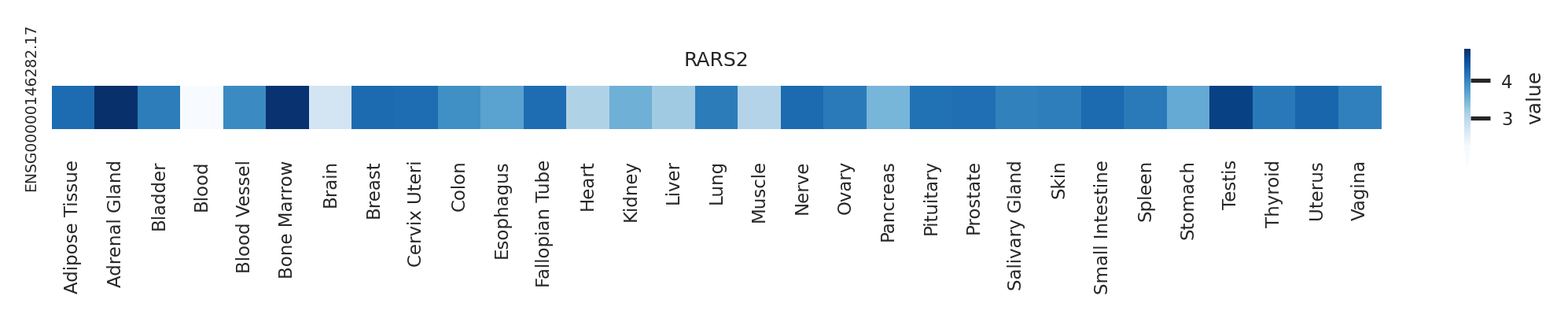
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer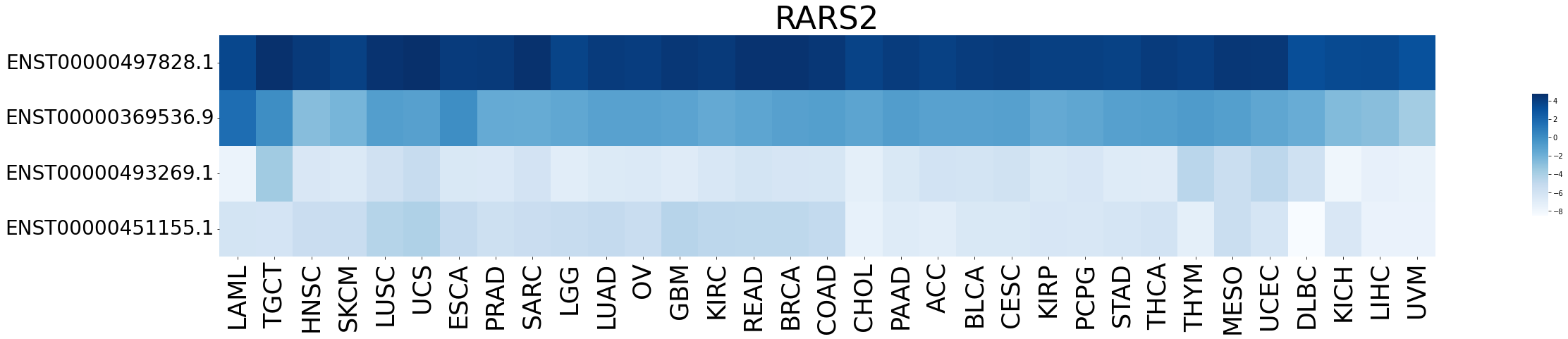
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
 Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation) Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor  Strongly correlated genes belong to cellular important gene groups with RARS2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with RARS2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
 Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))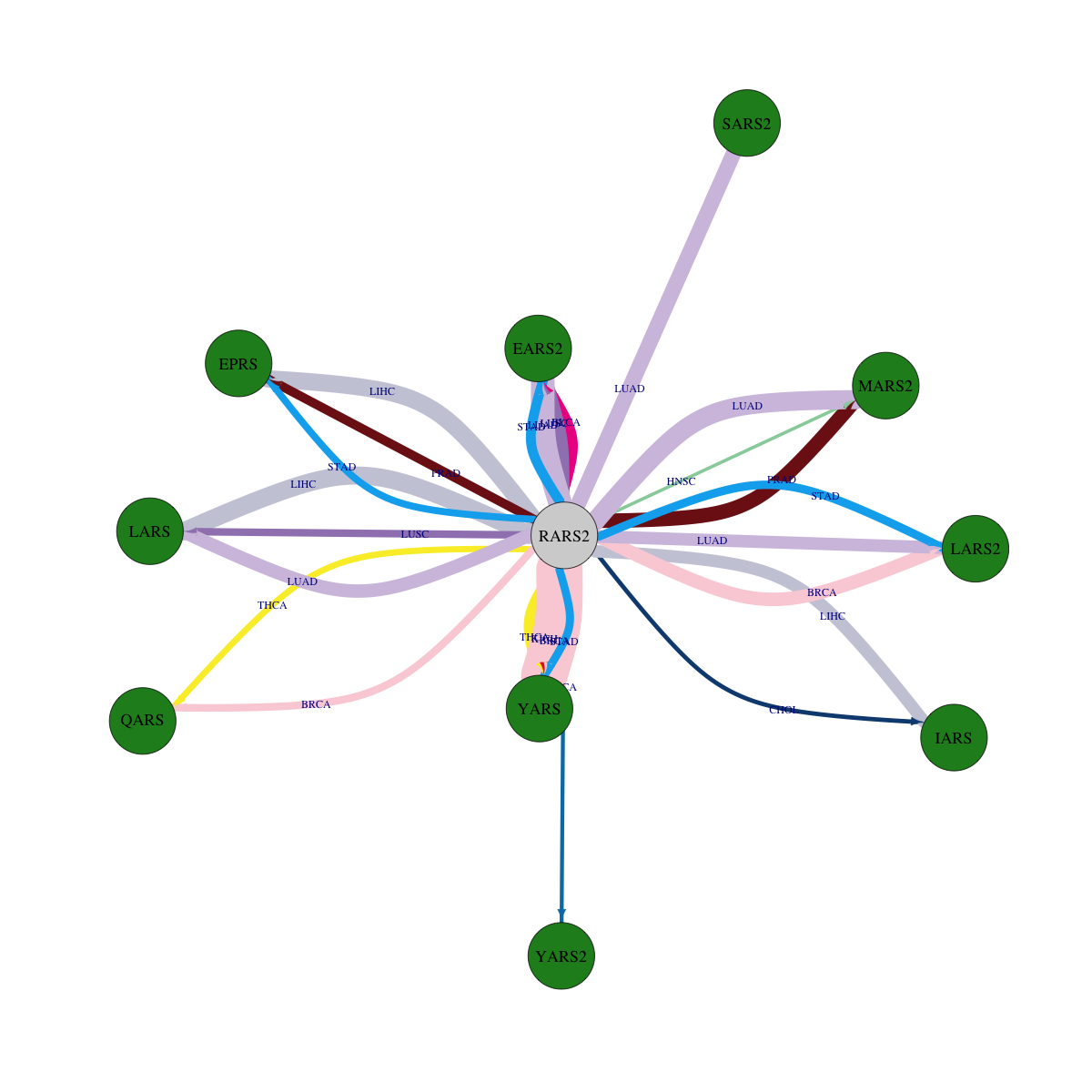
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of RARS2
Copy number variation (CNV) of RARS2 
 Fusion gene breakpoints (product of the structural variants (SVs)) across RARS2
Fusion gene breakpoints (product of the structural variants (SVs)) across RARS2 
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS) 
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)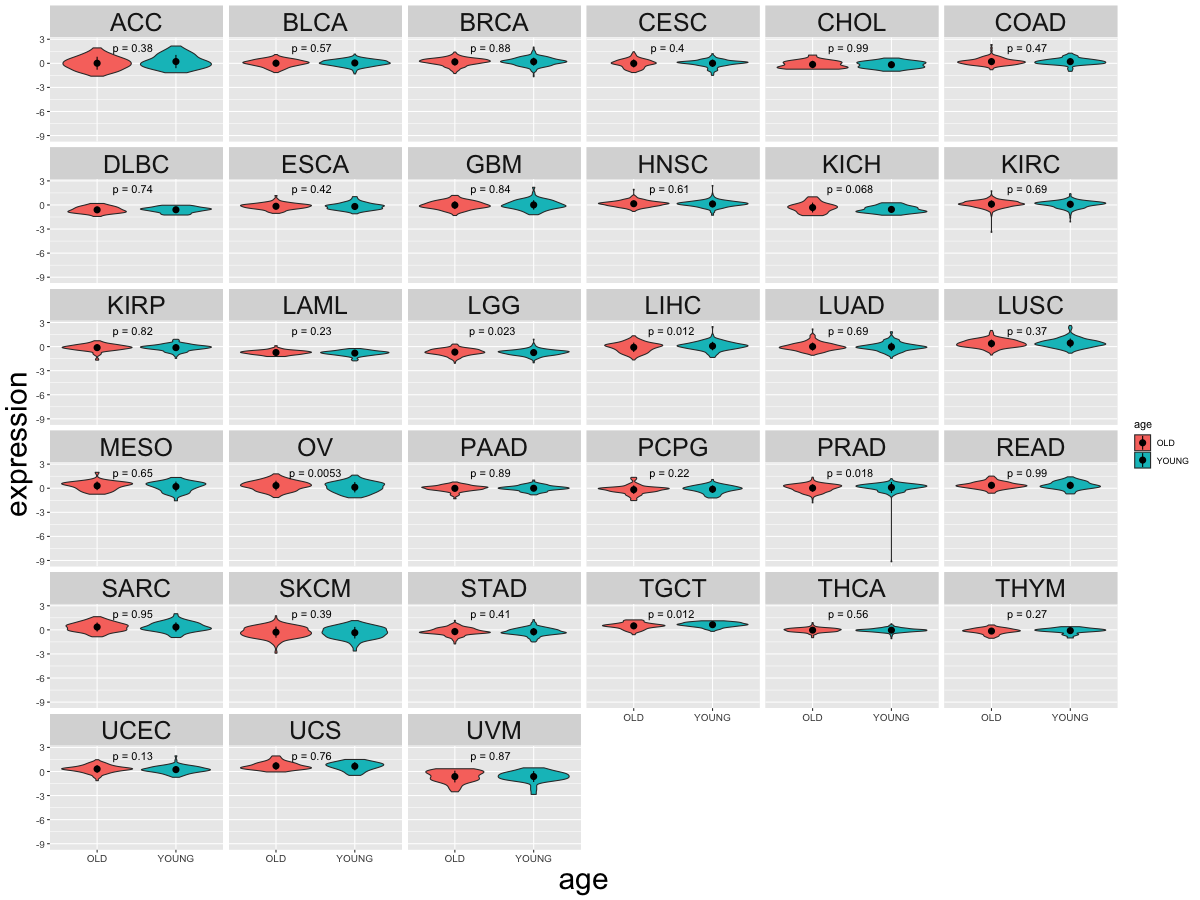
 Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
