| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| PURA | chr5 | 139493745 | CGCGGCAGCGGAGCGCAGCATCATGGCGGACCGAGACA | C | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001582|initiatior_codon_variant,SO:0001589|frameshift_variant | SO:0001582|initiatior_codon_variant,SO:0001589|frameshift_variant |
| PURA | chr5 | 139493767 | A | G | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant |
| PURA | chr5 | 139493767 | A | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant |
| PURA | chr5 | 139493769 | GGCGGA | G | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493770 | GCGGAC | G | Deletion | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493772 | G | GGACC | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493776 | C | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139493780 | AC | A | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493780 | ACAGCGGCAGCG | C | Indel | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493793 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493800 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493802 | TGCGGCGCTGGGTTCGG | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493803 | GC | G | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493806 | G | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493808 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493809 | C | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493810 | T | TGGGTTCGGGCGGCTCC | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493815 | TC | T | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493816 | C | CG | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493817 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493817 | G | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493824 | T | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493828 | T | TG | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493828 | TG | T | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493829 | G | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493836 | C | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493836 | C | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493838 | C | CGGCTCG | Duplication | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493839 | G | A | single_nucleotide_variant | Uncertain_significance | History_of_neurodevelopmental_disorder | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493843 | C | A | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139493848 | T | C | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493850 | AGGCTCC | A | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493855 | C | CCGGCGGGGGCGGTGGTGG | Duplication | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493855 | CCGGCGGGGGCGGTGGTGG | C | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493858 | GC | G | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493859 | CG | C | Deletion | Pathogenic | Epileptic_encephalopathy|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493862 | GGGCGGTGGTGGC | G | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493865 | CG | C | Deletion | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493866 | G | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493866 | GGTGGTGGCGGCGGGGGCGGCGGCGGCA | G | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493871 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493871 | T | TGGCGGCGGG | Duplication | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493871 | TGGCGGCGGG | T | Deletion | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493871 | TGGCGGCGGGGGCGGCGGCGGCAGTGGC | T | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493871 | TGGCGGCGGGGGCGGCGGCGGCAGTGGCGGCGGC | T | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493875 | G | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493876 | G | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493877 | C | CG | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493880 | G | GGGCGGCGGCGGCAGT | Duplication | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493880 | G | GGGCGGCGGCGGCAGTGGCGGC | Duplication | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493880 | GGGCGGCGGCGGCAGT | G | Deletion | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493880 | GGGCGGCGGCGGCAGTGGCGGCGGC | G | Deletion | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493883 | C | CGGCGGCGGCAGT | Duplication | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493883 | CG | C | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493883 | CGGCGGCGGCAGT | C | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493887 | G | GGCGGCA | Duplication | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|not_specified|not_provided | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493892 | CAGTG | C | Deletion | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493893 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493895 | T | TGGC | Microsatellite | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder|not_specified|not_provided | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493895 | T | TGGCGGC | Microsatellite | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139493895 | TGGC | T | Microsatellite | Benign/Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493895 | TGGCGGC | T | Microsatellite | Uncertain_significance | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493895 | TGGCGGCGGC | T | Microsatellite | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139493901 | CG | C | Deletion | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493904 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493907 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493909 | GC | G | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493910 | CG | C | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493911 | G | GGGGCCCCA | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493912 | G | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493918 | CA | C | Deletion | Pathogenic | Inborn_genetic_diseases|History_of_neurodevelopmental_disorder | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493919 | A | AG | Duplication | Pathogenic | Intellectual_disability|PURA-related_severe_neonatal_hypotonia-seizures-encephalopathy_syndrome_due_to_a_point_mutation|Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493921 | G | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493921 | G | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493929 | C | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139493931 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493934 | C | CGA | Microsatellite | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493935 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493939 | C | CT | Insertion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139493941 | C | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139493944 | G | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139493968 | A | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493971 | C | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139493977 | A | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493987 | A | G | single_nucleotide_variant | Likely_pathogenic | See_cases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493990 | T | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493990 | T | G | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139493994 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139493997 | G | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494006 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494007 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494011 | AG | A | Deletion | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494021 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494028 | ATCG | A | Deletion | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia | SO:0001820|inframe_indel | SO:0001820|inframe_indel |
| PURA | chr5 | 139494029 | T | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494029 | TC | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494031 | G | A | single_nucleotide_variant | Uncertain_significance | Neurodevelopmental_disorder | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494031 | G | C | single_nucleotide_variant | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494031 | GC | G | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494033 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494034 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494034 | G | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494036 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494044 | C | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494044 | CG | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494045 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494055 | A | G | single_nucleotide_variant | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia|Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494063 | CCTTACTCTCTCCATGTCA | C | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494065 | T | C | single_nucleotide_variant | Likely_pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia|Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494066 | T | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494066 | TACTCTCTCC | T | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494067 | A | ACT | Microsatellite | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494067 | ACT | A | Microsatellite | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Neonatal_hypotonia|Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494069 | T | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494071 | T | C | single_nucleotide_variant | Likely_pathogenic | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494071 | T | G | single_nucleotide_variant | Pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494072 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494077 | TG | T | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494081 | A | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494087 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494087 | C | T | single_nucleotide_variant | Benign/Likely_benign | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494091 | G | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494100 | G | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494105 | C | G | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494116 | T | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494128 | A | ACG | Microsatellite | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494129 | C | G | single_nucleotide_variant | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia|not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494133 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494135 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494141 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494142 | C | CCCAG | Microsatellite | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494144 | C | CA | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494146 | G | GC | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494151 | C | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494153 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494153 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494156 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494156 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494160 | C | G | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494165 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494168 | G | GGCGCAGGACGAGCC | Duplication | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494170 | CGCAGGACGAGCCGCGCCGGGCGCTCAAAA | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494172 | C | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494185 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494187 | C | A | single_nucleotide_variant | Likely_benign | History_of_neurodevelopmental_disorder | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494187 | C | CG | Duplication | Pathogenic | PURA_Syndrome | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494192 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494193 | C | G | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494196 | A | G | single_nucleotide_variant | Likely_pathogenic | Intellectual_disability|Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494198 | A | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494198 | A | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494213 | GCGCGAGAA | G | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494216 | C | CGAGAACCGCAAGTACTACATGGATCTCAAG | Duplication | Likely_pathogenic | Inborn_genetic_diseases | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139494216 | C | G | single_nucleotide_variant | Benign/Likely_benign | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494216 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494217 | G | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494224 | G | C | single_nucleotide_variant | Pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494224 | G | GC | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494228 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494231 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494236 | T | A | single_nucleotide_variant | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Neonatal_hypotonia | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494238 | G | T | single_nucleotide_variant | Likely_pathogenic | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494241 | C | G | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494243 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494243 | CAAG | C | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494244 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494246 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494250 | A | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494252 | C | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494253 | C | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494257 | G | GCGGC | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494258 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494259 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494259 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494260 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494261 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494262 | C | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494262 | C | T | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494264 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494266 | T | TC | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494267 | C | G | single_nucleotide_variant | Uncertain_significance | Global_developmental_delay-visual_anomalies-progressive_cerebellar_atrophy-truncal_hypotonia_syndrome | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494269 | T | C | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494270 | GC | TT | Indel | Uncertain_significance | not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494272 | G | C | single_nucleotide_variant | Pathogenic | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494272 | GCATCCGC | G | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494275 | T | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494277 | C | G | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494283 | A | G | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494284 | C | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494285 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494291 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494292 | CG | C | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494294 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494294 | G | GGGGCCTGGCCTGGGCTCCACGCA | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494297 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494297 | G | GC | Duplication | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494298 | C | CCTGGCCTGGGCTCCACGCAGGG | Duplication | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494300 | T | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494303 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494303 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494309 | C | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494309 | C | T | single_nucleotide_variant | Benign/Likely_benign | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494322 | C | T | single_nucleotide_variant | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia|Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494326 | CCATTGCGCTGCCCGCG | C | Deletion | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494329 | T | C | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31|Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494330 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494331 | G | C | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494338 | C | T | single_nucleotide_variant | Likely_pathogenic | Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494339 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494342 | G | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494348 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494348 | G | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494350 | T | A | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494351 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494361 | C | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494362 | G | C | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia|Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494363 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494369 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494369 | T | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494371 | T | C | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494374 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494379 | CTCA | C | Deletion | Likely_pathogenic | not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494380 | T | C | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494381 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494382 | A | T | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494384 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494384 | CGACGACT | C | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494387 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|History_of_neurodevelopmental_disorder|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494396 | A | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494400 | G | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494404 | A | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494405 | G | GGAGCC | Duplication | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494406 | G | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Generalized_hypotonia|Apnea|Limb_dystonia|Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494411 | GGCCGAGCTGCCCGA | G | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494414 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494423 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494435 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494440 | C | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494440 | CTG | C | Microsatellite | Pathogenic | Intellectual_disability|Mental_retardation,_autosomal_dominant_31|Inborn_genetic_diseases|not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494443 | T | TGGACTGA | Insertion | Pathogenic | Inborn_genetic_diseases | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494451 | A | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494455 | GCTT | G | Microsatellite | Pathogenic/Likely_pathogenic | Mental_retardation,_autosomal_dominant_31|Inborn_genetic_diseases|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494458 | T | C | single_nucleotide_variant | Pathogenic | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494459 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494462 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494468 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494468 | TG | T | Deletion | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494470 | T | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494475 | T | TC | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494476 | C | T | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494480 | CA | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494486 | C | G | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494489 | CGT | C | Microsatellite | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494490 | G | GT | Duplication | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494492 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494495 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494499 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494500 | G | C | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494501 | A | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494507 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494509 | AG | A | Deletion | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494525 | T | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494535 | A | G | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494543 | G | A | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494543 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494545 | C | G | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494545 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494549 | C | G | single_nucleotide_variant | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494551 | AGGT | A | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494555 | G | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494568 | G | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494569 | GAC | G | Microsatellite | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494570 | A | C | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494570 | A | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494575 | CCTT | C | Deletion | Pathogenic | Intellectual_disability|Delayed_speech_and_language_development|Global_developmental_delay|Seizures|Neonatal_hypotonia|Mental_retardation,_autosomal_dominant_31|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494592 | G | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| PURA | chr5 | 139494607 | AT | A | Deletion | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494645 | GCAGCTTCAC | G | Deletion | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| PURA | chr5 | 139494660 | G | GC | Duplication | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494663 | A | ACAG | Microsatellite | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001821|inframe_insertion | SO:0001821|inframe_insertion |
| PURA | chr5 | 139494663 | AC | A | Deletion | Likely_pathogenic | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494684 | C | G | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494684 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| PURA | chr5 | 139494685 | G | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494686 | C | T | single_nucleotide_variant | Likely_benign | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494688 | G | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494688 | GCTGCCACCCTGCTA | G | Deletion | Pathogenic | Inborn_genetic_diseases | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| PURA | chr5 | 139494691 | G | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494707 | A | C | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494716 | A | T | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_31 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494721 | G | C | single_nucleotide_variant | Uncertain_significance | Intellectual_disability | SO:0001583|missense_variant | SO:0001583|missense_variant |
| PURA | chr5 | 139494754 | C | CCA | Microsatellite | Benign | not_provided | SO:0001624|3_prime_UTR_variant | SO:0001624|3_prime_UTR_variant |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'PURA[title] AND translation [title] AND human.'
We searched PubMed using 'PURA[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
 Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
 Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor 
 Strongly correlated genes belong to cellular important gene groups with PURA (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with PURA (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)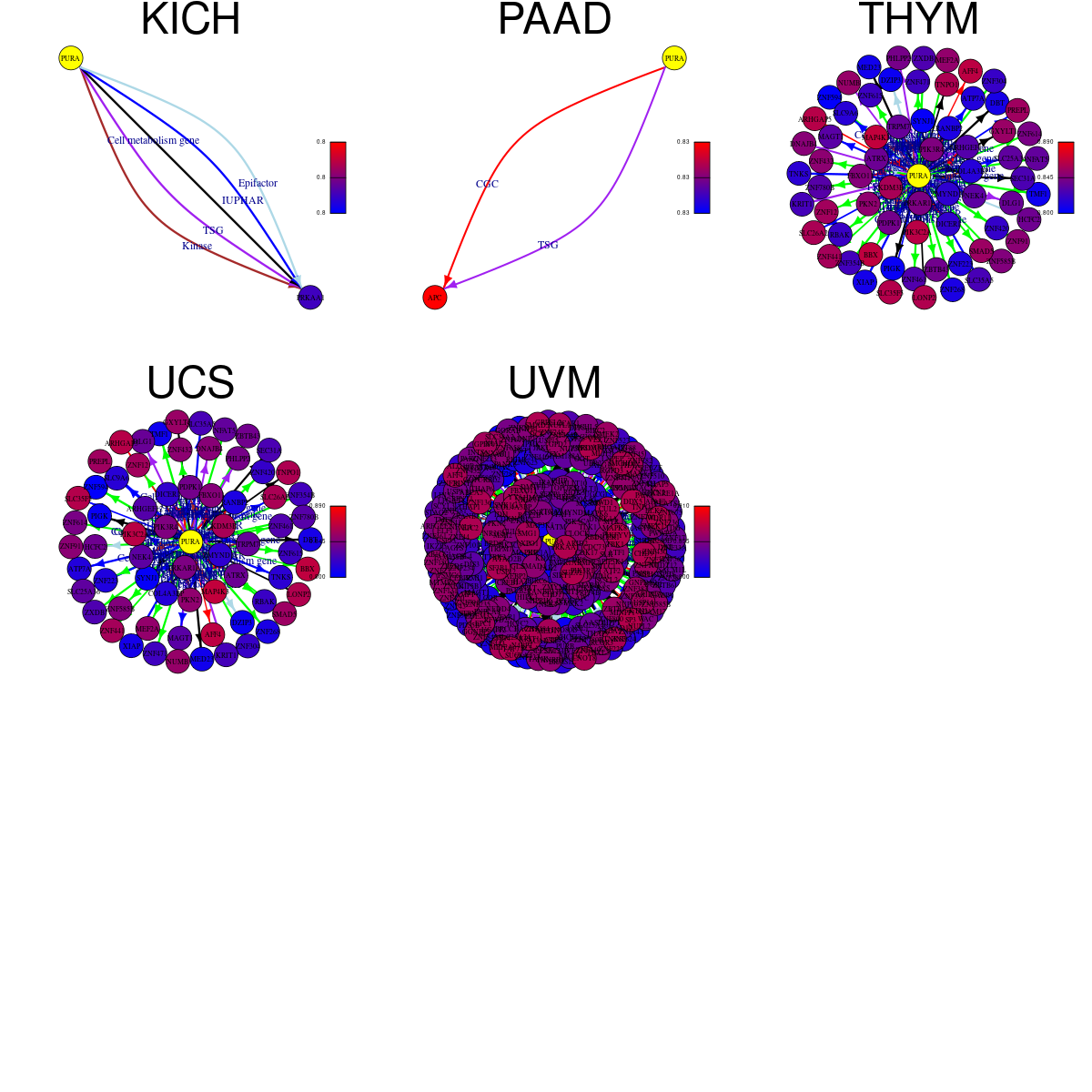
 Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 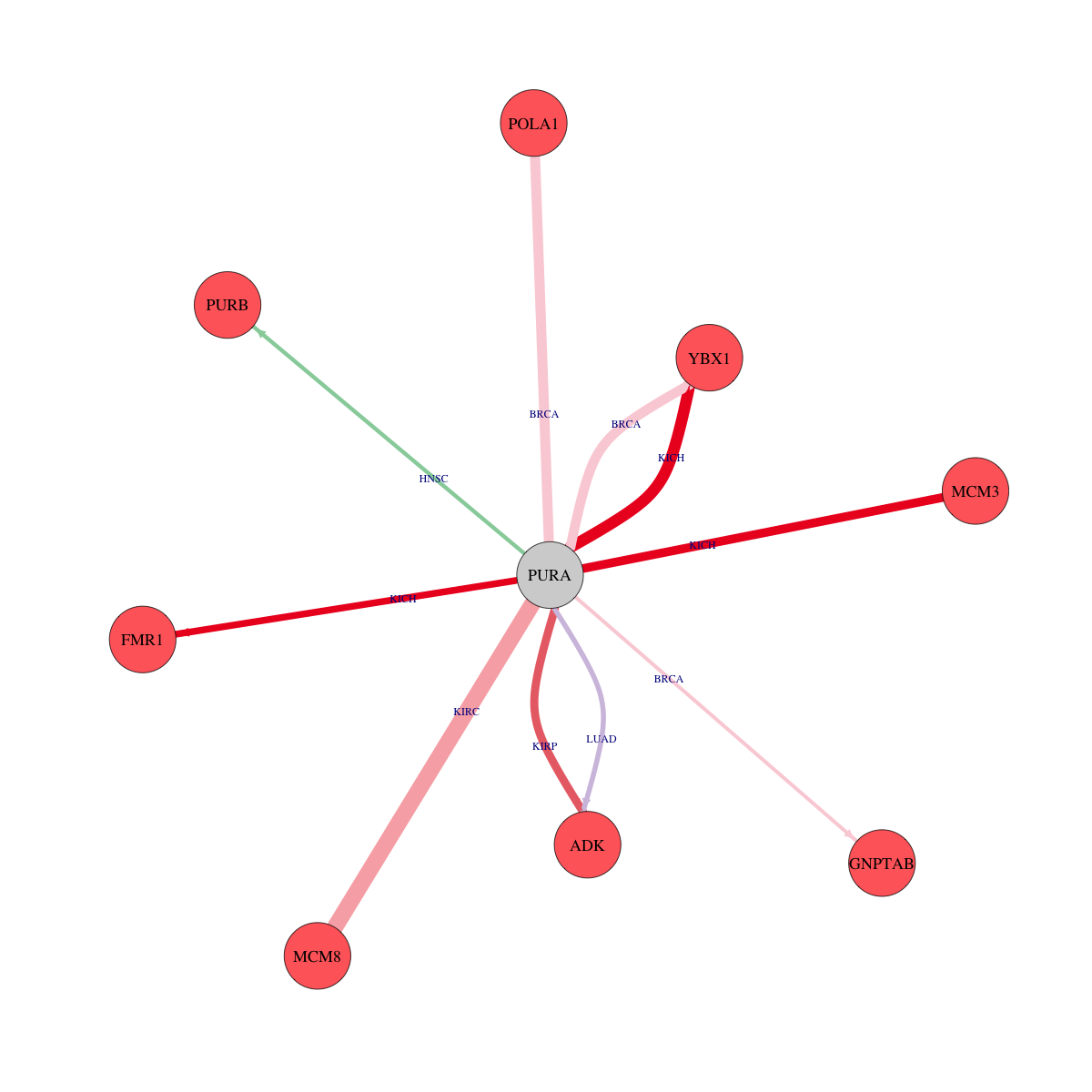
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))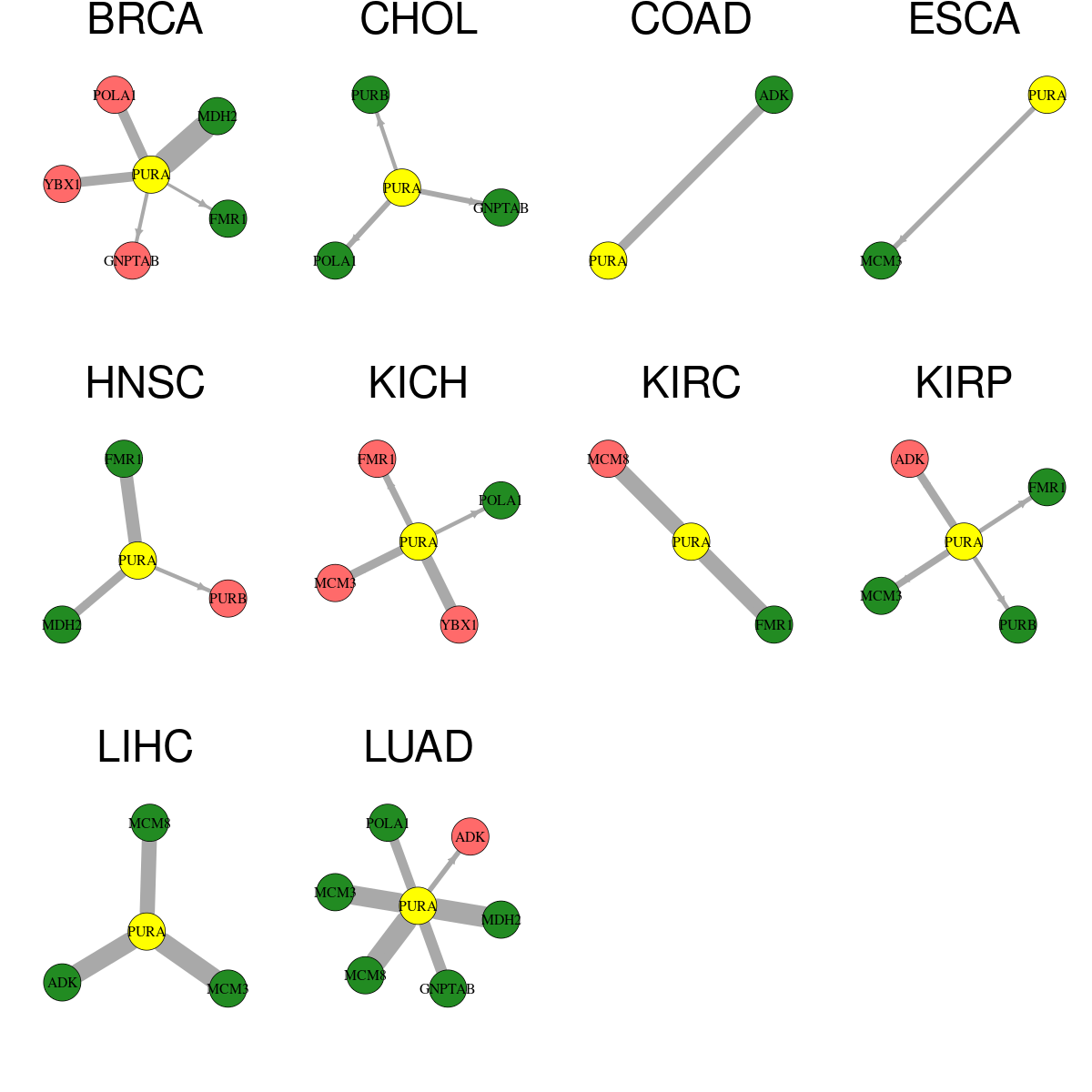
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of PURA
Copy number variation (CNV) of PURA 
 Fusion gene breakpoints (product of the structural variants (SVs)) across PURA
Fusion gene breakpoints (product of the structural variants (SVs)) across PURA 
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS) 
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
 Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
