| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| GFM1 | chr3 | 158362038 | G | T | single_nucleotide_variant | Benign | not_provided | | |
| GFM1 | chr3 | 158362339 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362351 | C | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362358 | C | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362386 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362391 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362393 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362399 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362413 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362426 | G | A | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362439 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362441 | T | C | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362444 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362462 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362472 | GC | G | Deletion | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362474 | C | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362477 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362477 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362479 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362489 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362500 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362505 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant,SO:0001623|5_prime_UTR_variant | SO:0001575|splice_donor_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158362509 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158362511 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158362514 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158362526 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158362588 | C | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158362769 | G | GT | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158362769 | GT | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158362769 | GTT | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158362769 | GTTT | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363220 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363300 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363416 | A | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| GFM1 | chr3 | 158363436 | C | T | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363438 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363463 | A | G | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363475 | C | T | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363477 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363498 | C | CATTG | Duplication | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363504 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363528 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363529 | C | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363529 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363556 | G | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363557 | C | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant |
| GFM1 | chr3 | 158363571 | G | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| GFM1 | chr3 | 158363582 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363587 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363757 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363869 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363940 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158363957 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158363964 | AAG | A | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158363967 | A | T | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158363984 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158363990 | TC | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158363998 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364008 | CAA | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364013 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364043 | C | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364043 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364063 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364082 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364309 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364333 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364355 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364498 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364514 | A | G | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364526 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364537 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364559 | A | C | single_nucleotide_variant | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364564 | G | GCCCT | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364572 | A | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364573 | G | A | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364587 | AG | A | Deletion | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364590 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364607 | T | C | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364619 | A | C | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364629 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364640 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364649 | T | TG | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364662 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364668 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364683 | T | TA | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364685 | A | G | single_nucleotide_variant | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364700 | TG | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364704 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364716 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364732 | A | C | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158364745 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158364745 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158366829 | G | C | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | not_provided | SO:0001574|splice_acceptor_variant,SO:0001627|intron_variant | SO:0001574|splice_acceptor_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366837 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366853 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366854 | G | A | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366864 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366869 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366873 | G | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366879 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366900 | G | A | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366918 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366924 | A | G | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366944 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158366945 | G | A | single_nucleotide_variant | Pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158367146 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158367557 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158367837 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158367854 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158368130 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158368157 | A | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369613 | AT | A | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369789 | T | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369798 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369845 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369878 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369880 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158369895 | C | T | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369896 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369897 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158369900 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158369914 | CT | C | Deletion | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369930 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158369930 | GGCCACTGACCAC | TTAATTAATTAG | Indel | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369933 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158369939 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158369941 | AC | A | Deletion | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369943 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency|Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369944 | G | A | single_nucleotide_variant | Uncertain_significance | Relative_macrocephaly|Severe_muscular_hypotonia|Developmental_regression | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369971 | A | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369983 | A | G | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158369987 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158370002 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158370020 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158370021 | A | AT | Duplication | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158370032 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158370045 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158370143 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158370352 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158370875 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158371101 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371119 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371131 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371139 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158371155 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371186 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371210 | C | T | single_nucleotide_variant | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158371216 | C | G | single_nucleotide_variant | | | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158371218 | A | C | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371218 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371245 | C | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371254 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158371254 | GGAGT | G | Deletion | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| GFM1 | chr3 | 158371263 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158371406 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158371546 | T | TA | Duplication | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158372115 | A | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158372335 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| GFM1 | chr3 | 158372342 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158372369 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158372372 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158372402 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158372411 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158372423 | A | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158372426 | T | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158372428 | G | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158372477 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158372502 | T | C | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158376412 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158376717 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376728 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376729 | T | C | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376737 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376741 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376743 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376745 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376770 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376771 | GACACCATCTATA | G | Deletion | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376779 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376788 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376806 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376807 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376808 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376815 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376825 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376836 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| GFM1 | chr3 | 158376856 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158376868 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158377051 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158377070 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158377171 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158378342 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158378653 | GT | G | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158378683 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378695 | T | TGCATTGTTTGGCATTGACTGTGC | Duplication | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158378697 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158378704 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378731 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378734 | CACAG | C | Deletion | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158378740 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378746 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378749 | C | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378752 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158378836 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158378847 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158380150 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158380402 | T | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158380417 | G | T | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158380419 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380422 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380436 | A | G | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158380437 | TC | T | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158380443 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380455 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380461 | G | A | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380462 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158380464 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380470 | C | T | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158380694 | T | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158383087 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158383117 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158383128 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158383129 | G | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency|Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383130 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383145 | CA | C | Deletion | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383151 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383167 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158383174 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383179 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158383202 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383209 | CAA | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383218 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158383232 | T | G | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158383239 | A | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158383264 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| GFM1 | chr3 | 158383269 | GA | G | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158383882 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158383882 | T | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158384060 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158384068 | A | G | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158384110 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158384120 | T | C | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158384145 | C | T | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158384150 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158384157 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158384167 | C | T | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158384168 | CCT | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158384169 | CT | C | Deletion | Pathogenic | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158384176 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001575|splice_donor_variant | SO:0001575|splice_donor_variant |
| GFM1 | chr3 | 158384184 | G | C | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158399757 | A | AT | Duplication | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158399808 | A | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399811 | A | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399838 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399847 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399865 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399868 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399886 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399904 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158399957 | A | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001578|stop_lost,SO:0001627|intron_variant | SO:0001578|stop_lost,SO:0001627|intron_variant |
| GFM1 | chr3 | 158402015 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158402219 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158402310 | CAG | C | Deletion | Pathogenic | not_provided | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| GFM1 | chr3 | 158402329 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158402330 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402362 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158402366 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402370 | C | T | single_nucleotide_variant | Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158402371 | G | A | single_nucleotide_variant | Pathogenic | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158402378 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402379 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402399 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402400 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158402405 | T | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402429 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402441 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158402470 | C | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158402470 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158402498 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158402588 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158402689 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158407930 | CT | C | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158407946 | A | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158407950 | A | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| GFM1 | chr3 | 158407951 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| GFM1 | chr3 | 158407964 | C | A | single_nucleotide_variant | | | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158407968 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158407978 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158407990 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158408032 | G | A | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158408050 | C | T | single_nucleotide_variant | Pathogenic | not_provided | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158408053 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158408054 | G | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158408058 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158408097 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158408101 | C | T | single_nucleotide_variant | Benign/Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158408103 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158408123 | T | G | single_nucleotide_variant | Benign/Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158408908 | T | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158408911 | A | G | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158409090 | AAAC | A | Deletion | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158409093 | C | CA | Duplication | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158409106 | C | CT | Duplication | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158409106 | CT | C | Deletion | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158409120 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| GFM1 | chr3 | 158409139 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409143 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158409163 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409165 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158409166 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409170 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409178 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409190 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409230 | GT | G | Deletion | Uncertain_significance | not_provided | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant |
| GFM1 | chr3 | 158409232 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| GFM1 | chr3 | 158409262 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_specified|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409265 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409372 | C | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409375 | G | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409420 | C | A | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409482 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1|not_provided | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409501 | A | C | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409685 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409707 | TTTG | T | Deletion | Likely_benign | Combined_oxidative_phosphorylation_deficiency | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409739 | T | TTA | Insertion | Likely_benign | Combined_oxidative_phosphorylation_deficiency | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409795 | C | T | single_nucleotide_variant | Likely_benign | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409815 | T | C | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409828 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409834 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409891 | A | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409947 | T | A | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409969 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158409987 | C | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158410022 | G | T | single_nucleotide_variant | Benign | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158410059 | T | C | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158410206 | C | T | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158410260 | T | C | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| GFM1 | chr3 | 158410289 | A | G | single_nucleotide_variant | Uncertain_significance | Combined_oxidative_phosphorylation_deficiency_1 | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'GFM1[title] AND translation [title] AND human.'
We searched PubMed using 'GFM1[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 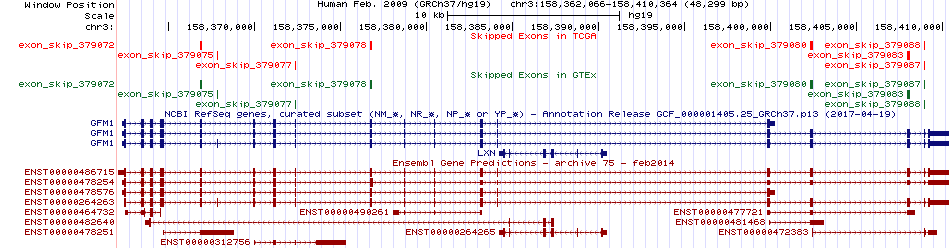
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue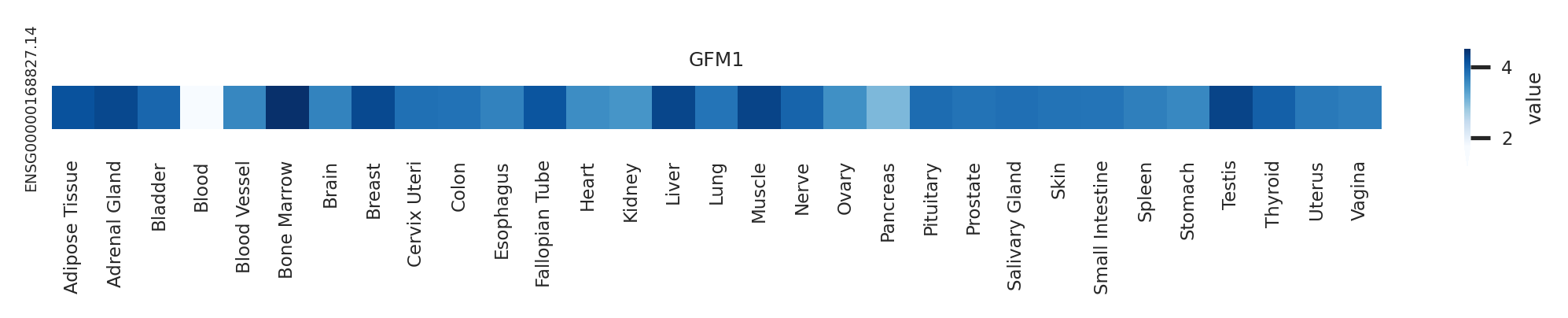
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue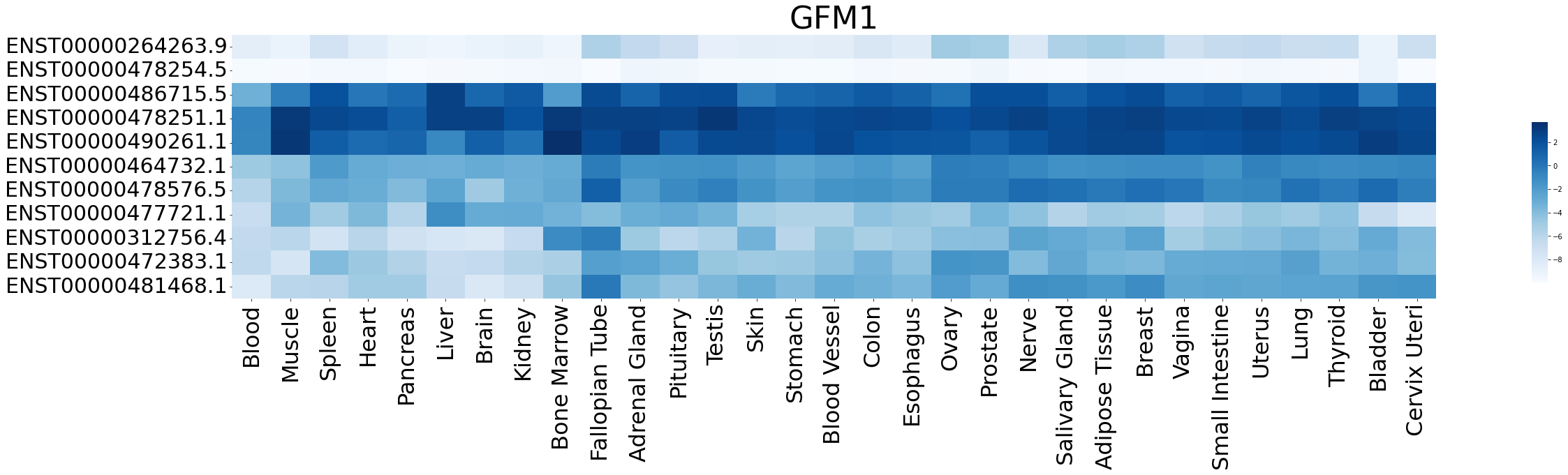
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution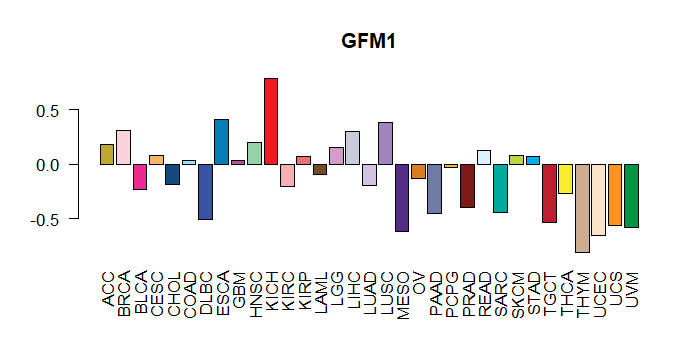
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)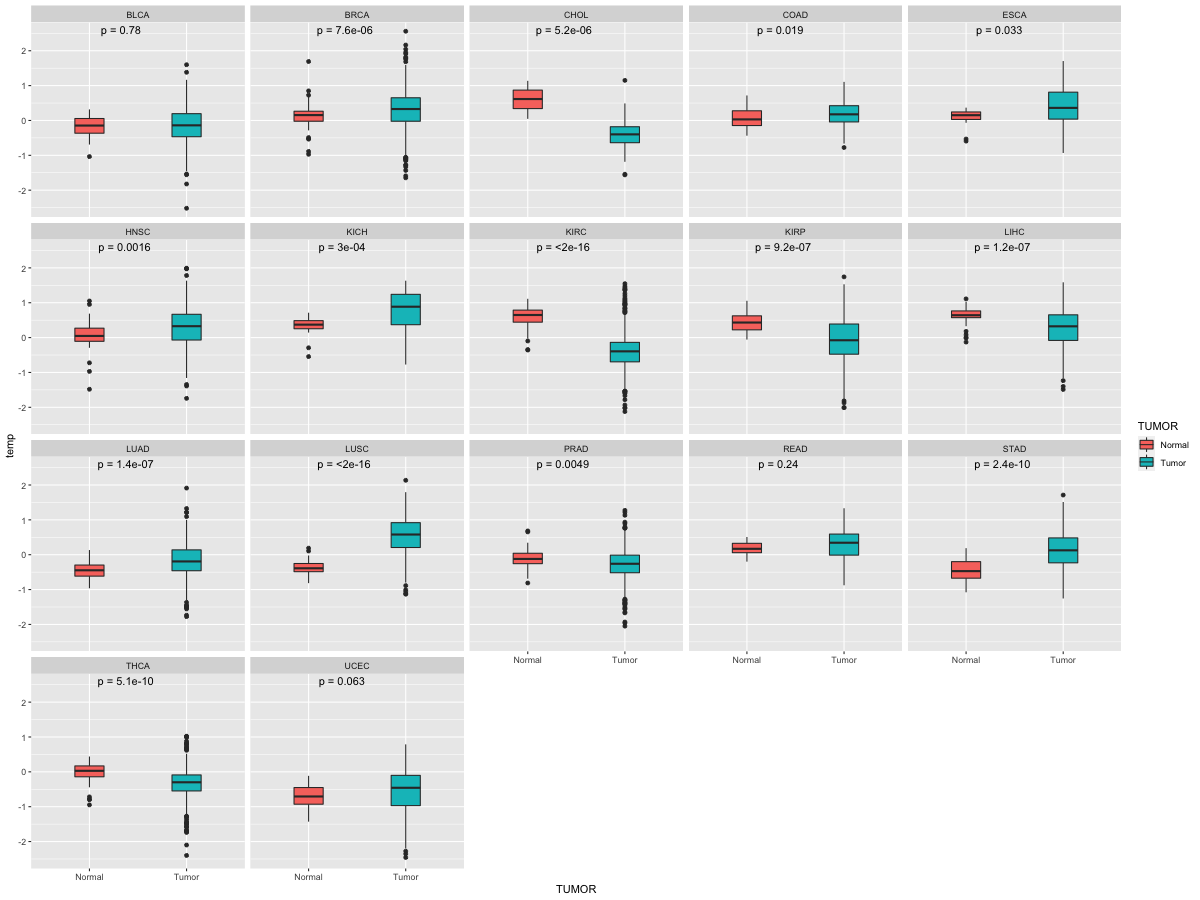
 Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
 Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor 
 Strongly correlated genes belong to cellular important gene groups with GFM1 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with GFM1 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)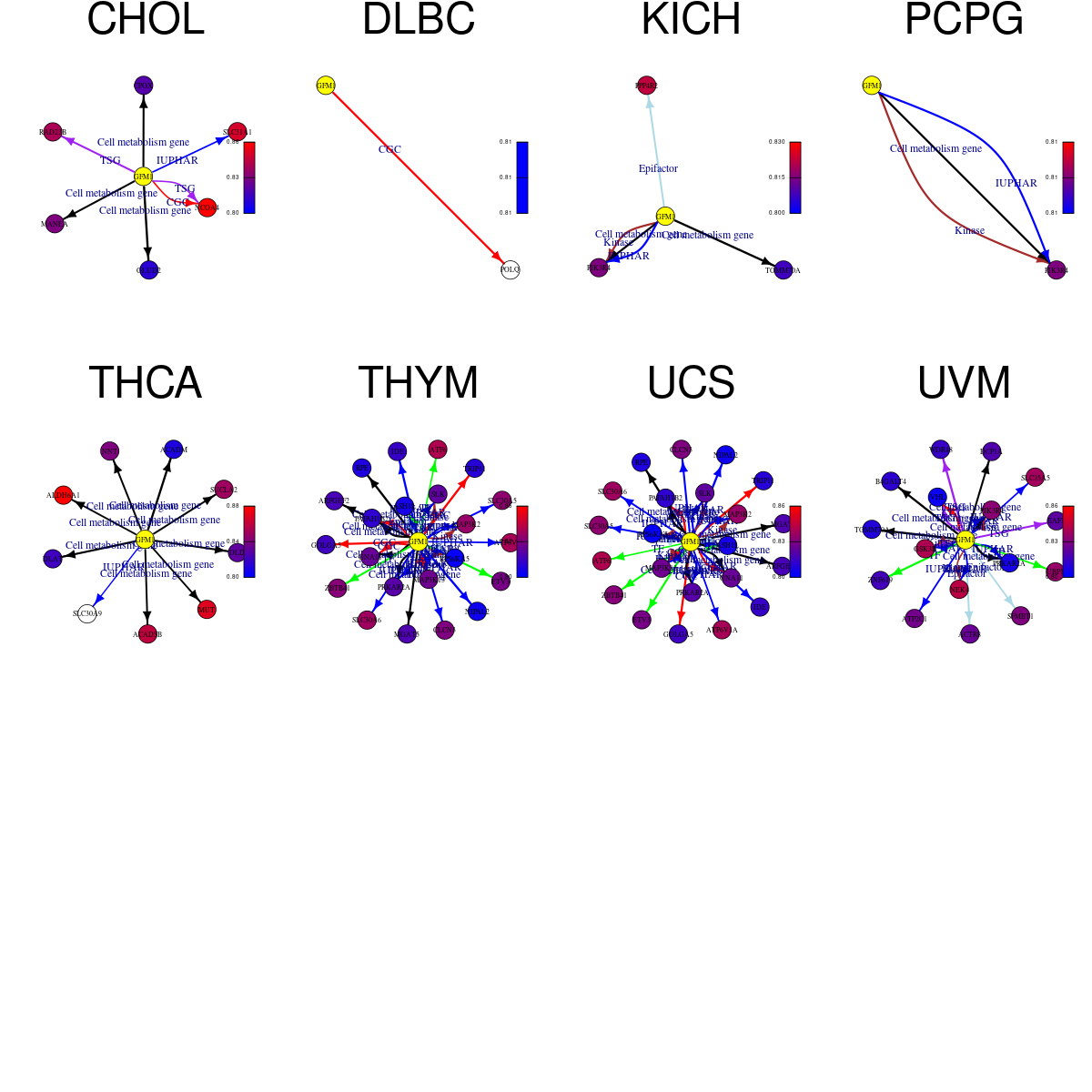
 Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of GFM1
Copy number variation (CNV) of GFM1 
 Fusion gene breakpoints (product of the structural variants (SVs)) across GFM1
Fusion gene breakpoints (product of the structural variants (SVs)) across GFM1 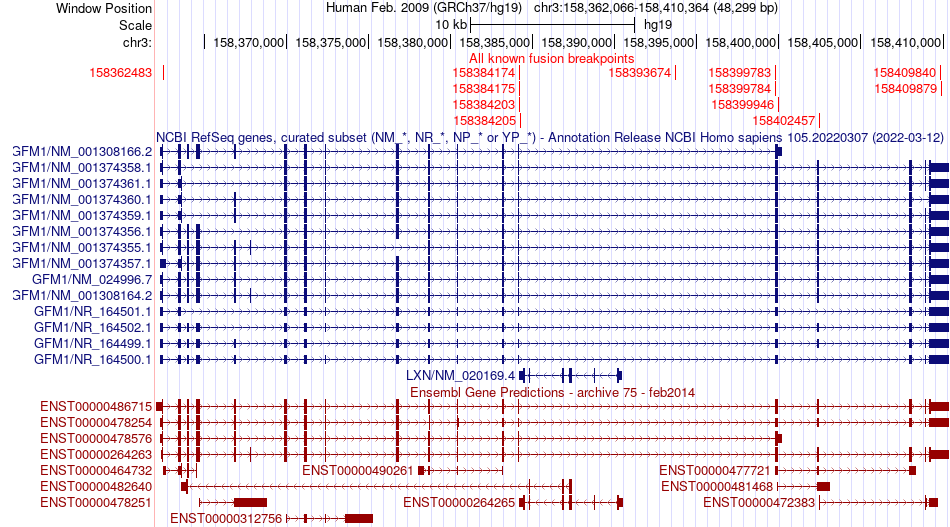
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS) 
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)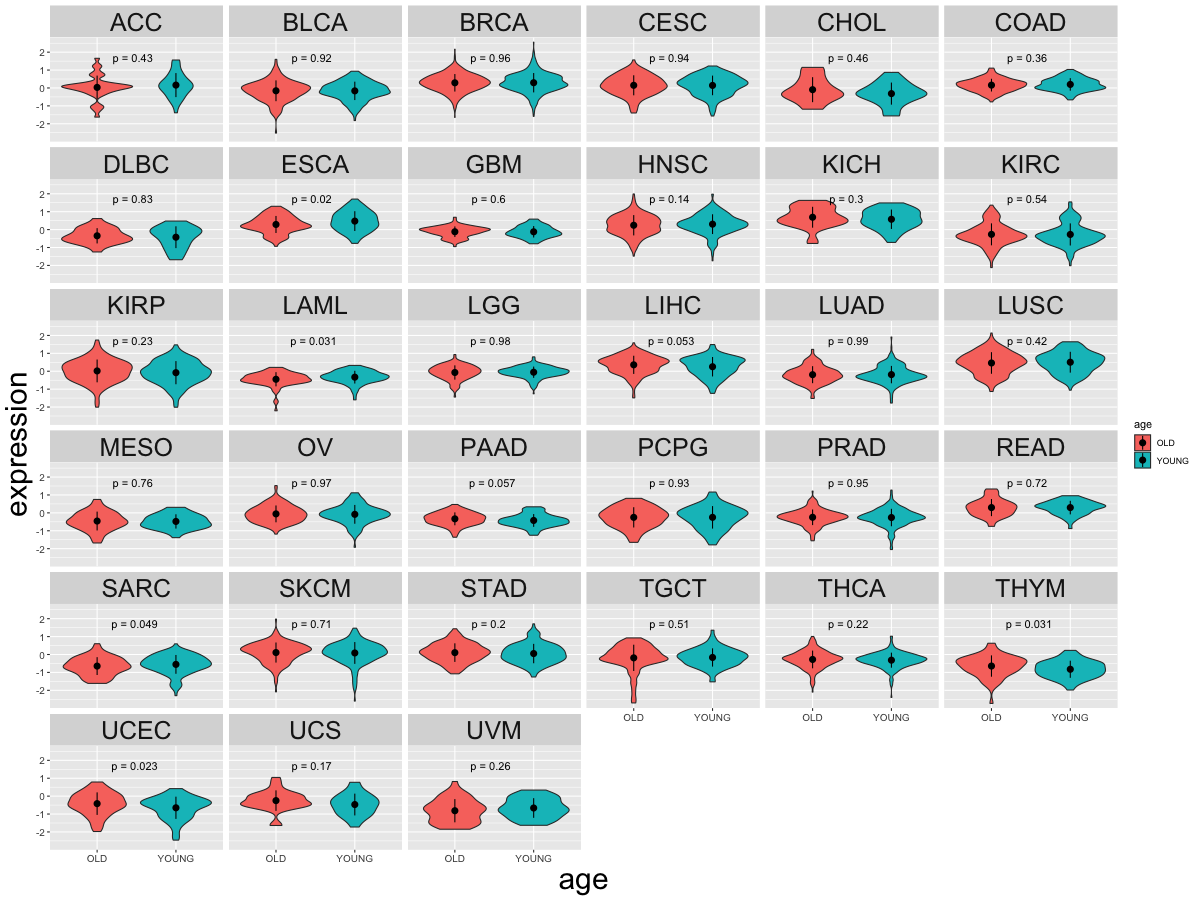
 Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
