
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:COL3A1-COL1A1 (FusionGDB2 ID:HG1281TG1277) |
Fusion Gene Summary for COL3A1-COL1A1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: COL3A1-COL1A1 | Fusion gene ID: hg1281tg1277 | Hgene | Tgene | Gene symbol | COL3A1 | COL1A1 | Gene ID | 1281 | 1277 |
| Gene name | collagen type III alpha 1 chain | collagen type I alpha 1 chain | |
| Synonyms | EDS4A|EDSVASC|PMGEDSV | CAFYD|EDSARTH1|EDSC|OI1|OI2|OI3|OI4 | |
| Cytomap | ('COL3A1')('COL1A1') 2q32.2 | 17q21.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | collagen alpha-1(III) chainEhlers-Danlos syndrome type IV, autosomal dominantalpha-1 type III collagenalpha1 (III) collagencollagen, fetalcollagen, type III, alpha 1 | collagen alpha-1(I) chainalpha-1 type I collagenalpha1(I) procollagencollagen alpha 1 chain type Icollagen alpha-1(I) chain preproproteincollagen of skin, tendon and bone, alpha-1 chaincollagen, type I, alpha 1pro-alpha-1 collagen type 1type I pro | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | P02461 | P02452 | |
| Ensembl transtripts involved in fusion gene | ENST00000304636, ENST00000317840, | ENST00000304636, ENST00000317840, | |
| Fusion gene scores | * DoF score | 46 X 40 X 15=27600 | 44 X 105 X 13=60060 |
| # samples | 56 | 80 | |
| ** MAII score | log2(56/27600*10)=-5.62309762960793 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(80/60060*10)=-6.23026066466979 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: COL3A1 [Title/Abstract] AND COL1A1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | COL3A1(189863444)-COL1A1(48268851), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | COL1A1-COL3A1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. COL1A1-COL3A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. COL3A1-COL1A1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. COL3A1-COL1A1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. COL3A1-COL1A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. COL3A1-COL1A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | COL3A1 | GO:0007160 | cell-matrix adhesion | 16912226 |
| Hgene | COL3A1 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 16360482 |
| Hgene | COL3A1 | GO:0009314 | response to radiation | 14736764 |
| Hgene | COL3A1 | GO:0018149 | peptide cross-linking | 16754721 |
| Hgene | COL3A1 | GO:0034097 | response to cytokine | 9076960|16360482 |
| Hgene | COL3A1 | GO:0042060 | wound healing | 1466622 |
| Tgene | COL1A1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 20018240 |
| Tgene | COL1A1 | GO:0030335 | positive regulation of cell migration | 20018240 |
| Tgene | COL1A1 | GO:0034504 | protein localization to nucleus | 20018240 |
| Tgene | COL1A1 | GO:0045893 | positive regulation of transcription, DNA-templated | 20018240 |
| Tgene | COL1A1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 20018240 |
 Fusion gene breakpoints across COL3A1 (5'-gene) Fusion gene breakpoints across COL3A1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
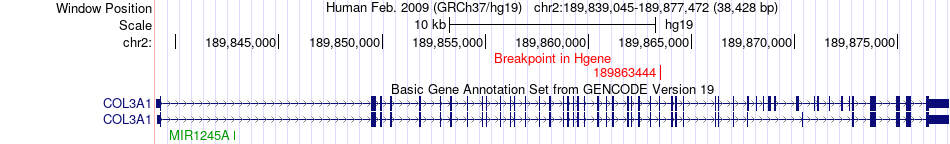 |
 Fusion gene breakpoints across COL1A1 (3'-gene) Fusion gene breakpoints across COL1A1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
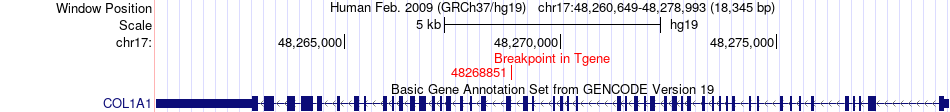 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 235N | COL3A1 | chr2 | 189863444 | + | COL1A1 | chr17 | 48268851 | - |
Top |
Fusion Gene ORF analysis for COL3A1-COL1A1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000304636 | ENST00000225964 | COL3A1 | chr2 | 189863444 | + | COL1A1 | chr17 | 48268851 | - |
| In-frame | ENST00000317840 | ENST00000225964 | COL3A1 | chr2 | 189863444 | + | COL1A1 | chr17 | 48268851 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000304636 | COL3A1 | chr2 | 189863444 | + | ENST00000225964 | COL1A1 | chr17 | 48268851 | - | 6673 | 2192 | 170 | 4459 | 1429 |
| ENST00000317840 | COL3A1 | chr2 | 189863444 | + | ENST00000225964 | COL1A1 | chr17 | 48268851 | - | 6620 | 2139 | 117 | 4406 | 1429 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000304636 | ENST00000225964 | COL3A1 | chr2 | 189863444 | + | COL1A1 | chr17 | 48268851 | - | 0.001841498 | 0.9981585 |
| ENST00000317840 | ENST00000225964 | COL3A1 | chr2 | 189863444 | + | COL1A1 | chr17 | 48268851 | - | 0.001736051 | 0.998264 |
Top |
Fusion Genomic Features for COL3A1-COL1A1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
Top |
Fusion Protein Features for COL3A1-COL1A1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:189863444/chr17:48268851) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| COL3A1 | COL1A1 |
| FUNCTION: Collagen type III occurs in most soft connective tissues along with type I collagen. Involved in regulation of cortical development. Is the major ligand of ADGRG1 in the developing brain and binding to ADGRG1 inhibits neuronal migration and activates the RhoA pathway by coupling ADGRG1 to GNA13 and possibly GNA12. | FUNCTION: Type I collagen is a member of group I collagen (fibrillar forming collagen). |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | COL3A1 | chr2:189863444 | chr17:48268851 | ENST00000304636 | + | 29 | 51 | 30_89 | 674 | 1467.0 | Domain | VWFC |
| Hgene | COL3A1 | chr2:189863444 | chr17:48268851 | ENST00000304636 | + | 29 | 51 | 149_167 | 674 | 1467.0 | Region | Note=Nonhelical region (N-terminal) |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 1229_1464 | 709 | 1465.0 | Domain | Fibrillar collagen NC1 | |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 1093_1095 | 709 | 1465.0 | Motif | Cell attachment site | |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 745_747 | 709 | 1465.0 | Motif | Cell attachment site | |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 1193_1218 | 709 | 1465.0 | Region | Note=Nonhelical region (C-terminal) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | COL3A1 | chr2:189863444 | chr17:48268851 | ENST00000304636 | + | 29 | 51 | 1232_1466 | 674 | 1467.0 | Domain | Fibrillar collagen NC1 |
| Hgene | COL3A1 | chr2:189863444 | chr17:48268851 | ENST00000304636 | + | 29 | 51 | 1197_1205 | 674 | 1467.0 | Region | Note=Nonhelical region (C-terminal) |
| Hgene | COL3A1 | chr2:189863444 | chr17:48268851 | ENST00000304636 | + | 29 | 51 | 168_1196 | 674 | 1467.0 | Region | Note=Triple-helical region |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 38_96 | 709 | 1465.0 | Domain | VWFC | |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 162_178 | 709 | 1465.0 | Region | Note=Nonhelical region (N-terminal) | |
| Tgene | COL1A1 | chr2:189863444 | chr17:48268851 | ENST00000225964 | 30 | 51 | 179_1192 | 709 | 1465.0 | Region | Note=Triple-helical region |
Top |
Fusion Gene Sequence for COL3A1-COL1A1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >18293_18293_1_COL3A1-COL1A1_COL3A1_chr2_189863444_ENST00000304636_COL1A1_chr17_48268851_ENST00000225964_length(transcript)=6673nt_BP=2192nt GTGAGGGAAGCCAAACTTTTTCCTATTTAAGGCCAAAGCAAAGGAATCTCAGTGGCTGAGTTTTATGACGGGCCCGGTGCTGAAGGGCAG GGAACAACTTGATGGTGCTACTTTGAACTGCTTTTCTTTTCTCCTTTTTGCACAAAGAGTCTCATGTCTGATATTTAGACATGATGAGCT TTGTGCAAAAGGGGAGCTGGCTACTTCTCGCTCTGCTTCATCCCACTATTATTTTGGCACAACAGGAAGCTGTTGAAGGAGGATGTTCCC ATCTTGGTCAGTCCTATGCGGATAGAGATGTCTGGAAGCCAGAACCATGCCAAATATGTGTCTGTGACTCAGGATCCGTTCTCTGCGATG ACATAATATGTGACGATCAAGAATTAGACTGCCCCAACCCAGAAATTCCATTTGGAGAATGTTGTGCAGTTTGCCCACAGCCTCCAACTG CTCCTACTCGCCCTCCTAATGGTCAAGGACCTCAAGGCCCCAAGGGAGATCCAGGCCCTCCTGGTATTCCTGGGAGAAATGGTGACCCTG GTATTCCAGGACAACCAGGGTCCCCTGGTTCTCCTGGCCCCCCTGGAATCTGTGAATCATGCCCTACTGGTCCTCAGAACTATTCTCCCC AGTATGATTCATATGATGTCAAGTCTGGAGTAGCAGTAGGAGGACTCGCAGGCTATCCTGGACCAGCTGGCCCCCCAGGCCCTCCCGGTC CCCCTGGTACATCTGGTCATCCTGGTTCCCCTGGATCTCCAGGATACCAAGGACCCCCTGGTGAACCTGGGCAAGCTGGTCCTTCAGGCC CTCCAGGACCTCCTGGTGCTATAGGTCCATCTGGTCCTGCTGGAAAAGATGGAGAATCAGGTAGACCCGGACGACCTGGAGAGCGAGGAT TGCCTGGACCTCCAGGTATCAAAGGTCCAGCTGGGATACCTGGATTCCCTGGTATGAAAGGACACAGAGGCTTCGATGGACGAAATGGAG AAAAGGGTGAAACAGGTGCTCCTGGATTAAAGGGTGAAAATGGTCTTCCAGGCGAAAATGGAGCTCCTGGACCCATGGGTCCAAGAGGGG CTCCTGGTGAGCGAGGACGGCCAGGACTTCCTGGGGCTGCAGGTGCTCGGGGTAATGACGGTGCTCGAGGCAGTGATGGTCAACCAGGCC CTCCTGGTCCTCCTGGAACTGCCGGATTCCCTGGATCCCCTGGTGCTAAGGGTGAAGTTGGACCTGCAGGGTCTCCTGGTTCAAATGGTG CCCCTGGACAAAGAGGAGAACCTGGACCTCAGGGACACGCTGGTGCTCAAGGTCCTCCTGGCCCTCCTGGGATTAATGGTAGTCCTGGTG GTAAAGGCGAAATGGGTCCCGCTGGCATTCCTGGAGCTCCTGGACTGATGGGAGCCCGGGGTCCTCCAGGACCAGCCGGTGCTAATGGTG CTCCTGGACTGCGAGGTGGTGCAGGTGAGCCTGGTAAGAATGGTGCCAAAGGAGAGCCCGGACCACGTGGTGAACGCGGTGAGGCTGGTA TTCCAGGTGTTCCAGGAGCTAAAGGCGAAGATGGCAAGGATGGATCACCTGGAGAACCTGGTGCAAATGGGCTTCCAGGAGCTGCAGGAG AAAGGGGTGCCCCTGGGTTCCGAGGACCTGCTGGACCAAATGGCATCCCAGGAGAAAAGGGTCCTGCTGGAGAGCGTGGTGCTCCAGGCC CTGCAGGGCCCAGAGGAGCTGCTGGAGAACCTGGCAGAGATGGCGTCCCTGGAGGTCCAGGAATGAGGGGCATGCCCGGAAGTCCAGGAG GACCAGGAAGTGATGGGAAACCAGGGCCTCCCGGAAGTCAAGGAGAAAGTGGTCGACCAGGTCCTCCTGGGCCATCTGGTCCCCGAGGTC AGCCTGGTGTCATGGGCTTCCCCGGTCCTAAAGGAAATGATGGTGCTCCTGGTAAGAATGGAGAACGAGGTGGCCCTGGAGGACCTGGCC CTCAGGGTCCTCCTGGAAAGAATGGTGAAACTGGACCTCAGGGACCCCCAGGGCCTACTGGGCCTGGTGGTGACAAAGGAGACACAGGAC CCCCTGGTCCACAAGGATTACAAGGCTTGCCTGGTACAGGTGGTCCTCCAGGAGAAAATGGAAAACCTGGGGAACCAGGTCCAAAGGGTG ATGCCGGTGCACCTGGAGCTCCAGGAGGCAAGGGTGATGCTGGTGCCCCTGGAGCTCCCGGTAGCCAGGGCGCCCCTGGCCTTCAGGGAA TGCCTGGTGAACGTGGTGCAGCTGGTCTTCCAGGGCCTAAGGGTGACAGAGGTGATGCTGGTCCCAAAGGTGCTGATGGCTCTCCTGGCA AAGATGGCGTCCGTGGTCTGACTGGCCCCATTGGTCCTCCTGGCCCTGCTGGTGCCCCTGGTGACAAGGGTGAAAGTGGTCCCAGCGGCC CTGCTGGTCCCACTGGAGCTCGTGGTGCCCCCGGAGACCGTGGTGAGCCTGGTCCCCCCGGCCCTGCTGGCTTTGCTGGCCCCCCTGGTG CTGACGGCCAACCTGGTGCTAAAGGCGAACCTGGTGATGCTGGTGCTAAAGGCGATGCTGGTCCCCCTGGCCCTGCCGGACCCGCTGGAC CCCCTGGCCCCATTGGTAATGTTGGTGCTCCTGGAGCCAAAGGTGCTCGCGGCAGCGCTGGTCCCCCTGGTGCTACTGGTTTCCCTGGTG CTGCTGGCCGAGTCGGTCCTCCTGGCCCCTCTGGAAATGCTGGACCCCCTGGCCCTCCTGGTCCTGCTGGCAAAGAAGGCGGCAAAGGTC CCCGTGGTGAGACTGGCCCTGCTGGACGTCCTGGTGAAGTTGGTCCCCCTGGTCCCCCTGGCCCTGCTGGCGAGAAAGGATCCCCTGGTG CTGATGGTCCTGCTGGTGCTCCTGGTACTCCCGGGCCTCAAGGTATTGCTGGACAGCGTGGTGTGGTCGGCCTGCCTGGTCAGAGAGGAG AGAGAGGCTTCCCTGGTCTTCCTGGCCCCTCTGGTGAACCTGGCAAACAAGGTCCCTCTGGAGCAAGTGGTGAACGTGGTCCCCCTGGTC CCATGGGCCCCCCTGGATTGGCTGGACCCCCTGGTGAATCTGGACGTGAGGGGGCTCCTGGTGCCGAAGGTTCCCCTGGACGAGACGGTT CTCCTGGCGCCAAGGGTGACCGTGGTGAGACCGGCCCCGCTGGACCCCCTGGTGCTCCTGGTGCTCCTGGTGCCCCTGGCCCCGTTGGCC CTGCTGGCAAGAGTGGTGATCGTGGTGAGACTGGTCCTGCTGGTCCCACCGGTCCTGTCGGCCCTGTTGGCGCCCGTGGCCCCGCCGGAC CCCAAGGCCCCCGTGGTGACAAGGGTGAGACAGGCGAACAGGGCGACAGAGGCATAAAGGGTCACCGTGGCTTCTCTGGCCTCCAGGGTC CCCCTGGCCCTCCTGGCTCTCCTGGTGAACAAGGTCCCTCTGGAGCCTCTGGTCCTGCTGGTCCCCGAGGTCCCCCTGGCTCTGCTGGTG CTCCTGGCAAAGATGGACTCAACGGTCTCCCTGGCCCCATTGGGCCCCCTGGTCCTCGCGGTCGCACTGGTGATGCTGGTCCTGTTGGTC CCCCCGGCCCTCCTGGACCTCCTGGTCCCCCTGGTCCTCCCAGCGCTGGTTTCGACTTCAGCTTCCTGCCCCAGCCACCTCAAGAGAAGG CTCACGATGGTGGCCGCTACTACCGGGCTGATGATGCCAATGTGGTTCGTGACCGTGACCTCGAGGTGGACACCACCCTCAAGAGCCTGA GCCAGCAGATCGAGAACATCCGGAGCCCAGAGGGCAGCCGCAAGAACCCCGCCCGCACCTGCCGTGACCTCAAGATGTGCCACTCTGACT GGAAGAGTGGAGAGTACTGGATTGACCCCAACCAAGGCTGCAACCTGGATGCCATCAAAGTCTTCTGCAACATGGAGACTGGTGAGACCT GCGTGTACCCCACTCAGCCCAGTGTGGCCCAGAAGAACTGGTACATCAGCAAGAACCCCAAGGACAAGAGGCATGTCTGGTTCGGCGAGA GCATGACCGATGGATTCCAGTTCGAGTATGGCGGCCAGGGCTCCGACCCTGCCGATGTGGCCATCCAGCTGACCTTCCTGCGCCTGATGT CCACCGAGGCCTCCCAGAACATCACCTACCACTGCAAGAACAGCGTGGCCTACATGGACCAGCAGACTGGCAACCTCAAGAAGGCCCTGC TCCTCCAGGGCTCCAACGAGATCGAGATCCGCGCCGAGGGCAACAGCCGCTTCACCTACAGCGTCACTGTCGATGGCTGCACGAGTCACA CCGGAGCCTGGGGCAAGACAGTGATTGAATACAAAACCACCAAGACCTCCCGCCTGCCCATCATCGATGTGGCCCCCTTGGACGTTGGTG CCCCAGACCAGGAATTCGGCTTCGACGTTGGCCCTGTCTGCTTCCTGTAAACTCCCTCCATCCCAACCTGGCTCCCTCCCACCCAACCAA CTTTCCCCCCAACCCGGAAACAGACAAGCAACCCAAACTGAACCCCCTCAAAAGCCAAAAAATGGGAGACAATTTCACATGGACTTTGGA AAATATTTTTTTCCTTTGCATTCATCTCTCAAACTTAGTTTTTATCTTTGACCAACCGAACATGACCAAAAACCAAAAGTGCATTCAACC TTACCAAAAAAAAAAAAAAAAAAAGAATAAATAAATAACTTTTTAAAAAAGGAAGCTTGGTCCACTTGCTTGAAGACCCATGCGGGGGTA AGTCCCTTTCTGCCCGTTGGGCTTATGAAACCCCAATGCTGCCCTTTCTGCTCCTTTCTCCACACCCCCCTTGGGGCCTCCCCTCCACTC CTTCCCAAATCTGTCTCCCCAGAAGACACAGGAAACAATGTATTGTCTGCCCAGCAATCAAAGGCAATGCTCAAACACCCAAGTGGCCCC CACCCTCAGCCCGCTCCTGCCCGCCCAGCACCCCCAGGCCCTGGGGGACCTGGGGTTCTCAGACTGCCAAAGAAGCCTTGCCATCTGGCG CTCCCATGGCTCTTGCAACATCTCCCCTTCGTTTTTGAGGGGGTCATGCCGGGGGAGCCACCAGCCCCTCACTGGGTTCGGAGGAGAGTC AGGAAGGGCCACGACAAAGCAGAAACATCGGATTTGGGGAACGCGTGTCAATCCCTTGTGCCGCAGGGCTGGGCGGGAGAGACTGTTCTG TTCCTTGTGTAACTGTGTTGCTGAAAGACTACCTCGTTCTTGTCTTGATGTGTCACCGGGGCAACTGCCTGGGGGCGGGGATGGGGGCAG GGTGGAAGCGGCTCCCCATTTTATACCAAAGGTGCTACATCTATGTGATGGGTGGGGTGGGGAGGGAATCACTGGTGCTATAGAAATTGA GATGCCCCCCCAGGCCAGCAAATGTTCCTTTTTGTTCAAAGTCTATTTTTATTCCTTGATATTTTTCTTTTTTTTTTTTTTTTTTTGTGG ATGGGGACTTGTGAATTTTTCTAAAGGTGCTATTTAACATGGGAGGAGAGCGTGTGCGGCTCCAGCCCAGCCCGCTGCTCACTTTCCACC CTCTCTCCACCTGCCTCTGGCTTCTCAGGCCTCTGCTCTCCGACCTCTCTCCTCTGAAACCCTCCTCCACAGCTGCAGCCCATCCTCCCG GCTCCCTCCTAGTCTGTCCTGCGTCCTCTGTCCCCGGGTTTCAGAGACAACTTCCCAAAGCACAAAGCAGTTTTTCCCCCTAGGGGTGGG AGGAAGCAAAAGACTCTGTACCTATTTTGTATGTGTATAATAATTTGAGATGTTTTTAATTATTTTGATTGCTGGAATAAAGCATGTGGA AATGACCCAAACATAATCCGCAGTGGCCTCCTAATTTCCTTCTTTGGAGTTGGGGGAGGGGTAGACATGGGGAAGGGGCTTTGGGGTGAT GGGCTTGCCTTCCATTCCTGCCCTTTCCCTCCCCACTATTCTCTTCTAGATCCCTCCATAACCCCACTCCCCTTTCTCTCACCCTTCTTA TACCGCAAACCTTTCTACTTCCTCTTTCATTTTCTATTCTTGCAATTTCCTTGCACCTTTTCCAAATCCTCTTCTCCCCTGCAATACCAT ACAGGCAATCCACGTGCACAACACACACACACACTCTTCACATCTGGGGTTGTCCAAACCTCATACCCACTCCCCTTCAAGCCCATCCAC TCTCCACCCCCTGGATGCCCTGCACTTGGTGGCGGTGGGATGCTCATGGATACTGGGAGGGTGAGGGGAGTGGAACCCGTGAGGAGGACC TGGGGGCCTCTCCTTGAACTGACATGAAGGGTCATCTGGCCTCTGCTCCCTTCTCACCCACGCTGACCTCCTGCCGAAGGAGCAACGCAA CAGGAGAGGGGTCTGCTGAGCCTGGCGAGGGTCTGGGAGGGACCAGGAGGAAGGCGTGCTCCCTGCTCGCTGTCCTGGCCCTGGGGGAGT GAGGGAGACAGACACCTGGGAGAGCTGTGGGGAAGGCACTCGCACCGTGCTCTTGGGAAGGAAGGAGACCTGGCCCTGCTCACCACGGAC TGGGTGCCTCGACCTCCTGAATCCCCAGAACACAACCCCCCTGGGCTGGGGTGGTCTGGGGAACCATCGTGCCCCCGCCTCCCGCCTACT >18293_18293_1_COL3A1-COL1A1_COL3A1_chr2_189863444_ENST00000304636_COL1A1_chr17_48268851_ENST00000225964_length(amino acids)=1429AA_BP=596 MMSFVQKGSWLLLALLHPTIILAQQEAVEGGCSHLGQSYADRDVWKPEPCQICVCDSGSVLCDDIICDDQELDCPNPEIPFGECCAVCPQ PPTAPTRPPNGQGPQGPKGDPGPPGIPGRNGDPGIPGQPGSPGSPGPPGICESCPTGPQNYSPQYDSYDVKSGVAVGGLAGYPGPAGPPG PPGPPGTSGHPGSPGSPGYQGPPGEPGQAGPSGPPGPPGAIGPSGPAGKDGESGRPGRPGERGLPGPPGIKGPAGIPGFPGMKGHRGFDG RNGEKGETGAPGLKGENGLPGENGAPGPMGPRGAPGERGRPGLPGAAGARGNDGARGSDGQPGPPGPPGTAGFPGSPGAKGEVGPAGSPG SNGAPGQRGEPGPQGHAGAQGPPGPPGINGSPGGKGEMGPAGIPGAPGLMGARGPPGPAGANGAPGLRGGAGEPGKNGAKGEPGPRGERG EAGIPGVPGAKGEDGKDGSPGEPGANGLPGAAGERGAPGFRGPAGPNGIPGEKGPAGERGAPGPAGPRGAAGEPGRDGVPGGPGMRGMPG SPGGPGSDGKPGPPGSQGESGRPGPPGPSGPRGQPGVMGFPGPKGNDGAPGKNGERGGPGGPGPQGPPGKNGETGPQGPPGPTGPGGDKG DTGPPGPQGLQGLPGTGGPPGENGKPGEPGPKGDAGAPGAPGGKGDAGAPGAPGSQGAPGLQGMPGERGAAGLPGPKGDRGDAGPKGADG SPGKDGVRGLTGPIGPPGPAGAPGDKGESGPSGPAGPTGARGAPGDRGEPGPPGPAGFAGPPGADGQPGAKGEPGDAGAKGDAGPPGPAG PAGPPGPIGNVGAPGAKGARGSAGPPGATGFPGAAGRVGPPGPSGNAGPPGPPGPAGKEGGKGPRGETGPAGRPGEVGPPGPPGPAGEKG SPGADGPAGAPGTPGPQGIAGQRGVVGLPGQRGERGFPGLPGPSGEPGKQGPSGASGERGPPGPMGPPGLAGPPGESGREGAPGAEGSPG RDGSPGAKGDRGETGPAGPPGAPGAPGAPGPVGPAGKSGDRGETGPAGPTGPVGPVGARGPAGPQGPRGDKGETGEQGDRGIKGHRGFSG LQGPPGPPGSPGEQGPSGASGPAGPRGPPGSAGAPGKDGLNGLPGPIGPPGPRGRTGDAGPVGPPGPPGPPGPPGPPSAGFDFSFLPQPP QEKAHDGGRYYRADDANVVRDRDLEVDTTLKSLSQQIENIRSPEGSRKNPARTCRDLKMCHSDWKSGEYWIDPNQGCNLDAIKVFCNMET GETCVYPTQPSVAQKNWYISKNPKDKRHVWFGESMTDGFQFEYGGQGSDPADVAIQLTFLRLMSTEASQNITYHCKNSVAYMDQQTGNLK -------------------------------------------------------------- >18293_18293_2_COL3A1-COL1A1_COL3A1_chr2_189863444_ENST00000317840_COL1A1_chr17_48268851_ENST00000225964_length(transcript)=6620nt_BP=2139nt GGCTGAGTTTTATGACGGGCCCGGTGCTGAAGGGCAGGGAACAACTTGATGGTGCTACTTTGAACTGCTTTTCTTTTCTCCTTTTTGCAC AAAGAGTCTCATGTCTGATATTTAGACATGATGAGCTTTGTGCAAAAGGGGAGCTGGCTACTTCTCGCTCTGCTTCATCCCACTATTATT TTGGCACAACAGGAAGCTGTTGAAGGAGGATGTTCCCATCTTGGTCAGTCCTATGCGGATAGAGATGTCTGGAAGCCAGAACCATGCCAA ATATGTGTCTGTGACTCAGGATCCGTTCTCTGCGATGACATAATATGTGACGATCAAGAATTAGACTGCCCCAACCCAGAAATTCCATTT GGAGAATGTTGTGCAGTTTGCCCACAGCCTCCAACTGCTCCTACTCGCCCTCCTAATGGTCAAGGACCTCAAGGCCCCAAGGGAGATCCA GGCCCTCCTGGTATTCCTGGGAGAAATGGTGACCCTGGTATTCCAGGACAACCAGGGTCCCCTGGTTCTCCTGGCCCCCCTGGAATCTGT GAATCATGCCCTACTGGTCCTCAGAACTATTCTCCCCAGTATGATTCATATGATGTCAAGTCTGGAGTAGCAGTAGGAGGACTCGCAGGC TATCCTGGACCAGCTGGCCCCCCAGGCCCTCCCGGTCCCCCTGGTACATCTGGTCATCCTGGTTCCCCTGGATCTCCAGGATACCAAGGA CCCCCTGGTGAACCTGGGCAAGCTGGTCCTTCAGGCCCTCCAGGACCTCCTGGTGCTATAGGTCCATCTGGTCCTGCTGGAAAAGATGGA GAATCAGGTAGACCCGGACGACCTGGAGAGCGAGGATTGCCTGGACCTCCAGGTATCAAAGGTCCAGCTGGGATACCTGGATTCCCTGGT ATGAAAGGACACAGAGGCTTCGATGGACGAAATGGAGAAAAGGGTGAAACAGGTGCTCCTGGATTAAAGGGTGAAAATGGTCTTCCAGGC GAAAATGGAGCTCCTGGACCCATGGGTCCAAGAGGGGCTCCTGGTGAGCGAGGACGGCCAGGACTTCCTGGGGCTGCAGGTGCTCGGGGT AATGACGGTGCTCGAGGCAGTGATGGTCAACCAGGCCCTCCTGGTCCTCCTGGAACTGCCGGATTCCCTGGATCCCCTGGTGCTAAGGGT GAAGTTGGACCTGCAGGGTCTCCTGGTTCAAATGGTGCCCCTGGACAAAGAGGAGAACCTGGACCTCAGGGACACGCTGGTGCTCAAGGT CCTCCTGGCCCTCCTGGGATTAATGGTAGTCCTGGTGGTAAAGGCGAAATGGGTCCCGCTGGCATTCCTGGAGCTCCTGGACTGATGGGA GCCCGGGGTCCTCCAGGACCAGCCGGTGCTAATGGTGCTCCTGGACTGCGAGGTGGTGCAGGTGAGCCTGGTAAGAATGGTGCCAAAGGA GAGCCCGGACCACGTGGTGAACGCGGTGAGGCTGGTATTCCAGGTGTTCCAGGAGCTAAAGGCGAAGATGGCAAGGATGGATCACCTGGA GAACCTGGTGCAAATGGGCTTCCAGGAGCTGCAGGAGAAAGGGGTGCCCCTGGGTTCCGAGGACCTGCTGGACCAAATGGCATCCCAGGA GAAAAGGGTCCTGCTGGAGAGCGTGGTGCTCCAGGCCCTGCAGGGCCCAGAGGAGCTGCTGGAGAACCTGGCAGAGATGGCGTCCCTGGA GGTCCAGGAATGAGGGGCATGCCCGGAAGTCCAGGAGGACCAGGAAGTGATGGGAAACCAGGGCCTCCCGGAAGTCAAGGAGAAAGTGGT CGACCAGGTCCTCCTGGGCCATCTGGTCCCCGAGGTCAGCCTGGTGTCATGGGCTTCCCCGGTCCTAAAGGAAATGATGGTGCTCCTGGT AAGAATGGAGAACGAGGTGGCCCTGGAGGACCTGGCCCTCAGGGTCCTCCTGGAAAGAATGGTGAAACTGGACCTCAGGGACCCCCAGGG CCTACTGGGCCTGGTGGTGACAAAGGAGACACAGGACCCCCTGGTCCACAAGGATTACAAGGCTTGCCTGGTACAGGTGGTCCTCCAGGA GAAAATGGAAAACCTGGGGAACCAGGTCCAAAGGGTGATGCCGGTGCACCTGGAGCTCCAGGAGGCAAGGGTGATGCTGGTGCCCCTGGA GCTCCCGGTAGCCAGGGCGCCCCTGGCCTTCAGGGAATGCCTGGTGAACGTGGTGCAGCTGGTCTTCCAGGGCCTAAGGGTGACAGAGGT GATGCTGGTCCCAAAGGTGCTGATGGCTCTCCTGGCAAAGATGGCGTCCGTGGTCTGACTGGCCCCATTGGTCCTCCTGGCCCTGCTGGT GCCCCTGGTGACAAGGGTGAAAGTGGTCCCAGCGGCCCTGCTGGTCCCACTGGAGCTCGTGGTGCCCCCGGAGACCGTGGTGAGCCTGGT CCCCCCGGCCCTGCTGGCTTTGCTGGCCCCCCTGGTGCTGACGGCCAACCTGGTGCTAAAGGCGAACCTGGTGATGCTGGTGCTAAAGGC GATGCTGGTCCCCCTGGCCCTGCCGGACCCGCTGGACCCCCTGGCCCCATTGGTAATGTTGGTGCTCCTGGAGCCAAAGGTGCTCGCGGC AGCGCTGGTCCCCCTGGTGCTACTGGTTTCCCTGGTGCTGCTGGCCGAGTCGGTCCTCCTGGCCCCTCTGGAAATGCTGGACCCCCTGGC CCTCCTGGTCCTGCTGGCAAAGAAGGCGGCAAAGGTCCCCGTGGTGAGACTGGCCCTGCTGGACGTCCTGGTGAAGTTGGTCCCCCTGGT CCCCCTGGCCCTGCTGGCGAGAAAGGATCCCCTGGTGCTGATGGTCCTGCTGGTGCTCCTGGTACTCCCGGGCCTCAAGGTATTGCTGGA CAGCGTGGTGTGGTCGGCCTGCCTGGTCAGAGAGGAGAGAGAGGCTTCCCTGGTCTTCCTGGCCCCTCTGGTGAACCTGGCAAACAAGGT CCCTCTGGAGCAAGTGGTGAACGTGGTCCCCCTGGTCCCATGGGCCCCCCTGGATTGGCTGGACCCCCTGGTGAATCTGGACGTGAGGGG GCTCCTGGTGCCGAAGGTTCCCCTGGACGAGACGGTTCTCCTGGCGCCAAGGGTGACCGTGGTGAGACCGGCCCCGCTGGACCCCCTGGT GCTCCTGGTGCTCCTGGTGCCCCTGGCCCCGTTGGCCCTGCTGGCAAGAGTGGTGATCGTGGTGAGACTGGTCCTGCTGGTCCCACCGGT CCTGTCGGCCCTGTTGGCGCCCGTGGCCCCGCCGGACCCCAAGGCCCCCGTGGTGACAAGGGTGAGACAGGCGAACAGGGCGACAGAGGC ATAAAGGGTCACCGTGGCTTCTCTGGCCTCCAGGGTCCCCCTGGCCCTCCTGGCTCTCCTGGTGAACAAGGTCCCTCTGGAGCCTCTGGT CCTGCTGGTCCCCGAGGTCCCCCTGGCTCTGCTGGTGCTCCTGGCAAAGATGGACTCAACGGTCTCCCTGGCCCCATTGGGCCCCCTGGT CCTCGCGGTCGCACTGGTGATGCTGGTCCTGTTGGTCCCCCCGGCCCTCCTGGACCTCCTGGTCCCCCTGGTCCTCCCAGCGCTGGTTTC GACTTCAGCTTCCTGCCCCAGCCACCTCAAGAGAAGGCTCACGATGGTGGCCGCTACTACCGGGCTGATGATGCCAATGTGGTTCGTGAC CGTGACCTCGAGGTGGACACCACCCTCAAGAGCCTGAGCCAGCAGATCGAGAACATCCGGAGCCCAGAGGGCAGCCGCAAGAACCCCGCC CGCACCTGCCGTGACCTCAAGATGTGCCACTCTGACTGGAAGAGTGGAGAGTACTGGATTGACCCCAACCAAGGCTGCAACCTGGATGCC ATCAAAGTCTTCTGCAACATGGAGACTGGTGAGACCTGCGTGTACCCCACTCAGCCCAGTGTGGCCCAGAAGAACTGGTACATCAGCAAG AACCCCAAGGACAAGAGGCATGTCTGGTTCGGCGAGAGCATGACCGATGGATTCCAGTTCGAGTATGGCGGCCAGGGCTCCGACCCTGCC GATGTGGCCATCCAGCTGACCTTCCTGCGCCTGATGTCCACCGAGGCCTCCCAGAACATCACCTACCACTGCAAGAACAGCGTGGCCTAC ATGGACCAGCAGACTGGCAACCTCAAGAAGGCCCTGCTCCTCCAGGGCTCCAACGAGATCGAGATCCGCGCCGAGGGCAACAGCCGCTTC ACCTACAGCGTCACTGTCGATGGCTGCACGAGTCACACCGGAGCCTGGGGCAAGACAGTGATTGAATACAAAACCACCAAGACCTCCCGC CTGCCCATCATCGATGTGGCCCCCTTGGACGTTGGTGCCCCAGACCAGGAATTCGGCTTCGACGTTGGCCCTGTCTGCTTCCTGTAAACT CCCTCCATCCCAACCTGGCTCCCTCCCACCCAACCAACTTTCCCCCCAACCCGGAAACAGACAAGCAACCCAAACTGAACCCCCTCAAAA GCCAAAAAATGGGAGACAATTTCACATGGACTTTGGAAAATATTTTTTTCCTTTGCATTCATCTCTCAAACTTAGTTTTTATCTTTGACC AACCGAACATGACCAAAAACCAAAAGTGCATTCAACCTTACCAAAAAAAAAAAAAAAAAAAGAATAAATAAATAACTTTTTAAAAAAGGA AGCTTGGTCCACTTGCTTGAAGACCCATGCGGGGGTAAGTCCCTTTCTGCCCGTTGGGCTTATGAAACCCCAATGCTGCCCTTTCTGCTC CTTTCTCCACACCCCCCTTGGGGCCTCCCCTCCACTCCTTCCCAAATCTGTCTCCCCAGAAGACACAGGAAACAATGTATTGTCTGCCCA GCAATCAAAGGCAATGCTCAAACACCCAAGTGGCCCCCACCCTCAGCCCGCTCCTGCCCGCCCAGCACCCCCAGGCCCTGGGGGACCTGG GGTTCTCAGACTGCCAAAGAAGCCTTGCCATCTGGCGCTCCCATGGCTCTTGCAACATCTCCCCTTCGTTTTTGAGGGGGTCATGCCGGG GGAGCCACCAGCCCCTCACTGGGTTCGGAGGAGAGTCAGGAAGGGCCACGACAAAGCAGAAACATCGGATTTGGGGAACGCGTGTCAATC CCTTGTGCCGCAGGGCTGGGCGGGAGAGACTGTTCTGTTCCTTGTGTAACTGTGTTGCTGAAAGACTACCTCGTTCTTGTCTTGATGTGT CACCGGGGCAACTGCCTGGGGGCGGGGATGGGGGCAGGGTGGAAGCGGCTCCCCATTTTATACCAAAGGTGCTACATCTATGTGATGGGT GGGGTGGGGAGGGAATCACTGGTGCTATAGAAATTGAGATGCCCCCCCAGGCCAGCAAATGTTCCTTTTTGTTCAAAGTCTATTTTTATT CCTTGATATTTTTCTTTTTTTTTTTTTTTTTTTGTGGATGGGGACTTGTGAATTTTTCTAAAGGTGCTATTTAACATGGGAGGAGAGCGT GTGCGGCTCCAGCCCAGCCCGCTGCTCACTTTCCACCCTCTCTCCACCTGCCTCTGGCTTCTCAGGCCTCTGCTCTCCGACCTCTCTCCT CTGAAACCCTCCTCCACAGCTGCAGCCCATCCTCCCGGCTCCCTCCTAGTCTGTCCTGCGTCCTCTGTCCCCGGGTTTCAGAGACAACTT CCCAAAGCACAAAGCAGTTTTTCCCCCTAGGGGTGGGAGGAAGCAAAAGACTCTGTACCTATTTTGTATGTGTATAATAATTTGAGATGT TTTTAATTATTTTGATTGCTGGAATAAAGCATGTGGAAATGACCCAAACATAATCCGCAGTGGCCTCCTAATTTCCTTCTTTGGAGTTGG GGGAGGGGTAGACATGGGGAAGGGGCTTTGGGGTGATGGGCTTGCCTTCCATTCCTGCCCTTTCCCTCCCCACTATTCTCTTCTAGATCC CTCCATAACCCCACTCCCCTTTCTCTCACCCTTCTTATACCGCAAACCTTTCTACTTCCTCTTTCATTTTCTATTCTTGCAATTTCCTTG CACCTTTTCCAAATCCTCTTCTCCCCTGCAATACCATACAGGCAATCCACGTGCACAACACACACACACACTCTTCACATCTGGGGTTGT CCAAACCTCATACCCACTCCCCTTCAAGCCCATCCACTCTCCACCCCCTGGATGCCCTGCACTTGGTGGCGGTGGGATGCTCATGGATAC TGGGAGGGTGAGGGGAGTGGAACCCGTGAGGAGGACCTGGGGGCCTCTCCTTGAACTGACATGAAGGGTCATCTGGCCTCTGCTCCCTTC TCACCCACGCTGACCTCCTGCCGAAGGAGCAACGCAACAGGAGAGGGGTCTGCTGAGCCTGGCGAGGGTCTGGGAGGGACCAGGAGGAAG GCGTGCTCCCTGCTCGCTGTCCTGGCCCTGGGGGAGTGAGGGAGACAGACACCTGGGAGAGCTGTGGGGAAGGCACTCGCACCGTGCTCT TGGGAAGGAAGGAGACCTGGCCCTGCTCACCACGGACTGGGTGCCTCGACCTCCTGAATCCCCAGAACACAACCCCCCTGGGCTGGGGTG >18293_18293_2_COL3A1-COL1A1_COL3A1_chr2_189863444_ENST00000317840_COL1A1_chr17_48268851_ENST00000225964_length(amino acids)=1429AA_BP=596 MMSFVQKGSWLLLALLHPTIILAQQEAVEGGCSHLGQSYADRDVWKPEPCQICVCDSGSVLCDDIICDDQELDCPNPEIPFGECCAVCPQ PPTAPTRPPNGQGPQGPKGDPGPPGIPGRNGDPGIPGQPGSPGSPGPPGICESCPTGPQNYSPQYDSYDVKSGVAVGGLAGYPGPAGPPG PPGPPGTSGHPGSPGSPGYQGPPGEPGQAGPSGPPGPPGAIGPSGPAGKDGESGRPGRPGERGLPGPPGIKGPAGIPGFPGMKGHRGFDG RNGEKGETGAPGLKGENGLPGENGAPGPMGPRGAPGERGRPGLPGAAGARGNDGARGSDGQPGPPGPPGTAGFPGSPGAKGEVGPAGSPG SNGAPGQRGEPGPQGHAGAQGPPGPPGINGSPGGKGEMGPAGIPGAPGLMGARGPPGPAGANGAPGLRGGAGEPGKNGAKGEPGPRGERG EAGIPGVPGAKGEDGKDGSPGEPGANGLPGAAGERGAPGFRGPAGPNGIPGEKGPAGERGAPGPAGPRGAAGEPGRDGVPGGPGMRGMPG SPGGPGSDGKPGPPGSQGESGRPGPPGPSGPRGQPGVMGFPGPKGNDGAPGKNGERGGPGGPGPQGPPGKNGETGPQGPPGPTGPGGDKG DTGPPGPQGLQGLPGTGGPPGENGKPGEPGPKGDAGAPGAPGGKGDAGAPGAPGSQGAPGLQGMPGERGAAGLPGPKGDRGDAGPKGADG SPGKDGVRGLTGPIGPPGPAGAPGDKGESGPSGPAGPTGARGAPGDRGEPGPPGPAGFAGPPGADGQPGAKGEPGDAGAKGDAGPPGPAG PAGPPGPIGNVGAPGAKGARGSAGPPGATGFPGAAGRVGPPGPSGNAGPPGPPGPAGKEGGKGPRGETGPAGRPGEVGPPGPPGPAGEKG SPGADGPAGAPGTPGPQGIAGQRGVVGLPGQRGERGFPGLPGPSGEPGKQGPSGASGERGPPGPMGPPGLAGPPGESGREGAPGAEGSPG RDGSPGAKGDRGETGPAGPPGAPGAPGAPGPVGPAGKSGDRGETGPAGPTGPVGPVGARGPAGPQGPRGDKGETGEQGDRGIKGHRGFSG LQGPPGPPGSPGEQGPSGASGPAGPRGPPGSAGAPGKDGLNGLPGPIGPPGPRGRTGDAGPVGPPGPPGPPGPPGPPSAGFDFSFLPQPP QEKAHDGGRYYRADDANVVRDRDLEVDTTLKSLSQQIENIRSPEGSRKNPARTCRDLKMCHSDWKSGEYWIDPNQGCNLDAIKVFCNMET GETCVYPTQPSVAQKNWYISKNPKDKRHVWFGESMTDGFQFEYGGQGSDPADVAIQLTFLRLMSTEASQNITYHCKNSVAYMDQQTGNLK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for COL3A1-COL1A1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for COL3A1-COL1A1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Hgene | COL3A1 | P02461 | DB00048 | Collagenase clostridium histolyticum | Binder | Biotech | Approved|Investigational |
| Tgene | COL1A1 | P02452 | DB00048 | Collagenase clostridium histolyticum | Binder | Biotech | Approved|Investigational |
| Tgene | COL1A1 | P02452 | DB12872 | Vonicog Alfa | Binder | Biotech | Approved|Investigational |
| Tgene | COL1A1 | P02452 | DB13133 | Von Willebrand Factor Human | Binder | Biotech | Approved|Investigational |
| Tgene | COL1A1 | P02452 | DB11338 | Clove oil | Biotech | Approved|Nutraceutical | |
| Tgene | COL1A1 | P02452 | DB04866 | Halofuginone | Small molecule | Investigational|Vet_approved |
Top |
Related Diseases for COL3A1-COL1A1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | COL3A1 | C0268338 | Ehlers-Danlos Syndrome, Type IV | 43 | CLINGEN;CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | COL3A1 | C4707243 | Familial thoracic aortic aneurysm and aortic dissection | 10 | CLINGEN;GENOMICS_ENGLAND |
| Hgene | COL3A1 | C0023890 | Liver Cirrhosis | 5 | CTD_human |
| Hgene | COL3A1 | C0239946 | Fibrosis, Liver | 5 | CTD_human |
| Hgene | COL3A1 | C0023893 | Liver Cirrhosis, Experimental | 2 | CTD_human |
| Hgene | COL3A1 | C0162871 | Aortic Aneurysm, Abdominal | 2 | ORPHANET |
| Hgene | COL3A1 | C1853365 | AORTIC ANEURYSM, FAMILIAL ABDOMINAL 1 | 2 | ORPHANET |
| Hgene | COL3A1 | C2697932 | Loeys-Dietz Syndrome | 2 | GENOMICS_ENGLAND |
| Hgene | COL3A1 | C0003504 | Aortic Valve Insufficiency | 1 | CTD_human |
| Hgene | COL3A1 | C0005398 | Cholestasis, Extrahepatic | 1 | CTD_human |
| Hgene | COL3A1 | C0015695 | Fatty Liver | 1 | CTD_human |
| Hgene | COL3A1 | C0016059 | Fibrosis | 1 | CTD_human |
| Hgene | COL3A1 | C0019193 | Hepatitis, Toxic | 1 | CTD_human |
| Hgene | COL3A1 | C0020443 | Hypercholesterolemia | 1 | CTD_human |
| Hgene | COL3A1 | C0020456 | Hyperglycemia | 1 | CTD_human |
| Hgene | COL3A1 | C0020459 | Hyperinsulinism | 1 | CTD_human |
| Hgene | COL3A1 | C0020538 | Hypertensive disease | 1 | CTD_human |
| Hgene | COL3A1 | C0022548 | Keloid | 1 | CTD_human |
| Hgene | COL3A1 | C0023891 | Liver Cirrhosis, Alcoholic | 1 | CTD_human |
| Hgene | COL3A1 | C0023895 | Liver diseases | 1 | CTD_human |
| Hgene | COL3A1 | C0027719 | Nephrosclerosis | 1 | CTD_human |
| Hgene | COL3A1 | C0034069 | Pulmonary Fibrosis | 1 | CTD_human |
| Hgene | COL3A1 | C0036341 | Schizophrenia | 1 | CTD_human |
| Hgene | COL3A1 | C0038454 | Cerebrovascular accident | 1 | GENOMICS_ENGLAND |
| Hgene | COL3A1 | C0041956 | Ureteral obstruction | 1 | CTD_human |
| Hgene | COL3A1 | C0086565 | Liver Dysfunction | 1 | CTD_human |
| Hgene | COL3A1 | C0149721 | Left Ventricular Hypertrophy | 1 | CTD_human |
| Hgene | COL3A1 | C0238288 | Muscular Dystrophy, Facioscapulohumeral | 1 | CTD_human |
| Hgene | COL3A1 | C0238590 | Acrogeria | 1 | ORPHANET |
| Hgene | COL3A1 | C0338586 | Vertebral Artery Dissection | 1 | GENOMICS_ENGLAND |
| Hgene | COL3A1 | C0406584 | Acrogeria, gottron type | 1 | ORPHANET |
| Hgene | COL3A1 | C0553692 | Brain hemorrhage | 1 | GENOMICS_ENGLAND |
| Hgene | COL3A1 | C0553980 | Endomyocardial Fibrosis | 1 | CTD_human |
| Hgene | COL3A1 | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human |
| Hgene | COL3A1 | C0948008 | Ischemic stroke | 1 | GENOMICS_ENGLAND |
| Hgene | COL3A1 | C1257963 | Endogenous Hyperinsulinism | 1 | CTD_human |
| Hgene | COL3A1 | C1257964 | Exogenous Hyperinsulinism | 1 | CTD_human |
| Hgene | COL3A1 | C1257965 | Compensatory Hyperinsulinemia | 1 | CTD_human |
| Hgene | COL3A1 | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human |
| Hgene | COL3A1 | C1623038 | Cirrhosis | 1 | CTD_human |
| Hgene | COL3A1 | C1855520 | Hyperglycemia, Postprandial | 1 | CTD_human |
| Hgene | COL3A1 | C1862932 | ANEURYSM, INTRACRANIAL BERRY, 1 (disorder) | 1 | ORPHANET |
| Hgene | COL3A1 | C2711227 | Steatohepatitis | 1 | CTD_human |
| Hgene | COL3A1 | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human |
| Hgene | COL3A1 | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human |
| Hgene | COL3A1 | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
| Hgene | COL3A1 | C4721507 | Alveolitis, Fibrosing | 1 | CTD_human |
| Tgene | C0268358 | Osteogenesis imperfecta, dominant perinatal lethal | 38 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0268362 | Osteogenesis imperfecta type III (disorder) | 17 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0023931 | Lobstein Disease | 15 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0268363 | Osteogenesis imperfecta type IV (disorder) | 12 | GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0023890 | Liver Cirrhosis | 4 | CTD_human | |
| Tgene | C0239946 | Fibrosis, Liver | 4 | CTD_human | |
| Tgene | C4551623 | EHLERS-DANLOS SYNDROME, ARTHROCHALASIA TYPE, 1 | 4 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C4552122 | EHLERS-DANLOS SYNDROME, CLASSIC TYPE, 1 | 4 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0020497 | Cortical Congenital Hyperostosis | 3 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 3 | CTD_human | |
| Tgene | C0268345 | EHLERS-DANLOS SYNDROME, ARTHROCHALASIA TYPE | 2 | ORPHANET | |
| Tgene | C0000786 | Spontaneous abortion | 1 | CTD_human | |
| Tgene | C0000822 | Abortion, Tubal | 1 | CTD_human | |
| Tgene | C0002949 | Aneurysm, Dissecting | 1 | CTD_human | |
| Tgene | C0003504 | Aortic Valve Insufficiency | 1 | CTD_human | |
| Tgene | C0004364 | Autoimmune Diseases | 1 | CTD_human | |
| Tgene | C0005398 | Cholestasis, Extrahepatic | 1 | CTD_human | |
| Tgene | C0005779 | Blood Coagulation Disorders | 1 | GENOMICS_ENGLAND | |
| Tgene | C0006663 | Calcinosis | 1 | CTD_human | |
| Tgene | C0008311 | Cholangitis | 1 | CTD_human | |
| Tgene | C0013720 | Ehlers-Danlos Syndrome | 1 | GENOMICS_ENGLAND | |
| Tgene | C0016059 | Fibrosis | 1 | CTD_human | |
| Tgene | C0018824 | Heart valve disease | 1 | CTD_human | |
| Tgene | C0020538 | Hypertensive disease | 1 | CTD_human | |
| Tgene | C0022548 | Keloid | 1 | CTD_human | |
| Tgene | C0027719 | Nephrosclerosis | 1 | CTD_human | |
| Tgene | C0027726 | Nephrotic Syndrome | 1 | CTD_human | |
| Tgene | C0029172 | Oral Submucous Fibrosis | 1 | CTD_human | |
| Tgene | C0029434 | Osteogenesis Imperfecta | 1 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C0149721 | Left Ventricular Hypertrophy | 1 | CTD_human | |
| Tgene | C0220679 | Ehlers-Danlos Syndrome, Autosomal Dominant, Type Unspecified | 1 | ORPHANET | |
| Tgene | C0263628 | Tumoral calcinosis | 1 | CTD_human | |
| Tgene | C0340643 | Dissection of aorta | 1 | CTD_human | |
| Tgene | C0521174 | Microcalcification | 1 | CTD_human | |
| Tgene | C1458140 | Bleeding tendency | 1 | GENOMICS_ENGLAND | |
| Tgene | C1619692 | Nephrogenic Fibrosing Dermopathy | 1 | CTD_human | |
| Tgene | C1623038 | Cirrhosis | 1 | CTD_human | |
| Tgene | C1846545 | Autoimmune Lymphoproliferative Syndrome Type 2B | 1 | GENOMICS_ENGLAND | |
| Tgene | C3830362 | Early Pregnancy Loss | 1 | CTD_human | |
| Tgene | C4277533 | Dissection, Blood Vessel | 1 | CTD_human | |
| Tgene | C4552766 | Miscarriage | 1 | CTD_human |
