
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NFATC2-DOK5 (FusionGDB2 ID:HG4773TG55816) |
Fusion Gene Summary for NFATC2-DOK5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NFATC2-DOK5 | Fusion gene ID: hg4773tg55816 | Hgene | Tgene | Gene symbol | NFATC2 | DOK5 | Gene ID | 4773 | 55816 |
| Gene name | nuclear factor of activated T cells 2 | docking protein 5 | |
| Synonyms | NFAT1|NFATP | C20orf180|IRS-6|IRS6 | |
| Cytomap | ('NFATC2')('DOK5') 20q13.2 | 20q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear factor of activated T-cells, cytoplasmic 2NF-ATc2NFAT pre-existing subunitNFAT transcription complex, preexisting componentT cell transcription factor NFAT1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2nuclear fact | docking protein 5downstream of tyrosine kinase 5insulin receptor substrate 6 | |
| Modification date | 20200329 | 20200320 | |
| UniProtAcc | Q13469 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000371564, ENST00000396009, ENST00000414705, ENST00000609507, ENST00000609943, ENST00000610033, | ||
| Fusion gene scores | * DoF score | 9 X 9 X 5=405 | 11 X 9 X 8=792 |
| # samples | 8 | 12 | |
| ** MAII score | log2(8/405*10)=-2.33985000288462 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/792*10)=-2.72246602447109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NFATC2 [Title/Abstract] AND DOK5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NFATC2(50133323)-DOK5(53171472), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. NFATC2-DOK5 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NFATC2 | GO:0016477 | cell migration | 21871017 |
| Hgene | NFATC2 | GO:0045893 | positive regulation of transcription, DNA-templated | 15790681 |
| Hgene | NFATC2 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation | 23853098 |
 Fusion gene breakpoints across NFATC2 (5'-gene) Fusion gene breakpoints across NFATC2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
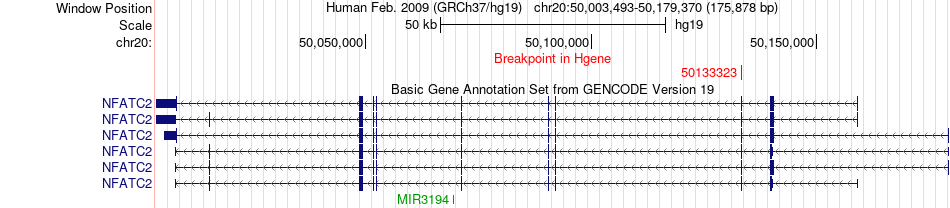 |
 Fusion gene breakpoints across DOK5 (3'-gene) Fusion gene breakpoints across DOK5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
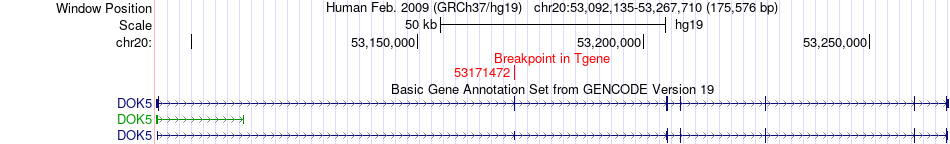 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-EE-A2MH-06A | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
Top |
Fusion Gene ORF analysis for NFATC2-DOK5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000371564 | ENST00000395939 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-5UTR | ENST00000396009 | ENST00000395939 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-5UTR | ENST00000414705 | ENST00000395939 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-5UTR | ENST00000609507 | ENST00000395939 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-5UTR | ENST00000609943 | ENST00000395939 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-5UTR | ENST00000610033 | ENST00000395939 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-intron | ENST00000371564 | ENST00000491469 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-intron | ENST00000396009 | ENST00000491469 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-intron | ENST00000414705 | ENST00000491469 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-intron | ENST00000609507 | ENST00000491469 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-intron | ENST00000609943 | ENST00000491469 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| 5CDS-intron | ENST00000610033 | ENST00000491469 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| Frame-shift | ENST00000371564 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| Frame-shift | ENST00000414705 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| In-frame | ENST00000396009 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| In-frame | ENST00000609507 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| In-frame | ENST00000609943 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
| In-frame | ENST00000610033 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000396009 | NFATC2 | chr20 | 50133323 | - | ENST00000262593 | DOK5 | chr20 | 53171472 | + | 3099 | 1552 | 211 | 2406 | 731 |
| ENST00000609943 | NFATC2 | chr20 | 50133323 | - | ENST00000262593 | DOK5 | chr20 | 53171472 | + | 3021 | 1474 | 202 | 2328 | 708 |
| ENST00000610033 | NFATC2 | chr20 | 50133323 | - | ENST00000262593 | DOK5 | chr20 | 53171472 | + | 2585 | 1038 | 363 | 1892 | 509 |
| ENST00000609507 | NFATC2 | chr20 | 50133323 | - | ENST00000262593 | DOK5 | chr20 | 53171472 | + | 2507 | 960 | 234 | 1814 | 526 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000396009 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + | 0.014642669 | 0.98535734 |
| ENST00000609943 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + | 0.018391458 | 0.98160857 |
| ENST00000610033 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + | 0.033968054 | 0.96603197 |
| ENST00000609507 | ENST00000262593 | NFATC2 | chr20 | 50133323 | - | DOK5 | chr20 | 53171472 | + | 0.027849523 | 0.9721505 |
Top |
Fusion Genomic Features for NFATC2-DOK5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| NFATC2 | chr20 | 50133322 | - | DOK5 | chr20 | 53171471 | + | 0.000343931 | 0.9996561 |
| NFATC2 | chr20 | 50133322 | - | DOK5 | chr20 | 53171471 | + | 0.000343931 | 0.9996561 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
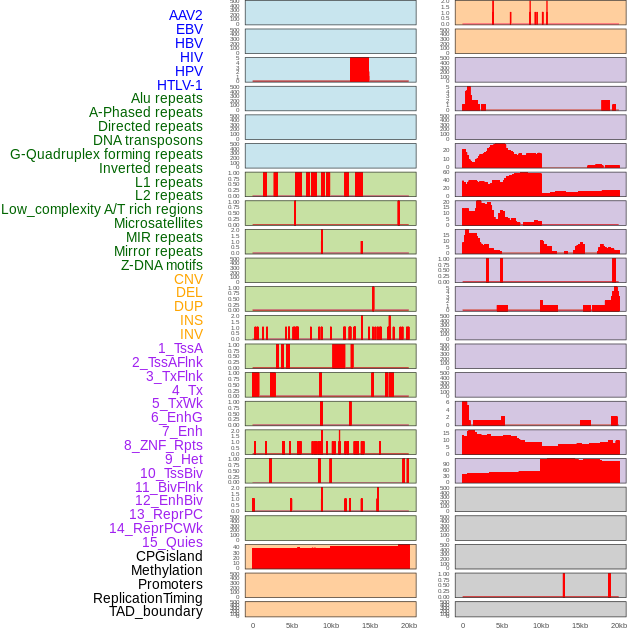 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for NFATC2-DOK5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:50133323/chr20:53171472) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFATC2 | . |
| FUNCTION: Plays a role in the inducible expression of cytokine genes in T-cells, especially in the induction of the IL-2, IL-3, IL-4, TNF-alpha or GM-CSF. Promotes invasive migration through the activation of GPC6 expression and WNT5A signaling pathway. {ECO:0000269|PubMed:15790681, ECO:0000269|PubMed:21871017}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 421_428 | 444 | 1989.6666666666667 | DNA binding | . |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 421_428 | 444 | 926.0 | DNA binding | . |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 251_253 | 444 | 1989.6666666666667 | Motif | Note=Nuclear localization signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 26_34 | 444 | 1989.6666666666667 | Motif | Note=9aaTAD |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 251_253 | 444 | 926.0 | Motif | Note=Nuclear localization signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 26_34 | 444 | 926.0 | Motif | Note=9aaTAD |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 251_253 | 424 | 639.3333333333334 | Motif | Note=Nuclear localization signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 26_34 | 424 | 639.3333333333334 | Motif | Note=9aaTAD |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 111_116 | 444 | 1989.6666666666667 | Region | Calcineurin-binding |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 119_199 | 444 | 1989.6666666666667 | Region | Note=Trans-activation domain A (TAD-A) |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 161_175 | 444 | 1989.6666666666667 | Region | Required for cytoplasmic retention of the phosphorylated form |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 184_286 | 444 | 1989.6666666666667 | Region | Note=3 X approximate SP repeats |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 111_116 | 444 | 926.0 | Region | Calcineurin-binding |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 119_199 | 444 | 926.0 | Region | Note=Trans-activation domain A (TAD-A) |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 161_175 | 444 | 926.0 | Region | Required for cytoplasmic retention of the phosphorylated form |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 184_286 | 444 | 926.0 | Region | Note=3 X approximate SP repeats |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 111_116 | 424 | 639.3333333333334 | Region | Calcineurin-binding |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 119_199 | 424 | 639.3333333333334 | Region | Note=Trans-activation domain A (TAD-A) |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 161_175 | 424 | 639.3333333333334 | Region | Required for cytoplasmic retention of the phosphorylated form |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 184_286 | 424 | 639.3333333333334 | Region | Note=3 X approximate SP repeats |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 184_200 | 444 | 1989.6666666666667 | Repeat | Note=1 |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 213_229 | 444 | 1989.6666666666667 | Repeat | Note=2 |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 272_286 | 444 | 1989.6666666666667 | Repeat | Note=3%3B approximate |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 184_200 | 444 | 926.0 | Repeat | Note=1 |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 213_229 | 444 | 926.0 | Repeat | Note=2 |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 272_286 | 444 | 926.0 | Repeat | Note=3%3B approximate |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 184_200 | 424 | 639.3333333333334 | Repeat | Note=1 |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 213_229 | 424 | 639.3333333333334 | Repeat | Note=2 |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 272_286 | 424 | 639.3333333333334 | Repeat | Note=3%3B approximate |
| Tgene | DOK5 | chr20:50133323 | chr20:53171472 | ENST00000262593 | 0 | 8 | 132_237 | 22 | 307.0 | Domain | IRS-type PTB | |
| Tgene | DOK5 | chr20:50133323 | chr20:53171472 | ENST00000395939 | 0 | 8 | 132_237 | 0 | 199.0 | Domain | IRS-type PTB | |
| Tgene | DOK5 | chr20:50133323 | chr20:53171472 | ENST00000395939 | 0 | 8 | 8_112 | 0 | 199.0 | Domain | Note=PH | |
| Tgene | DOK5 | chr20:50133323 | chr20:53171472 | ENST00000262593 | 0 | 8 | 263_273 | 22 | 307.0 | Motif | Note=DKFBH motif | |
| Tgene | DOK5 | chr20:50133323 | chr20:53171472 | ENST00000395939 | 0 | 8 | 263_273 | 0 | 199.0 | Motif | Note=DKFBH motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 421_428 | 424 | 639.3333333333334 | DNA binding | . |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 392_574 | 444 | 1989.6666666666667 | Domain | RHD |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 392_574 | 444 | 926.0 | Domain | RHD |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 392_574 | 424 | 639.3333333333334 | Domain | RHD |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 664_666 | 444 | 1989.6666666666667 | Motif | Note=Nuclear localization signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000371564 | - | 3 | 11 | 904_913 | 444 | 1989.6666666666667 | Motif | Note=Nuclear export signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 664_666 | 444 | 926.0 | Motif | Note=Nuclear localization signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000396009 | - | 3 | 10 | 904_913 | 444 | 926.0 | Motif | Note=Nuclear export signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 664_666 | 424 | 639.3333333333334 | Motif | Note=Nuclear localization signal |
| Hgene | NFATC2 | chr20:50133323 | chr20:53171472 | ENST00000414705 | - | 3 | 11 | 904_913 | 424 | 639.3333333333334 | Motif | Note=Nuclear export signal |
| Tgene | DOK5 | chr20:50133323 | chr20:53171472 | ENST00000262593 | 0 | 8 | 8_112 | 22 | 307.0 | Domain | Note=PH |
Top |
Fusion Gene Sequence for NFATC2-DOK5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >58789_58789_1_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000396009_DOK5_chr20_53171472_ENST00000262593_length(transcript)=3099nt_BP=1552nt AGCAGGAAGCTCGCGCCGCCGTCGCCGCCGCCGCTCAGCTTCCCCGGGCGCGTCCAGGACCCGCTGCGCCAGGCGCGCCGTCCCCGGACC CGGCGTGCGTCCCTACGAGGAAAGGGACCCCGCCGCTCGAGCCGCCTCCGCCAGCCCCACTGCGAGGGGTCCCAGAGCCAGCCGCGCCCG CCCTCGCCCCCGGCCCCGCAGCCTTCCCGCCCTGCGCGCCATGAACGCCCCCGAGCGGCAGCCCCAACCCGACGGCGGGGACGCCCCAGG CCACGAGCCTGGGGGCAGCCCCCAAGACGAGCTTGACTTCTCCATCCTCTTCGACTATGAGTATTTGAATCCGAACGAAGAAGAGCCGAA TGCACATAAGGTCGCCAGCCCACCCTCCGGACCCGCATACCCCGATGATGTCCTGGACTATGGCCTCAAGCCATACAGCCCCCTTGCTAG TCTCTCTGGCGAGCCCCCCGGCCGATTCGGAGAGCCGGATAGGGTAGGGCCGCAGAAGTTTCTGAGCGCGGCCAAGCCAGCAGGGGCCTC GGGCCTGAGCCCTCGGATCGAGATCACTCCGTCCCACGAACTGATCCAGGCAGTGGGGCCCCTCCGCATGAGAGACGCGGGCCTCCTGGT GGAGCAGCCGCCCCTGGCCGGGGTGGCCGCCAGCCCGAGGTTCACCCTGCCCGTGCCCGGCTTCGAGGGCTACCGCGAGCCGCTTTGCTT GAGCCCCGCTAGCAGCGGCTCCTCTGCCAGCTTCATTTCTGACACCTTCTCCCCCTACACCTCGCCCTGCGTCTCGCCCAATAACGGCGG GCCCGACGACCTGTGTCCGCAGTTTCAAAACATCCCTGCTCATTATTCCCCCAGAACCTCGCCAATAATGTCACCTCGAACCAGCCTCGC CGAGGACAGCTGCCTGGGCCGCCACTCGCCCGTGCCCCGTCCGGCCTCCCGCTCCTCATCGCCTGGTGCCAAGCGGAGGCATTCGTGCGC CGAGGCCTTGGTTGCCCTGCCGCCCGGAGCCTCACCCCAGCGCTCCCGGAGCCCCTCGCCGCAGCCCTCATCTCACGTGGCACCCCAGGA CCACGGCTCCCCGGCTGGGTACCCCCCTGTGGCTGGCTCTGCCGTGATCATGGATGCCCTGAACAGCCTCGCCACGGACTCGCCTTGTGG GATCCCCCCCAAGATGTGGAAGACCAGCCCTGACCCCTCGCCGGTGTCTGCCGCCCCATCCAAGGCCGGCCTGCCTCGCCACATCTACCC GGCCGTGGAGTTCCTGGGGCCCTGCGAGCAGGGCGAGAGGAGAAACTCGGCTCCAGAATCCATCCTGCTGGTTCCGCCCACTTGGCCCAA GCCGCTGGTGCCTGCCATTCCCATCTGCAGCATCCCAGTGACTGCATCCCTCCCTCCACTTGAGTGGCCGCTGTCCAGTCAGTCAGGCTC TTACGAGCTGCGGATCGAGGTGCAGCCCAAGCCACATCACCGGGCCCACTATGAGACAGAAGGCAGCCGAGGGGCTGTCAAAGCTCCAAC TGGAGGCCACCCTGTGGTTCAGATTTATCAGCGATGCTGGTTAGTATTCAAGAAAGCTTCAAGCAAAGGTCCAAAAAGACTGGAGAAATT TTCTGATGAACGTGCTGCATATTTCAGGTGTTATCATAAGGTTACAGAACTCAATAATGTGAAGAACGTAGCTCGATTGCCAAAAAGCAC CAAGAAACATGCCATAGGGATTTATTTCAATGACGATACCTCCAAGACTTTTGCTTGCGAATCAGATCTTGAGGCTGATGAGTGGTGCAA AGTACTCCAGATGGAGTGTGTAGGAACACGGATCAATGACATCAGCCTTGGAGAGCCTGACTTACTGGCCACTGGGGTTGAGAGAGAACA GAGTGAGAGATTCAATGTGTATTTGATGCCATCTCCTAACTTAGATGTACATGGCGAATGTGCCTTGCAGATTACATATGAGTATATCTG TCTTTGGGACGTCCAGAATCCCAGAGTCAAACTCATCTCTTGGCCGCTAAGCGCCCTGCGGCGGTATGGACGTGATACTACGTGGTTCAC TTTTGAGGCAGGGAGGATGTGTGAGACTGGTGAAGGGCTGTTTATCTTTCAGACCCGAGACGGGGAGGCCATCTATCAGAAAGTCCACTC TGCTGCCTTGGCCATAGCCGAGCAGCACGAGCGCTTGCTACAGAGTGTGAAAAACTCGATGCTCCAGATGAAGATGAGTGAGCGGGCCGC CTCGCTGAGCACCATGGTGCCCCTGCCTCGCAGCGCCTACTGGCAGCACATCACACGGCAGCACAGCACGGGACAGCTCTACCGCTTGCA AGATGTTTCCAGCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAA CACACTTGTGATGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTC CTGGCTAATTGTGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAA CGTGCTCTATAGATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTCTATTGTAGATAC TTTGTCCCTTGCACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCAGTTGTATGACAGTCAGTATTGA CAATAAAATGGCTTTTAATTATTTGTTATTTGTTTACACCCTATTCCTCAGTTATTATTACTGTGGTTCTGATTAACTACTGGAAATTAT ATTTGATTATATCACCAATTAGTTAAATCAGTGCTTCGACTCACTCTTATCTGTTCTGTTCAAAACTATTTGTTCAAAGAACCCGTTAGT GTTGTTTACAGGGTTACAGTTTCTCTCACATGCTTTCCTCACCCCTTTACCCCCCTTTTTGAAAGCCTTTATTTTGTTCGGAGTCTCTTC >58789_58789_1_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000396009_DOK5_chr20_53171472_ENST00000262593_length(amino acids)=731AA_BP=446 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQIYQ RCWLVFKKASSKGPKRLEKFSDERAAYFRCYHKVTELNNVKNVARLPKSTKKHAIGIYFNDDTSKTFACESDLEADEWCKVLQMECVGTR INDISLGEPDLLATGVEREQSERFNVYLMPSPNLDVHGECALQITYEYICLWDVQNPRVKLISWPLSALRRYGRDTTWFTFEAGRMCETG EGLFIFQTRDGEAIYQKVHSAALAIAEQHERLLQSVKNSMLQMKMSERAASLSTMVPLPRSAYWQHITRQHSTGQLYRLQDVSSPLKLHR -------------------------------------------------------------- >58789_58789_2_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000609507_DOK5_chr20_53171472_ENST00000262593_length(transcript)=2507nt_BP=960nt TCTGGAGTAAGCCGGATCGCGGAGCCGCGCCGACTCCGCCGAGCCGGGAGCCGGGAGGCGCGCAGCTCCCGGGTCGCTCCGAGGCTCCTC GGCCAGGGCAGCCCCGCGGGCACGCGGTAGAGAAGACGGCGTCCCCTCGGCTGCTGGTCGATACAAACAGATCCCCCTTTCCAAACACGC GCCAAGTCCCCGTGCCCTCCAGATGCAGAGAGAGGCTGCGTTCAGACTGGGGCACTGCCATCCCCTCCGCATCATGGGGTCTGTGGACCA AGAACCTCGCCAATAATGTCACCTCGAACCAGCCTCGCCGAGGACAGCTGCCTGGGCCGCCACTCGCCCGTGCCCCGTCCGGCCTCCCGC TCCTCATCGCCTGGTGCCAAGCGGAGGCATTCGTGCGCCGAGGCCTTGGTTGCCCTGCCGCCCGGAGCCTCACCCCAGCGCTCCCGGAGC CCCTCGCCGCAGCCCTCATCTCACGTGGCACCCCAGGACCACGGCTCCCCGGCTGGGTACCCCCCTGTGGCTGGCTCTGCCGTGATCATG GATGCCCTGAACAGCCTCGCCACGGACTCGCCTTGTGGGATCCCCCCCAAGATGTGGAAGACCAGCCCTGACCCCTCGCCGGTGTCTGCC GCCCCATCCAAGGCCGGCCTGCCTCGCCACATCTACCCGGCCGTGGAGTTCCTGGGGCCCTGCGAGCAGGGCGAGAGGAGAAACTCGGCT CCAGAATCCATCCTGCTGGTTCCGCCCACTTGGCCCAAGCCGCTGGTGCCTGCCATTCCCATCTGCAGCATCCCAGTGACTGCATCCCTC CCTCCACTTGAGTGGCCGCTGTCCAGTCAGTCAGGCTCTTACGAGCTGCGGATCGAGGTGCAGCCCAAGCCACATCACCGGGCCCACTAT GAGACAGAAGGCAGCCGAGGGGCTGTCAAAGCTCCAACTGGAGGCCACCCTGTGGTTCAGATTTATCAGCGATGCTGGTTAGTATTCAAG AAAGCTTCAAGCAAAGGTCCAAAAAGACTGGAGAAATTTTCTGATGAACGTGCTGCATATTTCAGGTGTTATCATAAGGTTACAGAACTC AATAATGTGAAGAACGTAGCTCGATTGCCAAAAAGCACCAAGAAACATGCCATAGGGATTTATTTCAATGACGATACCTCCAAGACTTTT GCTTGCGAATCAGATCTTGAGGCTGATGAGTGGTGCAAAGTACTCCAGATGGAGTGTGTAGGAACACGGATCAATGACATCAGCCTTGGA GAGCCTGACTTACTGGCCACTGGGGTTGAGAGAGAACAGAGTGAGAGATTCAATGTGTATTTGATGCCATCTCCTAACTTAGATGTACAT GGCGAATGTGCCTTGCAGATTACATATGAGTATATCTGTCTTTGGGACGTCCAGAATCCCAGAGTCAAACTCATCTCTTGGCCGCTAAGC GCCCTGCGGCGGTATGGACGTGATACTACGTGGTTCACTTTTGAGGCAGGGAGGATGTGTGAGACTGGTGAAGGGCTGTTTATCTTTCAG ACCCGAGACGGGGAGGCCATCTATCAGAAAGTCCACTCTGCTGCCTTGGCCATAGCCGAGCAGCACGAGCGCTTGCTACAGAGTGTGAAA AACTCGATGCTCCAGATGAAGATGAGTGAGCGGGCCGCCTCGCTGAGCACCATGGTGCCCCTGCCTCGCAGCGCCTACTGGCAGCACATC ACACGGCAGCACAGCACGGGACAGCTCTACCGCTTGCAAGATGTTTCCAGCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTAC AGATCTGAGCACTGACAGTAACTGCCAAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACA GCAAGGGAAATGACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTA TTTTCTTTTTCTTTTTTAAATTCTTAGTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAA GAAGTCAATGTCACGAAATGATTTTCTATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTT CCTTCCAAGTGTTTCAGTTGTATGACAGTCAGTATTGACAATAAAATGGCTTTTAATTATTTGTTATTTGTTTACACCCTATTCCTCAGT TATTATTACTGTGGTTCTGATTAACTACTGGAAATTATATTTGATTATATCACCAATTAGTTAAATCAGTGCTTCGACTCACTCTTATCT GTTCTGTTCAAAACTATTTGTTCAAAGAACCCGTTAGTGTTGTTTACAGGGTTACAGTTTCTCTCACATGCTTTCCTCACCCCTTTACCC >58789_58789_2_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000609507_DOK5_chr20_53171472_ENST00000262593_length(amino acids)=526AA_BP=2 MPSPPHHGVCGPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPA GYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPA IPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQIYQRCWLVFKKASSKGPKRLEKFSDERA AYFRCYHKVTELNNVKNVARLPKSTKKHAIGIYFNDDTSKTFACESDLEADEWCKVLQMECVGTRINDISLGEPDLLATGVEREQSERFN VYLMPSPNLDVHGECALQITYEYICLWDVQNPRVKLISWPLSALRRYGRDTTWFTFEAGRMCETGEGLFIFQTRDGEAIYQKVHSAALAI -------------------------------------------------------------- >58789_58789_3_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000609943_DOK5_chr20_53171472_ENST00000262593_length(transcript)=3021nt_BP=1474nt TCTGGAGTAAGCCGGATCGCGGAGCCGCGCCGACTCCGCCGAGCCGGGAGCCGGGAGGCGCGCAGCTCCCGGGTCGCTCCGAGGCTCCTC GGCCAGGGCAGCCCCGCGGGCACGCGGTAGAGAAGACGGCGTCCCCTCGGCTGCTGGTCGATACAAACAGATCCCCCTTTCCAAACACGC GCCAAGTCCCCGTGCCCTCCAGATGCAGAGAGAGGCTGCGTTCAGACTGGGGCACTGCCATCCCCTCCGCATCATGGGGTCTGTGGACCA AGAAGAGCCGAATGCACATAAGGTCGCCAGCCCACCCTCCGGACCCGCATACCCCGATGATGTCCTGGACTATGGCCTCAAGCCATACAG CCCCCTTGCTAGTCTCTCTGGCGAGCCCCCCGGCCGATTCGGAGAGCCGGATAGGGTAGGGCCGCAGAAGTTTCTGAGCGCGGCCAAGCC AGCAGGGGCCTCGGGCCTGAGCCCTCGGATCGAGATCACTCCGTCCCACGAACTGATCCAGGCAGTGGGGCCCCTCCGCATGAGAGACGC GGGCCTCCTGGTGGAGCAGCCGCCCCTGGCCGGGGTGGCCGCCAGCCCGAGGTTCACCCTGCCCGTGCCCGGCTTCGAGGGCTACCGCGA GCCGCTTTGCTTGAGCCCCGCTAGCAGCGGCTCCTCTGCCAGCTTCATTTCTGACACCTTCTCCCCCTACACCTCGCCCTGCGTCTCGCC CAATAACGGCGGGCCCGACGACCTGTGTCCGCAGTTTCAAAACATCCCTGCTCATTATTCCCCCAGAACCTCGCCAATAATGTCACCTCG AACCAGCCTCGCCGAGGACAGCTGCCTGGGCCGCCACTCGCCCGTGCCCCGTCCGGCCTCCCGCTCCTCATCGCCTGGTGCCAAGCGGAG GCATTCGTGCGCCGAGGCCTTGGTTGCCCTGCCGCCCGGAGCCTCACCCCAGCGCTCCCGGAGCCCCTCGCCGCAGCCCTCATCTCACGT GGCACCCCAGGACCACGGCTCCCCGGCTGGGTACCCCCCTGTGGCTGGCTCTGCCGTGATCATGGATGCCCTGAACAGCCTCGCCACGGA CTCGCCTTGTGGGATCCCCCCCAAGATGTGGAAGACCAGCCCTGACCCCTCGCCGGTGTCTGCCGCCCCATCCAAGGCCGGCCTGCCTCG CCACATCTACCCGGCCGTGGAGTTCCTGGGGCCCTGCGAGCAGGGCGAGAGGAGAAACTCGGCTCCAGAATCCATCCTGCTGGTTCCGCC CACTTGGCCCAAGCCGCTGGTGCCTGCCATTCCCATCTGCAGCATCCCAGTGACTGCATCCCTCCCTCCACTTGAGTGGCCGCTGTCCAG TCAGTCAGGCTCTTACGAGCTGCGGATCGAGGTGCAGCCCAAGCCACATCACCGGGCCCACTATGAGACAGAAGGCAGCCGAGGGGCTGT CAAAGCTCCAACTGGAGGCCACCCTGTGGTTCAGATTTATCAGCGATGCTGGTTAGTATTCAAGAAAGCTTCAAGCAAAGGTCCAAAAAG ACTGGAGAAATTTTCTGATGAACGTGCTGCATATTTCAGGTGTTATCATAAGGTTACAGAACTCAATAATGTGAAGAACGTAGCTCGATT GCCAAAAAGCACCAAGAAACATGCCATAGGGATTTATTTCAATGACGATACCTCCAAGACTTTTGCTTGCGAATCAGATCTTGAGGCTGA TGAGTGGTGCAAAGTACTCCAGATGGAGTGTGTAGGAACACGGATCAATGACATCAGCCTTGGAGAGCCTGACTTACTGGCCACTGGGGT TGAGAGAGAACAGAGTGAGAGATTCAATGTGTATTTGATGCCATCTCCTAACTTAGATGTACATGGCGAATGTGCCTTGCAGATTACATA TGAGTATATCTGTCTTTGGGACGTCCAGAATCCCAGAGTCAAACTCATCTCTTGGCCGCTAAGCGCCCTGCGGCGGTATGGACGTGATAC TACGTGGTTCACTTTTGAGGCAGGGAGGATGTGTGAGACTGGTGAAGGGCTGTTTATCTTTCAGACCCGAGACGGGGAGGCCATCTATCA GAAAGTCCACTCTGCTGCCTTGGCCATAGCCGAGCAGCACGAGCGCTTGCTACAGAGTGTGAAAAACTCGATGCTCCAGATGAAGATGAG TGAGCGGGCCGCCTCGCTGAGCACCATGGTGCCCCTGCCTCGCAGCGCCTACTGGCAGCACATCACACGGCAGCACAGCACGGGACAGCT CTACCGCTTGCAAGATGTTTCCAGCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCACTGACAGTAACTGCC AAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATGACGACCAAGAGAAG AAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCTTTTTTAAATTCTTA GTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTCACGAAATGATTTTC TATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGTTTCAGTTGTATGAC AGTCAGTATTGACAATAAAATGGCTTTTAATTATTTGTTATTTGTTTACACCCTATTCCTCAGTTATTATTACTGTGGTTCTGATTAACT ACTGGAAATTATATTTGATTATATCACCAATTAGTTAAATCAGTGCTTCGACTCACTCTTATCTGTTCTGTTCAAAACTATTTGTTCAAA GAACCCGTTAGTGTTGTTTACAGGGTTACAGTTTCTCTCACATGCTTTCCTCACCCCTTTACCCCCCTTTTTGAAAGCCTTTATTTTGTT >58789_58789_3_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000609943_DOK5_chr20_53171472_ENST00000262593_length(amino acids)=708AA_BP=423 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQIYQRCWLVFKKASSKGPKRLEKFSDE RAAYFRCYHKVTELNNVKNVARLPKSTKKHAIGIYFNDDTSKTFACESDLEADEWCKVLQMECVGTRINDISLGEPDLLATGVEREQSER FNVYLMPSPNLDVHGECALQITYEYICLWDVQNPRVKLISWPLSALRRYGRDTTWFTFEAGRMCETGEGLFIFQTRDGEAIYQKVHSAAL -------------------------------------------------------------- >58789_58789_4_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000610033_DOK5_chr20_53171472_ENST00000262593_length(transcript)=2585nt_BP=1038nt AGCAGGAAGCTCGCGCCGCCGTCGCCGCCGCCGCTCAGCTTCCCCGGGCGCGTCCAGGACCCGCTGCGCCAGGCGCGCCGTCCCCGGACC CGGCGTGCGTCCCTACGAGGAAAGGGACCCCGCCGCTCGAGCCGCCTCCGCCAGCCCCACTGCGAGGGGTCCCAGAGCCAGCCGCGCCCG CCCTCGCCCCCGGCCCCGCAGCCTTCCCGCCCTGCGCGCCATGAACGCCCCCGAGCGGCAGCCCCAACCCGACGGCGGGGACGCCCCAGG CCACGAGCCTGGGGGCAGCCCCCAAGACGAGCTTGACTTCTCCATCCTCTTCGACTATGAGTATTTGAATCCGAACGAAGAACCTCGCCA ATAATGTCACCTCGAACCAGCCTCGCCGAGGACAGCTGCCTGGGCCGCCACTCGCCCGTGCCCCGTCCGGCCTCCCGCTCCTCATCGCCT GGTGCCAAGCGGAGGCATTCGTGCGCCGAGGCCTTGGTTGCCCTGCCGCCCGGAGCCTCACCCCAGCGCTCCCGGAGCCCCTCGCCGCAG CCCTCATCTCACGTGGCACCCCAGGACCACGGCTCCCCGGCTGGGTACCCCCCTGTGGCTGGCTCTGCCGTGATCATGGATGCCCTGAAC AGCCTCGCCACGGACTCGCCTTGTGGGATCCCCCCCAAGATGTGGAAGACCAGCCCTGACCCCTCGCCGGTGTCTGCCGCCCCATCCAAG GCCGGCCTGCCTCGCCACATCTACCCGGCCGTGGAGTTCCTGGGGCCCTGCGAGCAGGGCGAGAGGAGAAACTCGGCTCCAGAATCCATC CTGCTGGTTCCGCCCACTTGGCCCAAGCCGCTGGTGCCTGCCATTCCCATCTGCAGCATCCCAGTGACTGCATCCCTCCCTCCACTTGAG TGGCCGCTGTCCAGTCAGTCAGGCTCTTACGAGCTGCGGATCGAGGTGCAGCCCAAGCCACATCACCGGGCCCACTATGAGACAGAAGGC AGCCGAGGGGCTGTCAAAGCTCCAACTGGAGGCCACCCTGTGGTTCAGATTTATCAGCGATGCTGGTTAGTATTCAAGAAAGCTTCAAGC AAAGGTCCAAAAAGACTGGAGAAATTTTCTGATGAACGTGCTGCATATTTCAGGTGTTATCATAAGGTTACAGAACTCAATAATGTGAAG AACGTAGCTCGATTGCCAAAAAGCACCAAGAAACATGCCATAGGGATTTATTTCAATGACGATACCTCCAAGACTTTTGCTTGCGAATCA GATCTTGAGGCTGATGAGTGGTGCAAAGTACTCCAGATGGAGTGTGTAGGAACACGGATCAATGACATCAGCCTTGGAGAGCCTGACTTA CTGGCCACTGGGGTTGAGAGAGAACAGAGTGAGAGATTCAATGTGTATTTGATGCCATCTCCTAACTTAGATGTACATGGCGAATGTGCC TTGCAGATTACATATGAGTATATCTGTCTTTGGGACGTCCAGAATCCCAGAGTCAAACTCATCTCTTGGCCGCTAAGCGCCCTGCGGCGG TATGGACGTGATACTACGTGGTTCACTTTTGAGGCAGGGAGGATGTGTGAGACTGGTGAAGGGCTGTTTATCTTTCAGACCCGAGACGGG GAGGCCATCTATCAGAAAGTCCACTCTGCTGCCTTGGCCATAGCCGAGCAGCACGAGCGCTTGCTACAGAGTGTGAAAAACTCGATGCTC CAGATGAAGATGAGTGAGCGGGCCGCCTCGCTGAGCACCATGGTGCCCCTGCCTCGCAGCGCCTACTGGCAGCACATCACACGGCAGCAC AGCACGGGACAGCTCTACCGCTTGCAAGATGTTTCCAGCCCTCTGAAGCTTCATCGAACAGAGACTTTTCCAGCCTACAGATCTGAGCAC TGACAGTAACTGCCAAGAATTGTTAACACACTTGTGATGTGTCAGCCACAGATTCACCCAGGAGGTCACAGAATGACAGCAAGGGAAATG ACGACCAAGAGAAGAAGCTTAAAGTCCTGGCTAATTGTGTGGTCATTGGAAAACTCTGCAATACAATAATTTTCTTTATTTTCTTTTTCT TTTTTAAATTCTTAGTGTAATTGAAACGTGCTCTATAGATATTGACTCTGTGTTCCCTCTTTTACAGCTGGACAGAAAGAAGTCAATGTC ACGAAATGATTTTCTATTGTAGATACTTTGTCCCTTGCACTTCTCTGAATCTGTCCTTTTGTGGATTCTTGTGATTTTCCTTCCAAGTGT TTCAGTTGTATGACAGTCAGTATTGACAATAAAATGGCTTTTAATTATTTGTTATTTGTTTACACCCTATTCCTCAGTTATTATTACTGT GGTTCTGATTAACTACTGGAAATTATATTTGATTATATCACCAATTAGTTAAATCAGTGCTTCGACTCACTCTTATCTGTTCTGTTCAAA ACTATTTGTTCAAAGAACCCGTTAGTGTTGTTTACAGGGTTACAGTTTCTCTCACATGCTTTCCTCACCCCTTTACCCCCCTTTTTGAAA >58789_58789_4_NFATC2-DOK5_NFATC2_chr20_50133323_ENST00000610033_DOK5_chr20_53171472_ENST00000262593_length(amino acids)=509AA_BP=224 MSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNS LATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEW PLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQIYQRCWLVFKKASSKGPKRLEKFSDERAAYFRCYHKVTELNNVKN VARLPKSTKKHAIGIYFNDDTSKTFACESDLEADEWCKVLQMECVGTRINDISLGEPDLLATGVEREQSERFNVYLMPSPNLDVHGECAL QITYEYICLWDVQNPRVKLISWPLSALRRYGRDTTWFTFEAGRMCETGEGLFIFQTRDGEAIYQKVHSAALAIAEQHERLLQSVKNSMLQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NFATC2-DOK5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NFATC2-DOK5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NFATC2-DOK5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | NFATC2 | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Hgene | NFATC2 | C0020542 | Pulmonary Hypertension | 1 | CTD_human |
| Hgene | NFATC2 | C0027765 | nervous system disorder | 1 | CTD_human |
| Tgene | C0005586 | Bipolar Disorder | 1 | PSYGENET |
