
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ASXL2-HADHA (FusionGDB2 ID:HG55252TG3030) |
Fusion Gene Summary for ASXL2-HADHA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ASXL2-HADHA | Fusion gene ID: hg55252tg3030 | Hgene | Tgene | Gene symbol | ASXL2 | HADHA | Gene ID | 55252 | 3030 |
| Gene name | ASXL transcriptional regulator 2 | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha | |
| Synonyms | ASXH2|SHAPNS | ECHA|GBP|HADH|LCEH|LCHAD|MTPA|TP-ALPHA | |
| Cytomap | ('ASXL2')('HADHA') 2p23.3 | 2p23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | putative Polycomb group protein ASXL2additional sex combs like 2, transcriptional regulatoradditional sex combs-like protein 2polycomb group protein ASXH2 | trifunctional enzyme subunit alpha, mitochondrial3-ketoacyl-Coenzyme A (CoA) thiolase, alpha subunit3-oxoacyl-CoA thiolase78 kDa gastrin-binding proteingastrin-binding proteinhydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase ( | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q76L83 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000336112, ENST00000435504, ENST00000272341, ENST00000497092, ENST00000404843, | ||
| Fusion gene scores | * DoF score | 18 X 11 X 10=1980 | 7 X 6 X 7=294 |
| # samples | 24 | 9 | |
| ** MAII score | log2(24/1980*10)=-3.04439411935845 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/294*10)=-1.70781924850669 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ASXL2 [Title/Abstract] AND HADHA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ASXL2(26101035)-HADHA(26432758), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. ASXL2-HADHA seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ASXL2-HADHA seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ASXL2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 21047783 |
| Hgene | ASXL2 | GO:0045600 | positive regulation of fat cell differentiation | 21047783 |
| Hgene | ASXL2 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 21047783 |
| Tgene | HADHA | GO:0035965 | cardiolipin acyl-chain remodeling | 23152787 |
 Fusion gene breakpoints across ASXL2 (5'-gene) Fusion gene breakpoints across ASXL2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
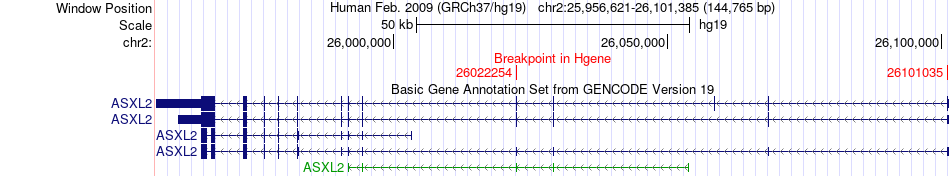 |
 Fusion gene breakpoints across HADHA (3'-gene) Fusion gene breakpoints across HADHA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
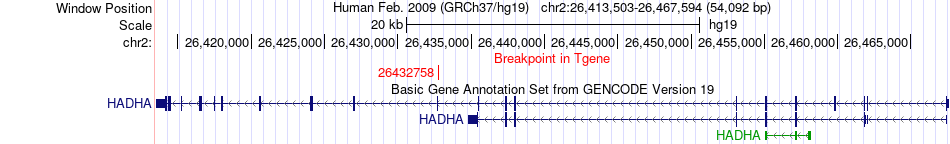 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-L5-A43M-01A | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| ChimerDB4 | ESCA | TCGA-L5-A43M | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| ChimerDB4 | UCEC | TCGA-AX-A3FW-01A | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
Top |
Fusion Gene ORF analysis for ASXL2-HADHA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000336112 | ENST00000457468 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5CDS-intron | ENST00000336112 | ENST00000461025 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5CDS-intron | ENST00000435504 | ENST00000457468 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5CDS-intron | ENST00000435504 | ENST00000457468 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5CDS-intron | ENST00000435504 | ENST00000461025 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5CDS-intron | ENST00000435504 | ENST00000461025 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-3CDS | ENST00000272341 | ENST00000380649 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-3CDS | ENST00000272341 | ENST00000380649 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-3CDS | ENST00000336112 | ENST00000380649 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-3CDS | ENST00000497092 | ENST00000380649 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000272341 | ENST00000457468 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000272341 | ENST00000457468 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000272341 | ENST00000461025 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000272341 | ENST00000461025 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000336112 | ENST00000457468 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000336112 | ENST00000461025 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000497092 | ENST00000457468 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| 5UTR-intron | ENST00000497092 | ENST00000461025 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| Frame-shift | ENST00000435504 | ENST00000380649 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| In-frame | ENST00000336112 | ENST00000380649 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| In-frame | ENST00000435504 | ENST00000380649 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| intron-3CDS | ENST00000404843 | ENST00000380649 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| intron-3CDS | ENST00000404843 | ENST00000380649 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| intron-3CDS | ENST00000497092 | ENST00000380649 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| intron-intron | ENST00000404843 | ENST00000457468 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| intron-intron | ENST00000404843 | ENST00000457468 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| intron-intron | ENST00000404843 | ENST00000461025 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| intron-intron | ENST00000404843 | ENST00000461025 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - |
| intron-intron | ENST00000497092 | ENST00000457468 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
| intron-intron | ENST00000497092 | ENST00000461025 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000435504 | ASXL2 | chr2 | 26101035 | - | ENST00000380649 | HADHA | chr2 | 26432758 | - | 2283 | 351 | 153 | 1667 | 504 |
| ENST00000336112 | ASXL2 | chr2 | 26022254 | - | ENST00000380649 | HADHA | chr2 | 26432758 | - | 2579 | 647 | 551 | 1963 | 470 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000435504 | ENST00000380649 | ASXL2 | chr2 | 26101035 | - | HADHA | chr2 | 26432758 | - | 0.001720573 | 0.99827945 |
| ENST00000336112 | ENST00000380649 | ASXL2 | chr2 | 26022254 | - | HADHA | chr2 | 26432758 | - | 0.001844894 | 0.99815506 |
Top |
Fusion Genomic Features for ASXL2-HADHA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
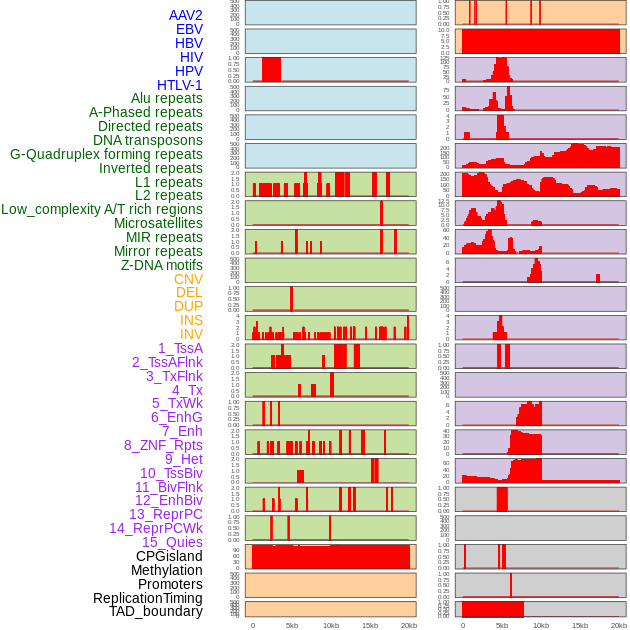 |
Top |
Fusion Protein Features for ASXL2-HADHA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:26101035/chr2:26432758) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ASXL2 | . |
| FUNCTION: Putative Polycomb group (PcG) protein. PcG proteins act by forming multiprotein complexes, which are required to maintain the transcriptionally repressive state of homeotic genes throughout development. PcG proteins are not required to initiate repression, but to maintain it during later stages of development. They probably act via methylation of histones, rendering chromatin heritably changed in its expressibility (By similarity). Involved in transcriptional regulation mediated by ligand-bound nuclear hormone receptors, such as peroxisome proliferator-activated receptor gamma (PPARG). Acts as coactivator for PPARG and enhances its adipocyte differentiation-inducing activity; the function seems to involve differential recruitment of acetylated and methylated histone H3. {ECO:0000250, ECO:0000269|PubMed:21047783}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 11_86 | 134 | 1436.0 | Domain | HTH HARE-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 1173_1176 | 0 | 919.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 303_307 | 0 | 919.0 | Compositional bias | Note=Poly-Leu |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 654_684 | 0 | 919.0 | Compositional bias | Note=Ala-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 687_739 | 0 | 919.0 | Compositional bias | Note=Gly-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 95_235 | 0 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 1173_1176 | 0 | 919.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 303_307 | 0 | 919.0 | Compositional bias | Note=Poly-Leu |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 654_684 | 0 | 919.0 | Compositional bias | Note=Ala-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 687_739 | 0 | 919.0 | Compositional bias | Note=Gly-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 95_235 | 0 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 1173_1176 | 134 | 1436.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 303_307 | 134 | 1436.0 | Compositional bias | Note=Poly-Leu |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 654_684 | 134 | 1436.0 | Compositional bias | Note=Ala-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 687_739 | 134 | 1436.0 | Compositional bias | Note=Gly-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 95_235 | 134 | 1436.0 | Compositional bias | Note=Ser-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 1173_1176 | 0 | 919.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 303_307 | 0 | 919.0 | Compositional bias | Note=Poly-Leu |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 654_684 | 0 | 919.0 | Compositional bias | Note=Ala-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 687_739 | 0 | 919.0 | Compositional bias | Note=Gly-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 95_235 | 0 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 1173_1176 | 0 | 919.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 303_307 | 0 | 919.0 | Compositional bias | Note=Poly-Leu |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 654_684 | 0 | 919.0 | Compositional bias | Note=Ala-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 687_739 | 0 | 919.0 | Compositional bias | Note=Gly-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 95_235 | 0 | 919.0 | Compositional bias | Note=Ser-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 1173_1176 | 19 | 1436.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 303_307 | 19 | 1436.0 | Compositional bias | Note=Poly-Leu |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 654_684 | 19 | 1436.0 | Compositional bias | Note=Ala-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 687_739 | 19 | 1436.0 | Compositional bias | Note=Gly-rich |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 95_235 | 19 | 1436.0 | Compositional bias | Note=Ser-rich |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 11_86 | 0 | 919.0 | Domain | HTH HARE-type |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 274_383 | 0 | 919.0 | Domain | DEUBAD |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 11_86 | 0 | 919.0 | Domain | HTH HARE-type |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 274_383 | 0 | 919.0 | Domain | DEUBAD |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 274_383 | 134 | 1436.0 | Domain | DEUBAD |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 11_86 | 0 | 919.0 | Domain | HTH HARE-type |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 274_383 | 0 | 919.0 | Domain | DEUBAD |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 11_86 | 0 | 919.0 | Domain | HTH HARE-type |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 274_383 | 0 | 919.0 | Domain | DEUBAD |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 11_86 | 19 | 1436.0 | Domain | HTH HARE-type |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 274_383 | 19 | 1436.0 | Domain | DEUBAD |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 174_178 | 0 | 919.0 | Motif | Nuclear localization signal |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 887_891 | 0 | 919.0 | Motif | Note=LXXLL motif 2 |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 174_178 | 0 | 919.0 | Motif | Nuclear localization signal |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 887_891 | 0 | 919.0 | Motif | Note=LXXLL motif 2 |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 174_178 | 134 | 1436.0 | Motif | Nuclear localization signal |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 887_891 | 134 | 1436.0 | Motif | Note=LXXLL motif 2 |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 174_178 | 0 | 919.0 | Motif | Nuclear localization signal |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 887_891 | 0 | 919.0 | Motif | Note=LXXLL motif 2 |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 174_178 | 0 | 919.0 | Motif | Nuclear localization signal |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 887_891 | 0 | 919.0 | Motif | Note=LXXLL motif 2 |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 174_178 | 19 | 1436.0 | Motif | Nuclear localization signal |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 887_891 | 19 | 1436.0 | Motif | Note=LXXLL motif 2 |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000272341 | - | 4 | 13 | 1397_1434 | 0 | 919.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 1397_1434 | 0 | 919.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASXL2 | chr2:26022254 | chr2:26432758 | ENST00000435504 | - | 5 | 13 | 1397_1434 | 134 | 1436.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000272341 | - | 1 | 13 | 1397_1434 | 0 | 919.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000404843 | - | 1 | 10 | 1397_1434 | 0 | 919.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASXL2 | chr2:26101035 | chr2:26432758 | ENST00000435504 | - | 1 | 13 | 1397_1434 | 19 | 1436.0 | Zinc finger | Note=PHD-type%3B atypical |
Top |
Fusion Gene Sequence for ASXL2-HADHA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >7307_7307_1_ASXL2-HADHA_ASXL2_chr2_26022254_ENST00000336112_HADHA_chr2_26432758_ENST00000380649_length(transcript)=2579nt_BP=647nt GTACAGTAAGAACCCAATATGGCAGCGGCGGTAGCAGTGGCAACGGCAGCAAGAGGACCGGCCGCCCCATAGACCCCCCCGCGCACCACC CCCGCCGCTTCCTCTGCTTGGGTGCCCCCCCGGTCTGACTTCCCTTTTCTCCCACCTTTCCCGGCACCGCGGGAAAAGCAGCGCAGGGCA GAGCAGGCAGGGAGGGCGGCCGGAGCCCGGACACGGGAGCCTCCCAGTCAGTTCAAGACCCGACATGAGGGAAAAGGGACGTAGGAAGAA GGGCAGGACCTGGGCGGAGGCCGCCAAGACGGTCTTAGAAAAATACCCCAATACACCCATGAGTCATAAAGAAATTCTTCAAGTTATCCA GAGAGAAGGACTAAAAGAAATCAGCAGTGGGACTTCTCCTCTTGCATGCCTGAATGCAATGCTTCACACAAACTCCAGAGGTGAAGAGGG CATCTTCTATAAGGTTCCAGGTAGAATGGGAGTATATACTTTGAAGAAAGATGTGCCGGATGGGGTGAAAGAGCTGTCAGAAGGTTCAGA AGAAAGCAGTGATGGTCAGTCAGATTCCCAGAGTTCTGAGAACAGCAGCAGCAGCAGTGATGGTGGCAGCAACAAGGAGGGAAAAAAGAG CAGGTGGAAAAGGAAAGAAATTTGGAGAGCTTGTAATGACCAAAGAATCAAAGGCCTTGATGGGACTCTACCATGGTCAGGTCCTGTGCA AGAAGAATAAATTTGGAGCTCCACAGAAGGATGTTAAGCATCTGGCTATTCTTGGTGCAGGGCTGATGGGAGCAGGCATCGCCCAAGTCT CCGTGGATAAGGGGCTAAAGACTATACTTAAAGATGCCACCCTCACTGCGCTAGACCGAGGACAGCAACAAGTGTTCAAAGGATTGAATG ACAAAGTGAAGAAGAAAGCTCTAACATCATTTGAAAGGGATTCCATCTTCAGCAACTTGACTGGGCAGCTTGATTACCAAGGTTTTGAAA AGGCCGACATGGTGATTGAAGCTGTGTTTGAGGACCTTAGTCTTAAGCACAGAGTGCTAAAGGAAGTAGAAGCGGTGATTCCAGATCACT GTATCTTTGCCAGTAACACATCTGCTCTCCCAATCAGTGAAATCGCTGCTGTCAGCAAAAGACCTGAGAAGGTGATTGGCATGCACTACT TCTCTCCCGTGGACAAGATGCAGCTGCTGGAGATTATCACGACCGAGAAAACTTCCAAAGACACCAGTGCTTCAGCTGTAGCAGTTGGTC TCAAGCAGGGGAAGGTCATCATTGTGGTTAAGGATGGACCTGGCTTCTATACTACCAGGTGTCTTGCGCCCATGATGTCTGAAGTCATCC GAATCCTCCAGGAAGGAGTTGACCCGAAGAAGCTGGATTCCCTGACCACAAGCTTTGGCTTTCCTGTGGGTGCCGCCACACTGGTGGATG AAGTTGGTGTGGATGTAGCGAAACATGTGGCGGAAGATCTGGGCAAAGTCTTTGGGGAGCGGTTTGGAGGTGGAAACCCAGAACTGCTGA CACAGATGGTGTCCAAGGGCTTCCTAGGTCGTAAATCTGGGAAGGGCTTTTACATCTATCAGGAGGGTGTGAAGAGGAAGGATTTGAATT CTGACATGGATAGTATTTTAGCGAGTCTGAAGCTGCCTCCTAAGTCTGAAGTCTCATCAGACGAAGACATCCAGTTCCGCCTGGTGACAA GATTTGTGAATGAGGCAGTCATGTGCCTGCAAGAGGGGATCTTGGCCACACCTGCAGAGGGAGACATCGGAGCCGTCTTTGGGCTTGGCT TCCCGCCTTGTCTGGGAGGGCCTTTCCGCTTTGTGGATCTGTATGGCGCCCAGAAGATAGTGGACCGGCTCAAGAAATATGAAGCTGCCT ATGGAAAACAGTTCACCCCATGCCAGCTGCTAGCTGACCATGCTAACAGCCCTAACAAGAAGTTCTACCAGTGAGCAGGCCTCATGCCTC GCTCAGTCAGTGCACTAACCCCAGCTGCCGGCAGTGCTGGTTCTCCAACAGAGTGGTGTCTAGATTTATCAGAGTAACGAGAAGACAAAC TCCGGCACTGGGTTTGCTCCCTGATTAAAGTGCCTTCAGCCAAGACCATCTCTCCCTCCTGGTGAAGTGTGACTTCGAATTAGTTTGCAC TTCCTGTTGGAAGGTAGAGCCCACTGCTCATTGTATAAGCCCCGAGGCCTAGAGTGGCAGCCAAGAGCCATCTGAAGCCACCTCTCTGCC TGTTCCTCCCAAGAGGCCAGGGTGGCCAGGGGTGGTGAGGGCAGTTCTGCACCCAGCCAAACACATAACAATAAAAACCAAACTCTGTGT CAGCATCTTTGCCCTTCTGGTTTAAACGCCTCCTTCAAAAAGCAATCTGGAAGAAAGCCCTGTGCTTTGGGGGAGTAAGAATGTGTGTGC AGAATTCTAGGCAGCACCTTAGGGAGGGACTGGGATGAGAGAAAGTGGGACCTGGTGGGCTCAACCACACACACCTGTCTGTGCAGATGC >7307_7307_1_ASXL2-HADHA_ASXL2_chr2_26022254_ENST00000336112_HADHA_chr2_26432758_ENST00000380649_length(amino acids)=470AA_BP=32 MVSQIPRVLRTAAAAVMVAATRREKRAGGKGKKFGELVMTKESKALMGLYHGQVLCKKNKFGAPQKDVKHLAILGAGLMGAGIAQVSVDK GLKTILKDATLTALDRGQQQVFKGLNDKVKKKALTSFERDSIFSNLTGQLDYQGFEKADMVIEAVFEDLSLKHRVLKEVEAVIPDHCIFA SNTSALPISEIAAVSKRPEKVIGMHYFSPVDKMQLLEIITTEKTSKDTSASAVAVGLKQGKVIIVVKDGPGFYTTRCLAPMMSEVIRILQ EGVDPKKLDSLTTSFGFPVGAATLVDEVGVDVAKHVAEDLGKVFGERFGGGNPELLTQMVSKGFLGRKSGKGFYIYQEGVKRKDLNSDMD SILASLKLPPKSEVSSDEDIQFRLVTRFVNEAVMCLQEGILATPAEGDIGAVFGLGFPPCLGGPFRFVDLYGAQKIVDRLKKYEAAYGKQ -------------------------------------------------------------- >7307_7307_2_ASXL2-HADHA_ASXL2_chr2_26101035_ENST00000435504_HADHA_chr2_26432758_ENST00000380649_length(transcript)=2283nt_BP=351nt ACAGTAGTGACGGCCGCGCGGGAGCGGTCACATGACCGTAGCGGCAGCCTGTACAGTAAGAACCCAATATGGCAGCGGCGGTAGCAGTGG CAACGGCAGCAAGAGGACCGGCCGCCCCATAGACCCCCCCGCGCACCACCCCCGCCGCTTCCTCTGCTTGGGTGCCCCCCCGGTCTGACT TCCCTTTTCTCCCACCTTTCCCGGCACCGCGGGAAAAGCAGCGCAGGGCAGAGCAGGCAGGGAGGGCGGCCGGAGCCCGGACACGGGAGC CTCCCAGTCAGTTCAAGACCCGACATGAGGGAAAAGGGACGTAGGAAGAAGGGCAGGACCTGGGCGGAGGCCGCCAAGACGAAATTTGGA GAGCTTGTAATGACCAAAGAATCAAAGGCCTTGATGGGACTCTACCATGGTCAGGTCCTGTGCAAGAAGAATAAATTTGGAGCTCCACAG AAGGATGTTAAGCATCTGGCTATTCTTGGTGCAGGGCTGATGGGAGCAGGCATCGCCCAAGTCTCCGTGGATAAGGGGCTAAAGACTATA CTTAAAGATGCCACCCTCACTGCGCTAGACCGAGGACAGCAACAAGTGTTCAAAGGATTGAATGACAAAGTGAAGAAGAAAGCTCTAACA TCATTTGAAAGGGATTCCATCTTCAGCAACTTGACTGGGCAGCTTGATTACCAAGGTTTTGAAAAGGCCGACATGGTGATTGAAGCTGTG TTTGAGGACCTTAGTCTTAAGCACAGAGTGCTAAAGGAAGTAGAAGCGGTGATTCCAGATCACTGTATCTTTGCCAGTAACACATCTGCT CTCCCAATCAGTGAAATCGCTGCTGTCAGCAAAAGACCTGAGAAGGTGATTGGCATGCACTACTTCTCTCCCGTGGACAAGATGCAGCTG CTGGAGATTATCACGACCGAGAAAACTTCCAAAGACACCAGTGCTTCAGCTGTAGCAGTTGGTCTCAAGCAGGGGAAGGTCATCATTGTG GTTAAGGATGGACCTGGCTTCTATACTACCAGGTGTCTTGCGCCCATGATGTCTGAAGTCATCCGAATCCTCCAGGAAGGAGTTGACCCG AAGAAGCTGGATTCCCTGACCACAAGCTTTGGCTTTCCTGTGGGTGCCGCCACACTGGTGGATGAAGTTGGTGTGGATGTAGCGAAACAT GTGGCGGAAGATCTGGGCAAAGTCTTTGGGGAGCGGTTTGGAGGTGGAAACCCAGAACTGCTGACACAGATGGTGTCCAAGGGCTTCCTA GGTCGTAAATCTGGGAAGGGCTTTTACATCTATCAGGAGGGTGTGAAGAGGAAGGATTTGAATTCTGACATGGATAGTATTTTAGCGAGT CTGAAGCTGCCTCCTAAGTCTGAAGTCTCATCAGACGAAGACATCCAGTTCCGCCTGGTGACAAGATTTGTGAATGAGGCAGTCATGTGC CTGCAAGAGGGGATCTTGGCCACACCTGCAGAGGGAGACATCGGAGCCGTCTTTGGGCTTGGCTTCCCGCCTTGTCTGGGAGGGCCTTTC CGCTTTGTGGATCTGTATGGCGCCCAGAAGATAGTGGACCGGCTCAAGAAATATGAAGCTGCCTATGGAAAACAGTTCACCCCATGCCAG CTGCTAGCTGACCATGCTAACAGCCCTAACAAGAAGTTCTACCAGTGAGCAGGCCTCATGCCTCGCTCAGTCAGTGCACTAACCCCAGCT GCCGGCAGTGCTGGTTCTCCAACAGAGTGGTGTCTAGATTTATCAGAGTAACGAGAAGACAAACTCCGGCACTGGGTTTGCTCCCTGATT AAAGTGCCTTCAGCCAAGACCATCTCTCCCTCCTGGTGAAGTGTGACTTCGAATTAGTTTGCACTTCCTGTTGGAAGGTAGAGCCCACTG CTCATTGTATAAGCCCCGAGGCCTAGAGTGGCAGCCAAGAGCCATCTGAAGCCACCTCTCTGCCTGTTCCTCCCAAGAGGCCAGGGTGGC CAGGGGTGGTGAGGGCAGTTCTGCACCCAGCCAAACACATAACAATAAAAACCAAACTCTGTGTCAGCATCTTTGCCCTTCTGGTTTAAA CGCCTCCTTCAAAAAGCAATCTGGAAGAAAGCCCTGTGCTTTGGGGGAGTAAGAATGTGTGTGCAGAATTCTAGGCAGCACCTTAGGGAG GGACTGGGATGAGAGAAAGTGGGACCTGGTGGGCTCAACCACACACACCTGTCTGTGCAGATGCTTTGCCCAGGCTTCTCACCACGGTGT >7307_7307_2_ASXL2-HADHA_ASXL2_chr2_26101035_ENST00000435504_HADHA_chr2_26432758_ENST00000380649_length(amino acids)=504AA_BP=66 MLGCPPGLTSLFSHLSRHRGKSSAGQSRQGGRPEPGHGSLPVSSRPDMREKGRRKKGRTWAEAAKTKFGELVMTKESKALMGLYHGQVLC KKNKFGAPQKDVKHLAILGAGLMGAGIAQVSVDKGLKTILKDATLTALDRGQQQVFKGLNDKVKKKALTSFERDSIFSNLTGQLDYQGFE KADMVIEAVFEDLSLKHRVLKEVEAVIPDHCIFASNTSALPISEIAAVSKRPEKVIGMHYFSPVDKMQLLEIITTEKTSKDTSASAVAVG LKQGKVIIVVKDGPGFYTTRCLAPMMSEVIRILQEGVDPKKLDSLTTSFGFPVGAATLVDEVGVDVAKHVAEDLGKVFGERFGGGNPELL TQMVSKGFLGRKSGKGFYIYQEGVKRKDLNSDMDSILASLKLPPKSEVSSDEDIQFRLVTRFVNEAVMCLQEGILATPAEGDIGAVFGLG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ASXL2-HADHA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ASXL2-HADHA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ASXL2-HADHA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ASXL2 | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Hgene | ASXL2 | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Hgene | ASXL2 | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | ASXL2 | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | ASXL2 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Hgene | ASXL2 | C4310672 | SHASHI-PENA SYNDROME | 1 | GENOMICS_ENGLAND |
| Tgene | C3711645 | Long chain 3-hydroxyacyl-CoA dehydrogenase deficiency | 9 | CLINGEN;CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C1969443 | Trifunctional Protein Deficiency With Myopathy And Neuropathy | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0019193 | Hepatitis, Toxic | 1 | CTD_human | |
| Tgene | C0029408 | Degenerative polyarthritis | 1 | CTD_human | |
| Tgene | C0086743 | Osteoarthrosis Deformans | 1 | CTD_human | |
| Tgene | C0151744 | Myocardial Ischemia | 1 | CTD_human | |
| Tgene | C0369183 | Erythrocyte Mean Corpuscular Hemoglobin Test | 1 | GENOMICS_ENGLAND | |
| Tgene | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human | |
| Tgene | C1261502 | Finding of Mean Corpuscular Hemoglobin | 1 | GENOMICS_ENGLAND | |
| Tgene | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human | |
| Tgene | C1455728 | Acute fatty liver of pregnancy | 1 | ORPHANET | |
| Tgene | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human | |
| Tgene | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human | |
| Tgene | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
