
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TGM2-COL4A1 (FusionGDB2 ID:HG7052TG1282) |
Fusion Gene Summary for TGM2-COL4A1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TGM2-COL4A1 | Fusion gene ID: hg7052tg1282 | Hgene | Tgene | Gene symbol | TGM2 | COL4A1 | Gene ID | 7052 | 1282 |
| Gene name | transglutaminase 2 | collagen type IV alpha 1 chain | |
| Synonyms | TG(C)|TGC | BSVD|BSVD1|PADMAL|RATOR | |
| Cytomap | ('TGM2')('COL4A1') 20q11.23 | 13q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein-glutamine gamma-glutamyltransferase 2C polypeptide, protein-glutamine-gamma-glutamyltransferaseTGase CTGase HTGase-2tissue transglutaminasetransglutaminase Ctransglutaminase H | collagen alpha-1(IV) chainCOL4A1 NC1 domainarrestencollagen IV, alpha-1 polypeptidecollagen of basement membrane, alpha-1 chain | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | P02462 | |
| Ensembl transtripts involved in fusion gene | ENST00000361475, ENST00000536701, ENST00000536724, | ||
| Fusion gene scores | * DoF score | 11 X 11 X 4=484 | 14 X 14 X 7=1372 |
| # samples | 13 | 16 | |
| ** MAII score | log2(13/484*10)=-1.89649542424614 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1372*10)=-3.10013667128545 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TGM2 [Title/Abstract] AND COL4A1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TGM2(36770466)-COL4A1(110802791), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TGM2-COL4A1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TGM2-COL4A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TGM2 | GO:0018149 | peptide cross-linking | 30458214 |
| Hgene | TGM2 | GO:0043277 | apoptotic cell clearance | 19628791 |
 Fusion gene breakpoints across TGM2 (5'-gene) Fusion gene breakpoints across TGM2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
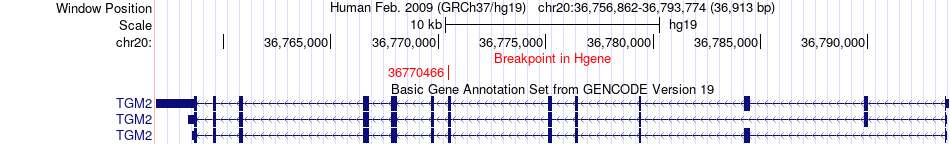 |
 Fusion gene breakpoints across COL4A1 (3'-gene) Fusion gene breakpoints across COL4A1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-8608-01A | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
Top |
Fusion Gene ORF analysis for TGM2-COL4A1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000361475 | ENST00000467182 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| 5CDS-intron | ENST00000361475 | ENST00000543140 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| 5CDS-intron | ENST00000536701 | ENST00000467182 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| 5CDS-intron | ENST00000536701 | ENST00000543140 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| 5CDS-intron | ENST00000536724 | ENST00000467182 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| 5CDS-intron | ENST00000536724 | ENST00000543140 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| In-frame | ENST00000361475 | ENST00000375820 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| In-frame | ENST00000536701 | ENST00000375820 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
| In-frame | ENST00000536724 | ENST00000375820 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000361475 | TGM2 | chr20 | 36770466 | - | ENST00000375820 | COL4A1 | chr13 | 110802791 | - | 2643 | 1169 | 126 | 1250 | 374 |
| ENST00000536701 | TGM2 | chr20 | 36770466 | - | ENST00000375820 | COL4A1 | chr13 | 110802791 | - | 2298 | 824 | 24 | 905 | 293 |
| ENST00000536724 | TGM2 | chr20 | 36770466 | - | ENST00000375820 | COL4A1 | chr13 | 110802791 | - | 2361 | 887 | 24 | 968 | 314 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000361475 | ENST00000375820 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - | 0.000383475 | 0.9996165 |
| ENST00000536701 | ENST00000375820 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - | 0.000751124 | 0.9992488 |
| ENST00000536724 | ENST00000375820 | TGM2 | chr20 | 36770466 | - | COL4A1 | chr13 | 110802791 | - | 0.002261272 | 0.9977387 |
Top |
Fusion Genomic Features for TGM2-COL4A1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
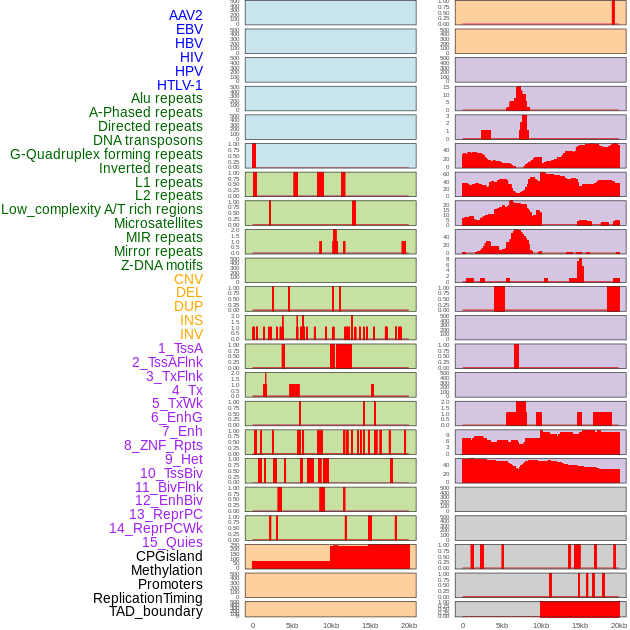 |
Top |
Fusion Protein Features for TGM2-COL4A1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:36770466/chr13:110802791) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | COL4A1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Type IV collagen is the major structural component of glomerular basement membranes (GBM), forming a 'chicken-wire' meshwork together with laminins, proteoglycans and entactin/nidogen. {ECO:0000250|UniProtKB:P02463}.; FUNCTION: Arresten, comprising the C-terminal NC1 domain, inhibits angiogenesis and tumor formation. The C-terminal half is found to possess the anti-angiogenic activity. Specifically inhibits endothelial cell proliferation, migration and tube formation. {ECO:0000269|PubMed:10811134, ECO:0000269|PubMed:18775695}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TGM2 | chr20:36770466 | chr13:110802791 | ENST00000361475 | - | 7 | 13 | 476_483 | 331 | 688.0 | Nucleotide binding | GTP |
| Hgene | TGM2 | chr20:36770466 | chr13:110802791 | ENST00000361475 | - | 7 | 13 | 580_583 | 331 | 688.0 | Nucleotide binding | GTP |
| Tgene | COL4A1 | chr20:36770466 | chr13:110802791 | ENST00000375820 | 50 | 52 | 1445_1669 | 1642 | 1670.0 | Domain | Collagen IV NC1 | |
| Tgene | COL4A1 | chr20:36770466 | chr13:110802791 | ENST00000375820 | 50 | 52 | 173_1440 | 1642 | 1670.0 | Region | Note=Triple-helical region |
Top |
Fusion Gene Sequence for TGM2-COL4A1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >90516_90516_1_TGM2-COL4A1_TGM2_chr20_36770466_ENST00000361475_COL4A1_chr13_110802791_ENST00000375820_length(transcript)=2643nt_BP=1169nt ACTGGGCAATGGGTGGCCTCCCAGGTCGCCGCCTTCCCGCGGGGCCCCGCCCCCGGCCCGCCCCAAAGCGGGCTATAAGTTAGCGCCGCT CTCCGCCTCGGCAGTGCCAGCCGCCAGTGGTCGCACTTGGAGGGTCTCGCCGCCAGTGGAAGGAGCCACCGCCCCCGCCCGACCATGGCC GAGGAGCTGGTCTTAGAGAGGTGTGATCTGGAGCTGGAGACCAATGGCCGAGACCACCACACGGCCGACCTGTGCCGGGAGAAGCTGGTG GTGCGACGGGGCCAGCCCTTCTGGCTGACCCTGCACTTTGAGGGCCGCAACTACGAGGCCAGTGTAGACAGTCTCACCTTCAGTGTCGTG ACCGGCCCAGCCCCTAGCCAGGAGGCCGGGACCAAGGCCCGTTTTCCACTAAGAGATGCTGTGGAGGAGGGTGACTGGACAGCCACCGTG GTGGACCAGCAAGACTGCACCCTCTCGCTGCAGCTCACCACCCCGGCCAACGCCCCCATCGGCCTGTATCGCCTCAGCCTGGAGGCCTCC ACTGGCTACCAGGGATCCAGCTTTGTGCTGGGCCACTTCATTTTGCTCTTCAACGCCTGGTGCCCAGCGGATGCTGTGTACCTGGACTCG GAAGAGGAGCGGCAGGAGTATGTCCTCACCCAGCAGGGCTTTATCTACCAGGGCTCGGCCAAGTTCATCAAGAACATACCTTGGAATTTT GGGCAGTTTGAAGATGGGATCCTAGACATCTGCCTGATCCTTCTAGATGTCAACCCCAAGTTCCTGAAGAACGCCGGCCGTGACTGCTCC CGCCGCAGCAGCCCCGTCTACGTGGGCCGGGTGGTGAGTGGCATGGTCAACTGCAACGATGACCAGGGTGTGCTGCTGGGACGCTGGGAC AACAACTACGGGGACGGCGTCAGCCCCATGTCCTGGATCGGCAGCGTGGACATCCTGCGGCGCTGGAAGAACCACGGCTGCCAGCGCGTC AAGTATGGCCAGTGCTGGGTCTTCGCCGCCGTGGCCTGCACAGTGCTGAGGTGCCTGGGCATCCCTACCCGCGTCGTGACCAACTACAAC TCGGCCCATGACCAGAACAGCAACCTTCTCATCGAGTACTTCCGCAATGAGTTTGGGGAGATCCAGGGTGACAAGAGCGAGATGATCTGG AAGCCTACGCCGTCCACCTTGAAGGCAGGGGAGCTGCGCACGCACGTCAGCCGCTGCCAAGTCTGTATGAGAAGAACATAATGAAGCCTG ACTCAGCTAATGTCACAACATGGTGCTACTTCTTCTTCTTTTTGTTAACAGCAACGAACCCTAGAAATATATCCTGTGTACCTCACTGTC CAATATGAAAACCGTAAAGTGCCTTATAGGAATTTGCGTAACTAACACACCCTGCTTCATTGACCTCTACTTGCTGAAGGAGAAAAAGAC AGCGATAAGCTTTCAATAGTGGCATACCAAATGGCACTTTTGATGAAATAAAATATCAATATTTTCTGCAATCCAATGCACTGATGTGTG AAGTGAGAACTCCATCAGAAAACCAAAGGGTGCTAGGAGGTGTGGGTGCCTTCCATACTGTTTGCCCATTTTCATTCTTGTATTATAATT AATTTTCTACCCCCAGAGATAAATGTTTGTTTATATCACTGTCTAGCTGTTTCAAAATTTAGGTCCCTTGGTCTGTACAAATAATAGCAA TGTAAAAATGGTTTTTTGAACCTCCAAATGGAATTACAGACTCAGTAGCCATATCTTCCAACCCCCCAGTATAAATTTCTGTCTTTCTGC TATGTGTGGTACTTTGCAGCTGCTTTTGCAGAAATCACAATTTTCCTGTGGAATAAAGATGGTCCAAAAATAGTCAAAAATTAAATATAT ATATATATTAGTAATTTATATAGATGTCAGCAATTAGGCAGATCAAGGTTTAGTTTAACTTCCACTGTTAAAATAAAGCTTACATAGTTT TCTTCCTTTGAAAGACTGTGCTGTCCTTTAACATAGGTTTTTAAAGACTAGGATATTGAATGTGAAACATCCGTTTTCATTGTTCACTTC TAAACCAAAAATTATGTGTTGCCAAAACCAAACCCAGGTTCATGAATATGGTGTCTATTATAGTGAAACATGTACTTTGAGCTTATTGTT TTTATTCTGTATTAAATATTTTCAGGGTTTTAAACACTAATCACAAACTGAATGACTTGACTTCAAAAGCAACAACCTTAAAGGCCGTCA TTTCATTAGTATTCCTCATTCTGCATCCTGGCTTGAAAAACAGCTCTGTTGAATCACAGTATCAGTATTTTCACACGTAAGCACATTCGG GCCATTTCCGTGGTTTCTCATGAGCTGTGTTCACAGACCTCAGCAGGGCATCGCATGGACCGCAGGAGGGCAGATTCGGACCACTAGGCC TGAAATGACATTTCACTAAAAGTCTCCAAAACATTTCTAAGACTACTAAGGCCTTTTATGTAATTTCTTTAAATGTGTATTTCTTAAGAA TTCAAATTTGTAATAAAACTATTTGTATAAAAATTAAGCTTTTATTAATTTGTTGCTAGTATTGCCACAGACGCATTAAAAGAAACTTAC >90516_90516_1_TGM2-COL4A1_TGM2_chr20_36770466_ENST00000361475_COL4A1_chr13_110802791_ENST00000375820_length(amino acids)=374AA_BP=348 MEGLAASGRSHRPRPTMAEELVLERCDLELETNGRDHHTADLCREKLVVRRGQPFWLTLHFEGRNYEASVDSLTFSVVTGPAPSQEAGTK ARFPLRDAVEEGDWTATVVDQQDCTLSLQLTTPANAPIGLYRLSLEASTGYQGSSFVLGHFILLFNAWCPADAVYLDSEEERQEYVLTQQ GFIYQGSAKFIKNIPWNFGQFEDGILDICLILLDVNPKFLKNAGRDCSRRSSPVYVGRVVSGMVNCNDDQGVLLGRWDNNYGDGVSPMSW IGSVDILRRWKNHGCQRVKYGQCWVFAAVACTVLRCLGIPTRVVTNYNSAHDQNSNLLIEYFRNEFGEIQGDKSEMIWKPTPSTLKAGEL -------------------------------------------------------------- >90516_90516_2_TGM2-COL4A1_TGM2_chr20_36770466_ENST00000536701_COL4A1_chr13_110802791_ENST00000375820_length(transcript)=2298nt_BP=824nt AGTGCCAGCCGCCAGTGGTCGCACTTGGAGGGTCTCGCCGCCAGTGGAAGGAGCCACCGCCCCCGCCCGACCATGGCCGAGGAGCTGGTC TTAGAGAGGTGTGATCTGGAGCTGGAGACCAATGGCCGAGACCACCACACGGCCGACCTGTGCCGGGAGAAGCTGGTGGTGCGACGGGGC CAGCCCTTCTGGCTGACCCTGCACTTTGAGGGCCGCAACTACGAGGCCAGTGTAGACAGTCTCACCTTCAGTGTCGTGACCGCGGATGCT GTGTACCTGGACTCGGAAGAGGAGCGGCAGGAGTATGTCCTCACCCAGCAGGGCTTTATCTACCAGGGCTCGGCCAAGTTCATCAAGAAC ATACCTTGGAATTTTGGGCAGTTTGAAGATGGGATCCTAGACATCTGCCTGATCCTTCTAGATGTCAACCCCAAGTTCCTGAAGAACGCC GGCCGTGACTGCTCCCGCCGCAGCAGCCCCGTCTACGTGGGCCGGGTGGTGAGTGGCATGGTCAACTGCAACGATGACCAGGGTGTGCTG CTGGGACGCTGGGACAACAACTACGGGGACGGCGTCAGCCCCATGTCCTGGATCGGCAGCGTGGACATCCTGCGGCGCTGGAAGAACCAC GGCTGCCAGCGCGTCAAGTATGGCCAGTGCTGGGTCTTCGCCGCCGTGGCCTGCACAGTGCTGAGGTGCCTGGGCATCCCTACCCGCGTC GTGACCAACTACAACTCGGCCCATGACCAGAACAGCAACCTTCTCATCGAGTACTTCCGCAATGAGTTTGGGGAGATCCAGGGTGACAAG AGCGAGATGATCTGGAAGCCTACGCCGTCCACCTTGAAGGCAGGGGAGCTGCGCACGCACGTCAGCCGCTGCCAAGTCTGTATGAGAAGA ACATAATGAAGCCTGACTCAGCTAATGTCACAACATGGTGCTACTTCTTCTTCTTTTTGTTAACAGCAACGAACCCTAGAAATATATCCT GTGTACCTCACTGTCCAATATGAAAACCGTAAAGTGCCTTATAGGAATTTGCGTAACTAACACACCCTGCTTCATTGACCTCTACTTGCT GAAGGAGAAAAAGACAGCGATAAGCTTTCAATAGTGGCATACCAAATGGCACTTTTGATGAAATAAAATATCAATATTTTCTGCAATCCA ATGCACTGATGTGTGAAGTGAGAACTCCATCAGAAAACCAAAGGGTGCTAGGAGGTGTGGGTGCCTTCCATACTGTTTGCCCATTTTCAT TCTTGTATTATAATTAATTTTCTACCCCCAGAGATAAATGTTTGTTTATATCACTGTCTAGCTGTTTCAAAATTTAGGTCCCTTGGTCTG TACAAATAATAGCAATGTAAAAATGGTTTTTTGAACCTCCAAATGGAATTACAGACTCAGTAGCCATATCTTCCAACCCCCCAGTATAAA TTTCTGTCTTTCTGCTATGTGTGGTACTTTGCAGCTGCTTTTGCAGAAATCACAATTTTCCTGTGGAATAAAGATGGTCCAAAAATAGTC AAAAATTAAATATATATATATATTAGTAATTTATATAGATGTCAGCAATTAGGCAGATCAAGGTTTAGTTTAACTTCCACTGTTAAAATA AAGCTTACATAGTTTTCTTCCTTTGAAAGACTGTGCTGTCCTTTAACATAGGTTTTTAAAGACTAGGATATTGAATGTGAAACATCCGTT TTCATTGTTCACTTCTAAACCAAAAATTATGTGTTGCCAAAACCAAACCCAGGTTCATGAATATGGTGTCTATTATAGTGAAACATGTAC TTTGAGCTTATTGTTTTTATTCTGTATTAAATATTTTCAGGGTTTTAAACACTAATCACAAACTGAATGACTTGACTTCAAAAGCAACAA CCTTAAAGGCCGTCATTTCATTAGTATTCCTCATTCTGCATCCTGGCTTGAAAAACAGCTCTGTTGAATCACAGTATCAGTATTTTCACA CGTAAGCACATTCGGGCCATTTCCGTGGTTTCTCATGAGCTGTGTTCACAGACCTCAGCAGGGCATCGCATGGACCGCAGGAGGGCAGAT TCGGACCACTAGGCCTGAAATGACATTTCACTAAAAGTCTCCAAAACATTTCTAAGACTACTAAGGCCTTTTATGTAATTTCTTTAAATG TGTATTTCTTAAGAATTCAAATTTGTAATAAAACTATTTGTATAAAAATTAAGCTTTTATTAATTTGTTGCTAGTATTGCCACAGACGCA >90516_90516_2_TGM2-COL4A1_TGM2_chr20_36770466_ENST00000536701_COL4A1_chr13_110802791_ENST00000375820_length(amino acids)=293AA_BP=267 MEGLAASGRSHRPRPTMAEELVLERCDLELETNGRDHHTADLCREKLVVRRGQPFWLTLHFEGRNYEASVDSLTFSVVTADAVYLDSEEE RQEYVLTQQGFIYQGSAKFIKNIPWNFGQFEDGILDICLILLDVNPKFLKNAGRDCSRRSSPVYVGRVVSGMVNCNDDQGVLLGRWDNNY GDGVSPMSWIGSVDILRRWKNHGCQRVKYGQCWVFAAVACTVLRCLGIPTRVVTNYNSAHDQNSNLLIEYFRNEFGEIQGDKSEMIWKPT -------------------------------------------------------------- >90516_90516_3_TGM2-COL4A1_TGM2_chr20_36770466_ENST00000536724_COL4A1_chr13_110802791_ENST00000375820_length(transcript)=2361nt_BP=887nt AGTGCCAGCCGCCAGTGGTCGCACTTGGAGGGTCTCGCCGCCAGTGGAAGGAGCCACCGCCCCCGCCCGACCATGGCCGAGGGCCCAGCC CCTAGCCAGGAGGCCGGGACCAAGGCCCGTTTTCCACTAAGAGATGCTGTGGAGGAGGGTGACTGGACAGCCACCGTGGTGGACCAGCAA GACTGCACCCTCTCGCTGCAGCTCACCACCCCGGCCAACGCCCCCATCGGCCTGTATCGCCTCAGCCTGGAGGCCTCCACTGGCTACCAG GGATCCAGCTTTGTGCTGGGCCACTTCATTTTGCTCTTCAACGCCTGGTGCCCAGCGGATGCTGTGTACCTGGACTCGGAAGAGGAGCGG CAGGAGTATGTCCTCACCCAGCAGGGCTTTATCTACCAGGGCTCGGCCAAGTTCATCAAGAACATACCTTGGAATTTTGGGCAGTTTGAA GATGGGATCCTAGACATCTGCCTGATCCTTCTAGATGTCAACCCCAAGTTCCTGAAGAACGCCGGCCGTGACTGCTCCCGCCGCAGCAGC CCCGTCTACGTGGGCCGGGTGGTGAGTGGCATGGTCAACTGCAACGATGACCAGGGTGTGCTGCTGGGACGCTGGGACAACAACTACGGG GACGGCGTCAGCCCCATGTCCTGGATCGGCAGCGTGGACATCCTGCGGCGCTGGAAGAACCACGGCTGCCAGCGCGTCAAGTATGGCCAG TGCTGGGTCTTCGCCGCCGTGGCCTGCACAGTGCTGAGGTGCCTGGGCATCCCTACCCGCGTCGTGACCAACTACAACTCGGCCCATGAC CAGAACAGCAACCTTCTCATCGAGTACTTCCGCAATGAGTTTGGGGAGATCCAGGGTGACAAGAGCGAGATGATCTGGAAGCCTACGCCG TCCACCTTGAAGGCAGGGGAGCTGCGCACGCACGTCAGCCGCTGCCAAGTCTGTATGAGAAGAACATAATGAAGCCTGACTCAGCTAATG TCACAACATGGTGCTACTTCTTCTTCTTTTTGTTAACAGCAACGAACCCTAGAAATATATCCTGTGTACCTCACTGTCCAATATGAAAAC CGTAAAGTGCCTTATAGGAATTTGCGTAACTAACACACCCTGCTTCATTGACCTCTACTTGCTGAAGGAGAAAAAGACAGCGATAAGCTT TCAATAGTGGCATACCAAATGGCACTTTTGATGAAATAAAATATCAATATTTTCTGCAATCCAATGCACTGATGTGTGAAGTGAGAACTC CATCAGAAAACCAAAGGGTGCTAGGAGGTGTGGGTGCCTTCCATACTGTTTGCCCATTTTCATTCTTGTATTATAATTAATTTTCTACCC CCAGAGATAAATGTTTGTTTATATCACTGTCTAGCTGTTTCAAAATTTAGGTCCCTTGGTCTGTACAAATAATAGCAATGTAAAAATGGT TTTTTGAACCTCCAAATGGAATTACAGACTCAGTAGCCATATCTTCCAACCCCCCAGTATAAATTTCTGTCTTTCTGCTATGTGTGGTAC TTTGCAGCTGCTTTTGCAGAAATCACAATTTTCCTGTGGAATAAAGATGGTCCAAAAATAGTCAAAAATTAAATATATATATATATTAGT AATTTATATAGATGTCAGCAATTAGGCAGATCAAGGTTTAGTTTAACTTCCACTGTTAAAATAAAGCTTACATAGTTTTCTTCCTTTGAA AGACTGTGCTGTCCTTTAACATAGGTTTTTAAAGACTAGGATATTGAATGTGAAACATCCGTTTTCATTGTTCACTTCTAAACCAAAAAT TATGTGTTGCCAAAACCAAACCCAGGTTCATGAATATGGTGTCTATTATAGTGAAACATGTACTTTGAGCTTATTGTTTTTATTCTGTAT TAAATATTTTCAGGGTTTTAAACACTAATCACAAACTGAATGACTTGACTTCAAAAGCAACAACCTTAAAGGCCGTCATTTCATTAGTAT TCCTCATTCTGCATCCTGGCTTGAAAAACAGCTCTGTTGAATCACAGTATCAGTATTTTCACACGTAAGCACATTCGGGCCATTTCCGTG GTTTCTCATGAGCTGTGTTCACAGACCTCAGCAGGGCATCGCATGGACCGCAGGAGGGCAGATTCGGACCACTAGGCCTGAAATGACATT TCACTAAAAGTCTCCAAAACATTTCTAAGACTACTAAGGCCTTTTATGTAATTTCTTTAAATGTGTATTTCTTAAGAATTCAAATTTGTA ATAAAACTATTTGTATAAAAATTAAGCTTTTATTAATTTGTTGCTAGTATTGCCACAGACGCATTAAAAGAAACTTACTGCACAAGCTGC >90516_90516_3_TGM2-COL4A1_TGM2_chr20_36770466_ENST00000536724_COL4A1_chr13_110802791_ENST00000375820_length(amino acids)=314AA_BP=288 MEGLAASGRSHRPRPTMAEGPAPSQEAGTKARFPLRDAVEEGDWTATVVDQQDCTLSLQLTTPANAPIGLYRLSLEASTGYQGSSFVLGH FILLFNAWCPADAVYLDSEEERQEYVLTQQGFIYQGSAKFIKNIPWNFGQFEDGILDICLILLDVNPKFLKNAGRDCSRRSSPVYVGRVV SGMVNCNDDQGVLLGRWDNNYGDGVSPMSWIGSVDILRRWKNHGCQRVKYGQCWVFAAVACTVLRCLGIPTRVVTNYNSAHDQNSNLLIE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TGM2-COL4A1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TGM2-COL4A1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TGM2-COL4A1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | TGM2 | C0005586 | Bipolar Disorder | 2 | PSYGENET |
| Hgene | TGM2 | C0017636 | Glioblastoma | 2 | CTD_human |
| Hgene | TGM2 | C0036341 | Schizophrenia | 2 | PSYGENET |
| Hgene | TGM2 | C0334588 | Giant Cell Glioblastoma | 2 | CTD_human |
| Hgene | TGM2 | C1621958 | Glioblastoma Multiforme | 2 | CTD_human |
| Hgene | TGM2 | C0006118 | Brain Neoplasms | 1 | CTD_human |
| Hgene | TGM2 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Hgene | TGM2 | C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| Hgene | TGM2 | C0007570 | Celiac Disease | 1 | CTD_human |
| Hgene | TGM2 | C0014859 | Esophageal Neoplasms | 1 | CTD_human |
| Hgene | TGM2 | C0020538 | Hypertensive disease | 1 | CTD_human |
| Hgene | TGM2 | C0029408 | Degenerative polyarthritis | 1 | CTD_human |
| Hgene | TGM2 | C0030297 | Pancreatic Neoplasm | 1 | CTD_human |
| Hgene | TGM2 | C0086743 | Osteoarthrosis Deformans | 1 | CTD_human |
| Hgene | TGM2 | C0153633 | Malignant neoplasm of brain | 1 | CTD_human |
| Hgene | TGM2 | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| Hgene | TGM2 | C0346647 | Malignant neoplasm of pancreas | 1 | CTD_human |
| Hgene | TGM2 | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human |
| Hgene | TGM2 | C0546837 | Malignant neoplasm of esophagus | 1 | CTD_human |
| Hgene | TGM2 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Hgene | TGM2 | C0750974 | Brain Tumor, Primary | 1 | CTD_human |
| Hgene | TGM2 | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human |
| Hgene | TGM2 | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human |
| Hgene | TGM2 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Hgene | TGM2 | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| Hgene | TGM2 | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| Hgene | TGM2 | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| Hgene | TGM2 | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| Hgene | TGM2 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Hgene | TGM2 | C1527390 | Neoplasms, Intracranial | 1 | CTD_human |
| Hgene | TGM2 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
| Tgene | C4551998 | Porencephaly, Type 1, Autosomal Dominant | 18 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C2673195 | Angiopathy, Hereditary, With Nephropathy, Aneurysms, And Muscle Cramps | 6 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C1843512 | BRAIN SMALL VESSEL DISEASE WITH HEMORRHAGE | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C0018965 | Hematuria | 3 | GENOMICS_ENGLAND | |
| Tgene | C0266548 | Axenfeld anomaly (disorder) | 3 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C1860475 | Retinal vascular tortuosity | 3 | GENOMICS_ENGLAND | |
| Tgene | C0011881 | Diabetic Nephropathy | 2 | CTD_human | |
| Tgene | C0017667 | Nodular glomerulosclerosis | 2 | CTD_human | |
| Tgene | C0017668 | Focal glomerulosclerosis | 2 | CTD_human | |
| Tgene | C0086432 | Hyalinosis, Segmental Glomerular | 2 | CTD_human | |
| Tgene | C0265341 | Rieger syndrome | 2 | CTD_human | |
| Tgene | C0266484 | Schizencephaly | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0302892 | Congenital porencephaly | 2 | CTD_human | |
| Tgene | C1280768 | Axenfeld syndrome | 2 | CTD_human | |
| Tgene | C1867983 | PORENCEPHALY, FAMILIAL | 2 | CTD_human;ORPHANET | |
| Tgene | C2675650 | Brain Small Vessel Disease With Axenfeld-Rieger Anomaly | 2 | CTD_human | |
| Tgene | C2678503 | AXENFELD-RIEGER SYNDROME, TYPE 3 | 2 | CTD_human | |
| Tgene | C3281105 | HEMORRHAGE, INTRACEREBRAL, SUSCEPTIBILITY TO | 2 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C3495488 | Axenfeld-Rieger syndrome | 2 | CTD_human | |
| Tgene | C3698507 | Post-traumatic Porencephaly | 2 | CTD_human | |
| Tgene | C3714873 | Axenfeld-Rieger Syndrome, Type 1 | 2 | CTD_human | |
| Tgene | C4082173 | Porencephaly | 2 | CTD_human | |
| Tgene | C4082301 | Developmental Porencephaly | 2 | CTD_human | |
| Tgene | C0002878 | Anemia, Hemolytic | 1 | CTD_human | |
| Tgene | C0002879 | Anemia, Hemolytic, Acquired | 1 | CTD_human | |
| Tgene | C0002889 | Anemia, Microangiopathic | 1 | CTD_human | |
| Tgene | C0015393 | Eye Abnormalities | 1 | CTD_human | |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human | |
| Tgene | C0026848 | Myopathy | 1 | GENOMICS_ENGLAND | |
| Tgene | C0027726 | Nephrotic Syndrome | 1 | CTD_human | |
| Tgene | C0036572 | Seizures | 1 | GENOMICS_ENGLAND | |
| Tgene | C0038454 | Cerebrovascular accident | 1 | GENOMICS_ENGLAND | |
| Tgene | C0149931 | Migraine Disorders | 1 | GENOMICS_ENGLAND | |
| Tgene | C0221021 | Microangiopathic hemolytic anemia | 1 | CTD_human | |
| Tgene | C0265221 | Walker-Warburg congenital muscular dystrophy | 1 | ORPHANET | |
| Tgene | C0270612 | Leukoencephalopathy | 1 | CTD_human | |
| Tgene | C0338656 | Impaired cognition | 1 | GENOMICS_ENGLAND | |
| Tgene | C0424605 | Developmental delay (disorder) | 1 | GENOMICS_ENGLAND | |
| Tgene | C0497327 | Dementia | 1 | GENOMICS_ENGLAND | |
| Tgene | C0557874 | Global developmental delay | 1 | GENOMICS_ENGLAND | |
| Tgene | C1135196 | Heart Failure, Diastolic | 1 | CTD_human | |
| Tgene | C1858991 | Childhood Ataxia with Central Nervous System Hypomyelinization | 1 | CTD_human | |
| Tgene | C1867327 | RETINAL ARTERIES, TORTUOSITY OF | 1 | CTD_human;ORPHANET;UNIPROT | |
| Tgene | C2733158 | Cerebral Small Vessel Diseases | 1 | GENOMICS_ENGLAND | |
| Tgene | C2930808 | Familial vascular leukoencephalopathy | 1 | ORPHANET | |
| Tgene | C2931870 | Familial schizencephaly | 1 | ORPHANET | |
| Tgene | C4013035 | BRAIN SMALL VESSEL DISEASE WITH OR WITHOUT OCULAR ANOMALIES | 1 | GENOMICS_ENGLAND |
