
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:VIM-DES (FusionGDB2 ID:HG7431TG1674) |
Fusion Gene Summary for VIM-DES |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: VIM-DES | Fusion gene ID: hg7431tg1674 | Hgene | Tgene | Gene symbol | VIM | DES | Gene ID | 7431 | 1674 |
| Gene name | vimentin | desmin | |
| Synonyms | - | CDCD3|CMD1F|CSM1|CSM2|LGMD1D|LGMD1E|LGMD2R | |
| Cytomap | ('VIM')('DES') 10p13 | 2q35 | |
| Type of gene | protein-coding | protein-coding | |
| Description | vimentinepididymis secretory sperm binding protein | desmincardiomyopathy, dilated 1F (autosomal dominant)epididymis secretory sperm binding proteinintermediate filament proteinmutant desmin p.K241E | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | . | P17661 | |
| Ensembl transtripts involved in fusion gene | ENST00000485947, ENST00000224237, ENST00000544301, | ENST00000485947, ENST00000224237, ENST00000544301, | |
| Fusion gene scores | * DoF score | 38 X 18 X 13=8892 | 6 X 8 X 5=240 |
| # samples | 42 | 9 | |
| ** MAII score | log2(42/8892*10)=-4.40404671536087 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/240*10)=-1.41503749927884 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: VIM [Title/Abstract] AND DES [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | VIM(17275681)-DES(220285217), # samples:1 VIM(17271757)-DES(220283497), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | DES-VIM seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. DES-VIM seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. VIM-DES seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. VIM-DES seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. VIM-DES seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. VIM-DES seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. VIM-DES seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. VIM-DES seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across VIM (5'-gene) Fusion gene breakpoints across VIM (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
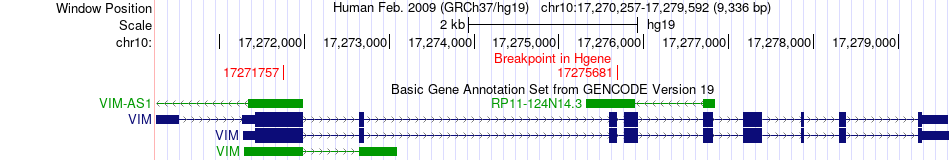 |
 Fusion gene breakpoints across DES (3'-gene) Fusion gene breakpoints across DES (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
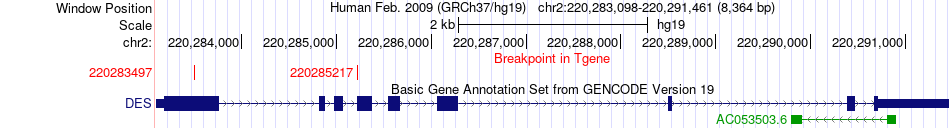 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-06-0644-01A | VIM | chr10 | 17275681 | + | DES | chr2 | 220285217 | + |
| ChimerDB4 | KICH | TCGA-KN-8427-01A | VIM | chr10 | 17271757 | + | DES | chr2 | 220283497 | + |
Top |
Fusion Gene ORF analysis for VIM-DES |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000485947 | ENST00000373960 | VIM | chr10 | 17271757 | + | DES | chr2 | 220283497 | + |
| Frame-shift | ENST00000224237 | ENST00000373960 | VIM | chr10 | 17271757 | + | DES | chr2 | 220283497 | + |
| Frame-shift | ENST00000544301 | ENST00000373960 | VIM | chr10 | 17271757 | + | DES | chr2 | 220283497 | + |
| In-frame | ENST00000224237 | ENST00000373960 | VIM | chr10 | 17275681 | + | DES | chr2 | 220285217 | + |
| In-frame | ENST00000544301 | ENST00000373960 | VIM | chr10 | 17275681 | + | DES | chr2 | 220285217 | + |
| intron-3CDS | ENST00000485947 | ENST00000373960 | VIM | chr10 | 17275681 | + | DES | chr2 | 220285217 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000544301 | VIM | chr10 | 17275681 | + | ENST00000373960 | DES | chr2 | 220285217 | + | 2560 | 1133 | 413 | 1810 | 465 |
| ENST00000224237 | VIM | chr10 | 17275681 | + | ENST00000373960 | DES | chr2 | 220285217 | + | 2292 | 865 | 145 | 1542 | 465 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000544301 | ENST00000373960 | VIM | chr10 | 17275681 | + | DES | chr2 | 220285217 | + | 0.008785206 | 0.99121475 |
| ENST00000224237 | ENST00000373960 | VIM | chr10 | 17275681 | + | DES | chr2 | 220285217 | + | 0.008548084 | 0.99145186 |
Top |
Fusion Genomic Features for VIM-DES |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| VIM | chr10 | 17275681 | + | DES | chr2 | 220285216 | + | 0.999481 | 0.000518976 |
| VIM | chr10 | 17275681 | + | DES | chr2 | 220285216 | + | 0.999481 | 0.000518976 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
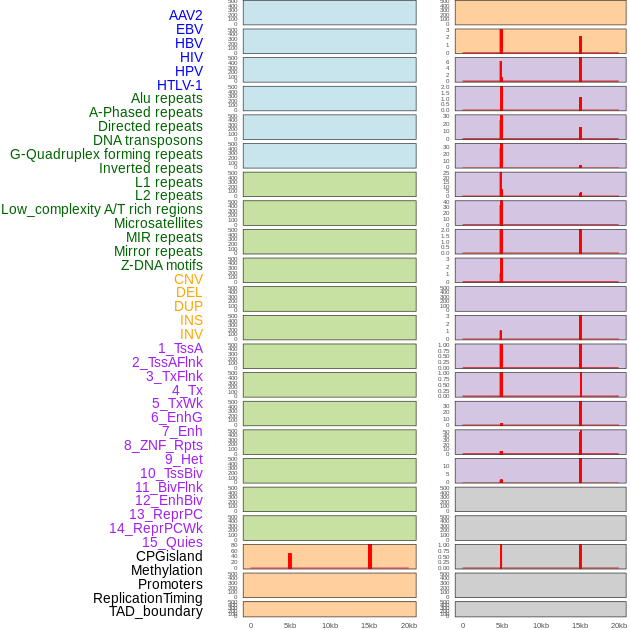 |
Top |
Fusion Protein Features for VIM-DES |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:17275681/chr2:220285217) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | DES |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Muscle-specific type III intermediate filament essential for proper muscular structure and function. Plays a crucial role in maintaining the structure of sarcomeres, inter-connecting the Z-disks and forming the myofibrils, linking them not only to the sarcolemmal cytoskeleton, but also to the nucleus and mitochondria, thus providing strength for the muscle fiber during activity (PubMed:25358400). In adult striated muscle they form a fibrous network connecting myofibrils to each other and to the plasma membrane from the periphery of the Z-line structures (PubMed:24200904, PubMed:25394388, PubMed:26724190). May act as a sarcomeric microtubule-anchoring protein: specifically associates with detyrosinated tubulin-alpha chains, leading to buckled microtubules and mechanical resistance to contraction. Contributes to the transcriptional regulation of the NKX2-5 gene in cardiac progenitor cells during a short period of cardiomyogenesis and in cardiac side population stem cells in the adult. Plays a role in maintaining an optimal conformation of nebulette (NEB) on heart muscle sarcomeres to bind and recruit cardiac alpha-actin (By similarity). {ECO:0000250|UniProtKB:P31001, ECO:0000269|PubMed:24200904, ECO:0000269|PubMed:25394388, ECO:0000269|PubMed:26724190, ECO:0000303|PubMed:25358400}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 96_131 | 240 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 96_131 | 240 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 132_153 | 240 | 467.0 | Region | Note=Linker 1 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 2_95 | 240 | 467.0 | Region | Note=Head |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 132_153 | 240 | 467.0 | Region | Note=Linker 1 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 2_95 | 240 | 467.0 | Region | Note=Head |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 253_268 | 245 | 471.0 | Region | Note=Linker 12 | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 269_287 | 245 | 471.0 | Region | Note=Coil 2A | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 288_295 | 245 | 471.0 | Region | Note=Linker 2 | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 296_412 | 245 | 471.0 | Region | Note=Coil 2B | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 413_470 | 245 | 471.0 | Region | Note=Tail |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 154_245 | 240 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 303_407 | 240 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 154_245 | 240 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 303_407 | 240 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 103_411 | 240 | 467.0 | Domain | IF rod |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 103_411 | 240 | 467.0 | Domain | IF rod |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 326_329 | 240 | 467.0 | Motif | [IL]-x-C-x-x-[DE] motif |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 326_329 | 240 | 467.0 | Motif | [IL]-x-C-x-x-[DE] motif |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 246_268 | 240 | 467.0 | Region | Note=Linker 12 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 269_407 | 240 | 467.0 | Region | Note=Coil 2 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000224237 | + | 3 | 9 | 408_466 | 240 | 467.0 | Region | Note=Tail |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 246_268 | 240 | 467.0 | Region | Note=Linker 12 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 269_407 | 240 | 467.0 | Region | Note=Coil 2 |
| Hgene | VIM | chr10:17275681 | chr2:220285217 | ENST00000544301 | + | 4 | 10 | 408_466 | 240 | 467.0 | Region | Note=Tail |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 108_416 | 245 | 471.0 | Domain | IF rod | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 109_141 | 245 | 471.0 | Region | Note=Coil 1A | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 142_151 | 245 | 471.0 | Region | Note=Linker 1 | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 152_252 | 245 | 471.0 | Region | Note=Coil 1B | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 2_108 | 245 | 471.0 | Region | Note=Head |
Top |
Fusion Gene Sequence for VIM-DES |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >98108_98108_1_VIM-DES_VIM_chr10_17275681_ENST00000224237_DES_chr2_220285217_ENST00000373960_length(transcript)=2292nt_BP=865nt CTCTGCCACTCTCGCTCCGAGGTCCCCGCGCCAGAGACGCAGCCGCGCTCCCACCACCCACACCCACCGCGCCCTCGTTCGCCTCTTCTC CGGGAGCCAGTCCGCGCCACCGCCGCCGCCCAGGCCATCGCCACCCTCCGCAGCCATGTCCACCAGGTCCGTGTCCTCGTCCTCCTACCG CAGGATGTTCGGCGGCCCGGGCACCGCGAGCCGGCCGAGCTCCAGCCGGAGCTACGTGACTACGTCCACCCGCACCTACAGCCTGGGCAG CGCGCTGCGCCCCAGCACCAGCCGCAGCCTCTACGCCTCGTCCCCGGGCGGCGTGTATGCCACGCGCTCCTCTGCCGTGCGCCTGCGGAG CAGCGTGCCCGGGGTGCGGCTCCTGCAGGACTCGGTGGACTTCTCGCTGGCCGACGCCATCAACACCGAGTTCAAGAACACCCGCACCAA CGAGAAGGTGGAGCTGCAGGAGCTGAATGACCGCTTCGCCAACTACATCGACAAGGTGCGCTTCCTGGAGCAGCAGAATAAGATCCTGCT GGCCGAGCTCGAGCAGCTCAAGGGCCAAGGCAAGTCGCGCCTGGGGGACCTCTACGAGGAGGAGATGCGGGAGCTGCGCCGGCAGGTGGA CCAGCTAACCAACGACAAAGCCCGCGTCGAGGTGGAGCGCGACAACCTGGCCGAGGACATCATGCGCCTCCGGGAGAAATTGCAGGAGGA GATGCTTCAGAGAGAGGAAGCCGAAAACACCCTGCAATCTTTCAGACAGGATGTTGACAATGCGTCTCTGGCACGTCTTGACCTTGAACG CAAAGTGGAATCTTTGCAAGAAGAGATTGCCTTTTTGAAGAAACTCCACGAAGAGGAGATCCGTGAGTTGCAGGCTCAGCTTCAGGAACA GCAGGTCCAGGTGGAGATGGACATGTCTAAGCCAGACCTCACTGCCGCCCTCAGGGACATCCGGGCTCAGTATGAGACCATCGCGGCTAA GAACATTTCTGAAGCTGAGGAGTGGTACAAGTCGAAGGTGTCAGACCTGACCCAGGCAGCCAACAAGAACAACGACGCCCTGCGCCAGGC CAAGCAGGAGATGATGGAATACCGACACCAGATCCAGTCCTACACCTGCGAGATTGACGCCCTGAAGGGCACTAACGATTCCCTGATGAG GCAGATGCGGGAATTGGAGGACCGATTTGCCAGTGAGGCCAGTGGCTACCAGGACAACATTGCGCGCCTGGAGGAGGAAATCCGGCACCT CAAGGATGAGATGGCCCGCCATCTGCGCGAGTACCAGGACCTGCTCAACGTGAAGATGGCCCTGGATGTGGAGATTGCCACCTACCGGAA GCTGCTGGAGGGAGAGGAGAGCCGGATCAATCTCCCCATCCAGACCTACTCTGCCCTCAACTTCCGAGAAACCAGCCCTGAGCAAAGGGG TTCTGAGGTCCATACCAAGAAGACGGTGATGATCAAGACCATCGAGACACGGGATGGGGAGGTCGTCAGTGAGGCCACACAGCAGCAGCA TGAAGTGCTCTAAAGACAGAGACCCTCTGCCACCAGAGACCGTCCTCACCCCTGTCCTCACTGCTCCCTGAAGCCAGCCTTCTTCCATCC CAGGACACCACACCCAGCCTCAGTCCTCCCCTCACAGCCTCTGACCCCTCCTCACTGGCCATCCCTCGTGGTCCCCAACAGCGACATAGC CCATCCCTGCCTGGTCACAGGGCATGCCCCGGCCACCTCTGCGGACCCCAGCTGTGAGCCTTGGCTGTTGGCAGTGAGTGAGCCTGGCTC TTGTGCTGGATGGAGCCCAGGCGGGAGCGGTGGCCCTGTCCCTCCCACCTCTGTGACCTCAGGCACTAGCCTTTGGCTCTGGAGACAGCC CCAGAGCAGGGTGTTGGGATACTGCAGGGCCAGGACTGAGCCCCGCAGACCTCCCCAGCCCCTAGCCCAGGAGAGAGAAAGCCAGGCAGG TAGCCAGGGGGACTAGCCCCTGTGGAGACTGGGGGGCTTGAAATTGTCCCCGTGGTCTCTTACTTTCCTTTCCCCAGCCCAGGGTGGACT TAGAAAGCAGGGGCTACAAGAGGGAATCCCCGAAGGTGCTGGAGGTGGGAGCAGGAGATTGAGAAGGAGAGAAAGTGGGTGAGATGCTGG AGAAGAGAGGAGAGGAGAGAGGCAGAGAGCGGTCTCAGGCTGGTGGGAGGGGCGCCCACCTCCCCACGCCCTCCCCTCCCCTGCTGCAGG >98108_98108_1_VIM-DES_VIM_chr10_17275681_ENST00000224237_DES_chr2_220285217_ENST00000373960_length(amino acids)=465AA_BP=84 MSTRSVSSSSYRRMFGGPGTASRPSSSRSYVTTSTRTYSLGSALRPSTSRSLYASSPGGVYATRSSAVRLRSSVPGVRLLQDSVDFSLAD AINTEFKNTRTNEKVELQELNDRFANYIDKVRFLEQQNKILLAELEQLKGQGKSRLGDLYEEEMRELRRQVDQLTNDKARVEVERDNLAE DIMRLREKLQEEMLQREEAENTLQSFRQDVDNASLARLDLERKVESLQEEIAFLKKLHEEEIRELQAQLQEQQVQVEMDMSKPDLTAALR DIRAQYETIAAKNISEAEEWYKSKVSDLTQAANKNNDALRQAKQEMMEYRHQIQSYTCEIDALKGTNDSLMRQMRELEDRFASEASGYQD NIARLEEEIRHLKDEMARHLREYQDLLNVKMALDVEIATYRKLLEGEESRINLPIQTYSALNFRETSPEQRGSEVHTKKTVMIKTIETRD -------------------------------------------------------------- >98108_98108_2_VIM-DES_VIM_chr10_17275681_ENST00000544301_DES_chr2_220285217_ENST00000373960_length(transcript)=2560nt_BP=1133nt GCCTCTCCAAAGGCTGCAGAAGTTTCTTGCTAACAAAAAGTCCGCACATTCGAGCAAAGACAGGCTTTAGCGAGTTATTAAAAACTTAGG GGCGCTCTTGTCCCCCACAGGGCCCGACCGCACACAGCAAGGCGATGGCCCAGCTGTAAGTTGGTAGCACTGAGAACTAGCAGCGCGCGC GGAGCCCGCTGAGACTTGAATCAATCTGGTCTAACGGTTTCCCCTAAACCGCTAGGAGCCCTCAATCGGCGGGACAGCAGGGCGCGTCCT CTGCCACTCTCGCTCCGAGGTCCCCGCGCCAGAGACGCAGCCGCGCTCCCACCACCCACACCCACCGCGCCCTCGTTCGCCTCTTCTCCG GGAGCCAGTCCGCGCCACCGCCGCCGCCCAGGCCATCGCCACCCTCCGCAGCCATGTCCACCAGGTCCGTGTCCTCGTCCTCCTACCGCA GGATGTTCGGCGGCCCGGGCACCGCGAGCCGGCCGAGCTCCAGCCGGAGCTACGTGACTACGTCCACCCGCACCTACAGCCTGGGCAGCG CGCTGCGCCCCAGCACCAGCCGCAGCCTCTACGCCTCGTCCCCGGGCGGCGTGTATGCCACGCGCTCCTCTGCCGTGCGCCTGCGGAGCA GCGTGCCCGGGGTGCGGCTCCTGCAGGACTCGGTGGACTTCTCGCTGGCCGACGCCATCAACACCGAGTTCAAGAACACCCGCACCAACG AGAAGGTGGAGCTGCAGGAGCTGAATGACCGCTTCGCCAACTACATCGACAAGGTGCGCTTCCTGGAGCAGCAGAATAAGATCCTGCTGG CCGAGCTCGAGCAGCTCAAGGGCCAAGGCAAGTCGCGCCTGGGGGACCTCTACGAGGAGGAGATGCGGGAGCTGCGCCGGCAGGTGGACC AGCTAACCAACGACAAAGCCCGCGTCGAGGTGGAGCGCGACAACCTGGCCGAGGACATCATGCGCCTCCGGGAGAAATTGCAGGAGGAGA TGCTTCAGAGAGAGGAAGCCGAAAACACCCTGCAATCTTTCAGACAGGATGTTGACAATGCGTCTCTGGCACGTCTTGACCTTGAACGCA AAGTGGAATCTTTGCAAGAAGAGATTGCCTTTTTGAAGAAACTCCACGAAGAGGAGATCCGTGAGTTGCAGGCTCAGCTTCAGGAACAGC AGGTCCAGGTGGAGATGGACATGTCTAAGCCAGACCTCACTGCCGCCCTCAGGGACATCCGGGCTCAGTATGAGACCATCGCGGCTAAGA ACATTTCTGAAGCTGAGGAGTGGTACAAGTCGAAGGTGTCAGACCTGACCCAGGCAGCCAACAAGAACAACGACGCCCTGCGCCAGGCCA AGCAGGAGATGATGGAATACCGACACCAGATCCAGTCCTACACCTGCGAGATTGACGCCCTGAAGGGCACTAACGATTCCCTGATGAGGC AGATGCGGGAATTGGAGGACCGATTTGCCAGTGAGGCCAGTGGCTACCAGGACAACATTGCGCGCCTGGAGGAGGAAATCCGGCACCTCA AGGATGAGATGGCCCGCCATCTGCGCGAGTACCAGGACCTGCTCAACGTGAAGATGGCCCTGGATGTGGAGATTGCCACCTACCGGAAGC TGCTGGAGGGAGAGGAGAGCCGGATCAATCTCCCCATCCAGACCTACTCTGCCCTCAACTTCCGAGAAACCAGCCCTGAGCAAAGGGGTT CTGAGGTCCATACCAAGAAGACGGTGATGATCAAGACCATCGAGACACGGGATGGGGAGGTCGTCAGTGAGGCCACACAGCAGCAGCATG AAGTGCTCTAAAGACAGAGACCCTCTGCCACCAGAGACCGTCCTCACCCCTGTCCTCACTGCTCCCTGAAGCCAGCCTTCTTCCATCCCA GGACACCACACCCAGCCTCAGTCCTCCCCTCACAGCCTCTGACCCCTCCTCACTGGCCATCCCTCGTGGTCCCCAACAGCGACATAGCCC ATCCCTGCCTGGTCACAGGGCATGCCCCGGCCACCTCTGCGGACCCCAGCTGTGAGCCTTGGCTGTTGGCAGTGAGTGAGCCTGGCTCTT GTGCTGGATGGAGCCCAGGCGGGAGCGGTGGCCCTGTCCCTCCCACCTCTGTGACCTCAGGCACTAGCCTTTGGCTCTGGAGACAGCCCC AGAGCAGGGTGTTGGGATACTGCAGGGCCAGGACTGAGCCCCGCAGACCTCCCCAGCCCCTAGCCCAGGAGAGAGAAAGCCAGGCAGGTA GCCAGGGGGACTAGCCCCTGTGGAGACTGGGGGGCTTGAAATTGTCCCCGTGGTCTCTTACTTTCCTTTCCCCAGCCCAGGGTGGACTTA GAAAGCAGGGGCTACAAGAGGGAATCCCCGAAGGTGCTGGAGGTGGGAGCAGGAGATTGAGAAGGAGAGAAAGTGGGTGAGATGCTGGAG AAGAGAGGAGAGGAGAGAGGCAGAGAGCGGTCTCAGGCTGGTGGGAGGGGCGCCCACCTCCCCACGCCCTCCCCTCCCCTGCTGCAGGGG >98108_98108_2_VIM-DES_VIM_chr10_17275681_ENST00000544301_DES_chr2_220285217_ENST00000373960_length(amino acids)=465AA_BP=84 MSTRSVSSSSYRRMFGGPGTASRPSSSRSYVTTSTRTYSLGSALRPSTSRSLYASSPGGVYATRSSAVRLRSSVPGVRLLQDSVDFSLAD AINTEFKNTRTNEKVELQELNDRFANYIDKVRFLEQQNKILLAELEQLKGQGKSRLGDLYEEEMRELRRQVDQLTNDKARVEVERDNLAE DIMRLREKLQEEMLQREEAENTLQSFRQDVDNASLARLDLERKVESLQEEIAFLKKLHEEEIRELQAQLQEQQVQVEMDMSKPDLTAALR DIRAQYETIAAKNISEAEEWYKSKVSDLTQAANKNNDALRQAKQEMMEYRHQIQSYTCEIDALKGTNDSLMRQMRELEDRFASEASGYQD NIARLEEEIRHLKDEMARHLREYQDLLNVKMALDVEIATYRKLLEGEESRINLPIQTYSALNFRETSPEQRGSEVHTKKTVMIKTIETRD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for VIM-DES |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 438_453 | 245.0 | 471.0 | CRYAB | |
| Tgene | DES | chr10:17275681 | chr2:220285217 | ENST00000373960 | 2 | 9 | 268_415 | 245.0 | 471.0 | NEB |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for VIM-DES |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for VIM-DES |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | VIM | C0006142 | Malignant neoplasm of breast | 4 | CTD_human |
| Hgene | VIM | C0678222 | Breast Carcinoma | 4 | CTD_human |
| Hgene | VIM | C1257931 | Mammary Neoplasms, Human | 4 | CTD_human |
| Hgene | VIM | C1458155 | Mammary Neoplasms | 4 | CTD_human |
| Hgene | VIM | C3805411 | CATARACT 30 | 4 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | VIM | C4704874 | Mammary Carcinoma, Human | 4 | CTD_human |
| Hgene | VIM | C0023890 | Liver Cirrhosis | 2 | CTD_human |
| Hgene | VIM | C0029408 | Degenerative polyarthritis | 2 | CTD_human |
| Hgene | VIM | C0033578 | Prostatic Neoplasms | 2 | CTD_human |
| Hgene | VIM | C0086743 | Osteoarthrosis Deformans | 2 | CTD_human |
| Hgene | VIM | C0239946 | Fibrosis, Liver | 2 | CTD_human |
| Hgene | VIM | C0376358 | Malignant neoplasm of prostate | 2 | CTD_human |
| Hgene | VIM | C0007140 | Carcinosarcoma | 1 | CTD_human |
| Hgene | VIM | C0007621 | Neoplastic Cell Transformation | 1 | CTD_human |
| Hgene | VIM | C0019193 | Hepatitis, Toxic | 1 | CTD_human |
| Hgene | VIM | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Hgene | VIM | C0027626 | Neoplasm Invasiveness | 1 | CTD_human |
| Hgene | VIM | C0027627 | Neoplasm Metastasis | 1 | CTD_human |
| Hgene | VIM | C0027720 | Nephrosis | 1 | CTD_human |
| Hgene | VIM | C0031149 | Peritoneal Neoplasms | 1 | CTD_human |
| Hgene | VIM | C0035126 | Reperfusion Injury | 1 | CTD_human |
| Hgene | VIM | C0035309 | Retinal Diseases | 1 | CTD_human |
| Hgene | VIM | C0039101 | synovial sarcoma | 1 | CTD_human |
| Hgene | VIM | C0043094 | Weight Gain | 1 | CTD_human |
| Hgene | VIM | C0085084 | Motor Neuron Disease | 1 | CTD_human |
| Hgene | VIM | C0086543 | Cataract | 1 | CTD_human |
| Hgene | VIM | C0154681 | Anterior Horn Cell Disease | 1 | CTD_human |
| Hgene | VIM | C0154682 | Lateral Sclerosis | 1 | CTD_human |
| Hgene | VIM | C0270715 | Degenerative Diseases, Central Nervous System | 1 | CTD_human |
| Hgene | VIM | C0270763 | Familial Motor Neuron Disease | 1 | CTD_human |
| Hgene | VIM | C0270764 | Motor Neuron Disease, Lower | 1 | CTD_human |
| Hgene | VIM | C0345967 | Malignant mesothelioma | 1 | CTD_human |
| Hgene | VIM | C0346990 | Carcinomatosis of peritoneal cavity | 1 | CTD_human |
| Hgene | VIM | C0521659 | Motor Neuron Disease, Upper | 1 | CTD_human |
| Hgene | VIM | C0524524 | Pseudoaphakia | 1 | CTD_human |
| Hgene | VIM | C0524851 | Neurodegenerative Disorders | 1 | CTD_human |
| Hgene | VIM | C0543858 | Motor Neuron Disease, Secondary | 1 | CTD_human |
| Hgene | VIM | C0751733 | Degenerative Diseases, Spinal Cord | 1 | CTD_human |
| Hgene | VIM | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human |
| Hgene | VIM | C0948089 | Acute Coronary Syndrome | 1 | CTD_human |
| Hgene | VIM | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human |
| Hgene | VIM | C1510497 | Lens Opacities | 1 | CTD_human |
| Hgene | VIM | C1833118 | Cataract, Pulverulent | 1 | ORPHANET |
| Hgene | VIM | C1852438 | CATARACT, COPPOCK-LIKE | 1 | ORPHANET |
| Hgene | VIM | C1862939 | AMYOTROPHIC LATERAL SCLEROSIS 1 | 1 | CTD_human |
| Hgene | VIM | C1862941 | Amyotrophic Lateral Sclerosis, Sporadic | 1 | CTD_human |
| Hgene | VIM | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human |
| Hgene | VIM | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human |
| Hgene | VIM | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
| Hgene | VIM | C4551993 | Amyotrophic Lateral Sclerosis, Familial | 1 | CTD_human |
| Tgene | C1832370 | MYOPATHY, MYOFIBRILLAR, DESMIN-RELATED | 38 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C1858154 | CARDIOMYOPATHY, DILATED, 1I | 6 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C1867005 | Scapuloperoneal Syndrome, Neurogenic, Kaeser Type | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0686353 | Muscular Dystrophies, Limb-Girdle | 3 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C2678065 | Myofibrillar Myopathy | 3 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C0004238 | Atrial Fibrillation | 1 | CTD_human | |
| Tgene | C0006142 | Malignant neoplasm of breast | 1 | CTD_human | |
| Tgene | C0007097 | Carcinoma | 1 | CTD_human | |
| Tgene | C0015934 | Fetal Growth Retardation | 1 | CTD_human | |
| Tgene | C0017658 | Glomerulonephritis | 1 | CTD_human | |
| Tgene | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human | |
| Tgene | C0024667 | Animal Mammary Neoplasms | 1 | CTD_human | |
| Tgene | C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human | |
| Tgene | C0027720 | Nephrosis | 1 | CTD_human | |
| Tgene | C0038356 | Stomach Neoplasms | 1 | CTD_human | |
| Tgene | C0205696 | Anaplastic carcinoma | 1 | CTD_human | |
| Tgene | C0205697 | Carcinoma, Spindle-Cell | 1 | CTD_human | |
| Tgene | C0205698 | Undifferentiated carcinoma | 1 | CTD_human | |
| Tgene | C0205699 | Carcinomatosis | 1 | CTD_human | |
| Tgene | C0235480 | Paroxysmal atrial fibrillation | 1 | CTD_human | |
| Tgene | C0235833 | Congenital diaphragmatic hernia | 1 | CTD_human | |
| Tgene | C0265699 | Congenital hernia of foramen of Morgagni | 1 | CTD_human | |
| Tgene | C0265700 | Congenital hernia of foramen of Bochdalek | 1 | CTD_human | |
| Tgene | C0340427 | Familial dilated cardiomyopathy | 1 | ORPHANET | |
| Tgene | C0598608 | Hyperhomocysteinemia | 1 | CTD_human | |
| Tgene | C0678222 | Breast Carcinoma | 1 | CTD_human | |
| Tgene | C0878544 | Cardiomyopathies | 1 | GENOMICS_ENGLAND | |
| Tgene | C1257925 | Mammary Carcinoma, Animal | 1 | CTD_human | |
| Tgene | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human | |
| Tgene | C1458155 | Mammary Neoplasms | 1 | CTD_human | |
| Tgene | C1704377 | Bright Disease | 1 | CTD_human | |
| Tgene | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human | |
| Tgene | C2585653 | Persistent atrial fibrillation | 1 | CTD_human | |
| Tgene | C3148763 | MUSCULAR DYSTROPHY, LIMB-GIRDLE, TYPE 1E | 1 | ORPHANET | |
| Tgene | C3468561 | familial atrial fibrillation | 1 | CTD_human | |
| Tgene | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
