
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TOX2-EIF5A (FusionGDB2 ID:HG84969TG1984) |
Fusion Gene Summary for TOX2-EIF5A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TOX2-EIF5A | Fusion gene ID: hg84969tg1984 | Hgene | Tgene | Gene symbol | TOX2 | EIF5A | Gene ID | 84969 | 1984 |
| Gene name | TOX high mobility group box family member 2 | eukaryotic translation initiation factor 5A | |
| Synonyms | C20orf100|GCX-1|GCX1|dJ1108D11.2|dJ495O3.1 | EIF-5A|EIF5A1|eIF-4D|eIF5AI | |
| Cytomap | ('TOX2')('EIF5A') 20q13.12 | 17p13.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | TOX high mobility group box family member 2granulosa cell HMG box 1 | eukaryotic translation initiation factor 5A-1eukaryotic initiation factor 5Arev-binding factor | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | P63241 | |
| Ensembl transtripts involved in fusion gene | ENST00000341197, ENST00000358131, ENST00000372999, ENST00000423191, ENST00000435864, | ||
| Fusion gene scores | * DoF score | 5 X 5 X 3=75 | 9 X 5 X 8=360 |
| # samples | 5 | 9 | |
| ** MAII score | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/360*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TOX2 [Title/Abstract] AND EIF5A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TOX2(42635432)-EIF5A(7212934), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TOX2-EIF5A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TOX2-EIF5A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TOX2-EIF5A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. TOX2-EIF5A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TOX2 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 25352127 |
| Tgene | EIF5A | GO:0006913 | nucleocytoplasmic transport | 12210765 |
| Tgene | EIF5A | GO:0006915 | apoptotic process | 15371445|17187778 |
 Fusion gene breakpoints across TOX2 (5'-gene) Fusion gene breakpoints across TOX2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
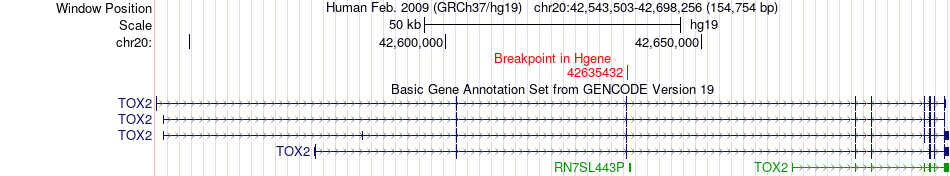 |
 Fusion gene breakpoints across EIF5A (3'-gene) Fusion gene breakpoints across EIF5A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
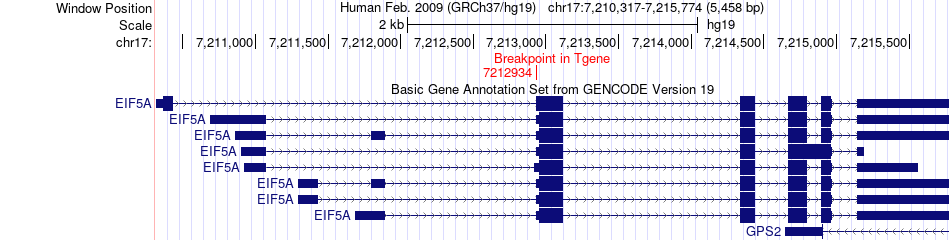 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-J4-A67N-01A | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
Top |
Fusion Gene ORF analysis for TOX2-EIF5A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000341197 | ENST00000336458 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000341197 | ENST00000416016 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000341197 | ENST00000419711 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000341197 | ENST00000571955 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000341197 | ENST00000572815 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000341197 | ENST00000573542 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000341197 | ENST00000576930 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000336458 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000416016 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000419711 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000571955 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000572815 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000573542 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000358131 | ENST00000576930 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000336458 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000416016 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000419711 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000571955 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000572815 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000573542 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000372999 | ENST00000576930 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000336458 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000416016 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000419711 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000571955 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000572815 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000573542 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| 5CDS-5UTR | ENST00000423191 | ENST00000576930 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| In-frame | ENST00000341197 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| In-frame | ENST00000358131 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| In-frame | ENST00000372999 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| In-frame | ENST00000423191 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-3CDS | ENST00000435864 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000336458 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000416016 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000419711 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000571955 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000572815 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000573542 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
| intron-5UTR | ENST00000435864 | ENST00000576930 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000341197 | TOX2 | chr20 | 42635432 | + | ENST00000336452 | EIF5A | chr17 | 7212934 | + | 1570 | 439 | 28 | 924 | 298 |
| ENST00000423191 | TOX2 | chr20 | 42635432 | + | ENST00000336452 | EIF5A | chr17 | 7212934 | + | 1502 | 371 | 86 | 856 | 256 |
| ENST00000372999 | TOX2 | chr20 | 42635432 | + | ENST00000336452 | EIF5A | chr17 | 7212934 | + | 1610 | 479 | 140 | 964 | 274 |
| ENST00000358131 | TOX2 | chr20 | 42635432 | + | ENST00000336452 | EIF5A | chr17 | 7212934 | + | 1777 | 646 | 169 | 1131 | 320 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000341197 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + | 0.001130666 | 0.99886936 |
| ENST00000423191 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + | 0.006505524 | 0.99349445 |
| ENST00000372999 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + | 0.006356164 | 0.9936439 |
| ENST00000358131 | ENST00000336452 | TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212934 | + | 0.003068025 | 0.9969319 |
Top |
Fusion Genomic Features for TOX2-EIF5A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212933 | + | 5.09E-08 | 1 |
| TOX2 | chr20 | 42635432 | + | EIF5A | chr17 | 7212933 | + | 5.09E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
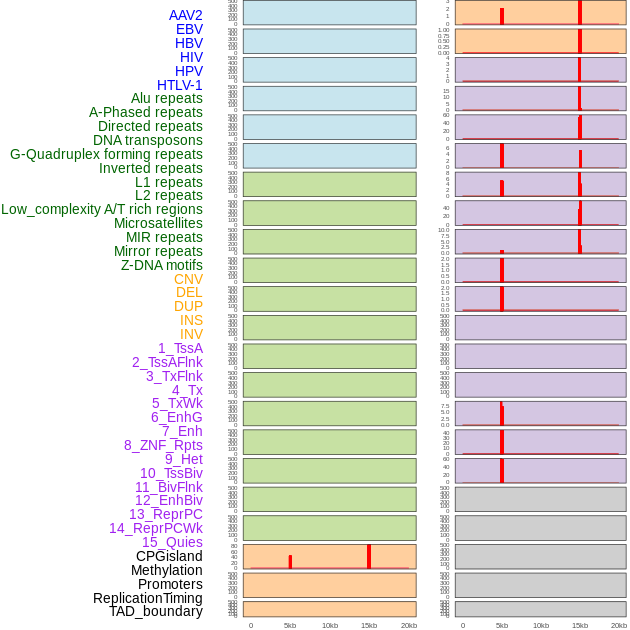 |
Top |
Fusion Protein Features for TOX2-EIF5A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:42635432/chr17:7212934) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | EIF5A |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: mRNA-binding protein involved in translation elongation. Has an important function at the level of mRNA turnover, probably acting downstream of decapping. Involved in actin dynamics and cell cycle progression, mRNA decay and probably in a pathway involved in stress response and maintenance of cell wall integrity. With syntenin SDCBP, functions as a regulator of p53/TP53 and p53/TP53-dependent apoptosis. Regulates also TNF-alpha-mediated apoptosis. Mediates effects of polyamines on neuronal process extension and survival. May play an important role in brain development and function, and in skeletal muscle stem cell differentiation. Also described as a cellular cofactor of human T-cell leukemia virus type I (HTLV-1) Rex protein and of human immunodeficiency virus type 1 (HIV-1) Rev protein, essential for mRNA export of retroviral transcripts. {ECO:0000269|PubMed:15371445, ECO:0000269|PubMed:15452064, ECO:0000269|PubMed:16987817, ECO:0000269|PubMed:17187778, ECO:0000269|PubMed:17360499}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000341197 | + | 3 | 9 | 76_114 | 137 | 507.0 | Region | Required for transcriptional activation |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000358131 | + | 3 | 8 | 76_114 | 146 | 489.0 | Region | Required for transcriptional activation |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000336452 | 0 | 6 | 20_90 | 23 | 357.3333333333333 | Region | Note=DOHH-binding | |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000336458 | 0 | 6 | 20_90 | 0 | 239.66666666666666 | Region | Note=DOHH-binding | |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000416016 | 0 | 6 | 20_90 | 0 | 298.0 | Region | Note=DOHH-binding | |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000419711 | 0 | 6 | 20_90 | 0 | 319.6666666666667 | Region | Note=DOHH-binding | |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000571955 | 1 | 7 | 20_90 | 0 | 319.6666666666667 | Region | Note=DOHH-binding | |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000573542 | 0 | 6 | 20_90 | 0 | 245.66666666666666 | Region | Note=DOHH-binding | |
| Tgene | EIF5A | chr20:42635432 | chr17:7212934 | ENST00000576930 | 1 | 7 | 20_90 | 0 | 295.3333333333333 | Region | Note=DOHH-binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000341197 | + | 3 | 9 | 245_250 | 137 | 507.0 | Compositional bias | Note=Poly-Lys |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000341197 | + | 3 | 9 | 372_456 | 137 | 507.0 | Compositional bias | Note=Pro-rich |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000358131 | + | 3 | 8 | 245_250 | 146 | 489.0 | Compositional bias | Note=Poly-Lys |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000358131 | + | 3 | 8 | 372_456 | 146 | 489.0 | Compositional bias | Note=Pro-rich |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000372999 | + | 4 | 10 | 245_250 | 95 | 465.0 | Compositional bias | Note=Poly-Lys |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000372999 | + | 4 | 10 | 372_456 | 95 | 465.0 | Compositional bias | Note=Pro-rich |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000423191 | + | 3 | 9 | 245_250 | 95 | 465.0 | Compositional bias | Note=Poly-Lys |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000423191 | + | 3 | 9 | 372_456 | 95 | 465.0 | Compositional bias | Note=Pro-rich |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000341197 | + | 3 | 9 | 255_323 | 137 | 507.0 | DNA binding | HMG box |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000358131 | + | 3 | 8 | 255_323 | 146 | 489.0 | DNA binding | HMG box |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000372999 | + | 4 | 10 | 255_323 | 95 | 465.0 | DNA binding | HMG box |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000423191 | + | 3 | 9 | 255_323 | 95 | 465.0 | DNA binding | HMG box |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000341197 | + | 3 | 9 | 223_252 | 137 | 507.0 | Motif | Nuclear localization signal |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000358131 | + | 3 | 8 | 223_252 | 146 | 489.0 | Motif | Nuclear localization signal |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000372999 | + | 4 | 10 | 223_252 | 95 | 465.0 | Motif | Nuclear localization signal |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000423191 | + | 3 | 9 | 223_252 | 95 | 465.0 | Motif | Nuclear localization signal |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000372999 | + | 4 | 10 | 76_114 | 95 | 465.0 | Region | Required for transcriptional activation |
| Hgene | TOX2 | chr20:42635432 | chr17:7212934 | ENST00000423191 | + | 3 | 9 | 76_114 | 95 | 465.0 | Region | Required for transcriptional activation |
Top |
Fusion Gene Sequence for TOX2-EIF5A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >93149_93149_1_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000341197_EIF5A_chr17_7212934_ENST00000336452_length(transcript)=1570nt_BP=439nt AGCCGCCGCCGCCGCCGCCGCGCCCGCCATGGACGTCCGCCTGTACCCCTCGGCGCCCGCGGTGGGCGCGCGGCCCGGGGCCGAGCCGGC CGGCCTGGCGCACCTGGACTATTACCACGGCGGCAAGTTTGATGGTGACAGTGCCTACGTGGGGATGAGTGACGGAAACCCAGAGCTCCT GTCAACCAGCCAGACCTACAACGGCCAGAGCGAGAACAACGAAGACTATGAGATCCCCCCGATAACACCTCCCAACCTCCCGGAGCCATC CCTCCTGCACCTGGGGGACCACGAAGCCAGCTACCACTCGCTGTGCCACGGCCTCACCCCCAACGGTCTGCTCCCTGCCTACTCCTATCA GGCCATGGACCTCCCAGCCATCATGGTGTCCAACATGCTAGCACAGGACAGCCACCTGCTGTCGGGCCAGCTGCCCACGTTGGAATCGAA GCCTCTTAAAATGGCAGATGACTTGGACTTCGAGACAGGAGATGCAGGGGCCTCAGCCACCTTCCCAATGCAGTGCTCAGCATTACGTAA GAATGGCTTTGTGGTGCTCAAAGGCCGGCCATGTAAGATCGTCGAGATGTCTACTTCGAAGACTGGCAAGCACGGCCACGCCAAGGTCCA TCTGGTTGGTATTGACATCTTTACTGGGAAGAAATATGAAGATATCTGCCCGTCAACTCATAATATGGATGTCCCCAACATCAAAAGGAA TGACTTCCAGCTGATTGGCATCCAGGATGGGTACCTATCACTGCTCCAGGACAGCGGGGAGGTACGAGAGGACCTTCGTCTCCCTGAGGG AGACCTTGGCAAGGAGATTGAGCAGAAGTACGACTGTGGAGAAGAGATCCTGATCACGGTGCTGTCTGCCATGACAGAGGAGGCAGCTGT TGCAATCAAGGCCATGGCAAAATAACTGGCTCCCAGGGTGGCGGTGGTGGCAGCAGTGATCCTCTGAACCTGCAGAGGCCCCCTCCCCGA GCCTGGCCTGGCTCTGGCCCGGTCCTAAGCTGGACTCCTCCTACACAATTTATTTGACGTTTTATTTTGGTTTTCCCCACCCCCTCAATC TGTCGGGGAGCCCCTGCCCTTCACCTAGCTCCCTTGGCCAGGAGCGAGCGAAGCTGTGGCCTTGGTGAAGCTGCCCTCCTCTTCTCCCCT CACACTACAGCCCTGGTGGGGGAGAAGGGGGTGGGTGCTGCTTGTGGTTTAGTCTTTTTTTTTTTTTTTTTTTTAATTCAATCTGGAATC AGAAAGCGGTGGATTCTGGCAAATGGTCCTTGTGCCCTCCCCACTCATCCCTGGTCTGGTCCCCTGTTGCCTATAGCCCTTTACCCTGAG CACCACCCCAACAGACTGGGGACCAGCCCCCTCGCCTGCCTGTGTCTCTCCCCAAACCCCTTTAGATGGGGAGGGAAGAGGAGGAGAGGG GAGGGGACCTGCCCCCTCCTCAGGCATCTGGGAGGGCCCTGCCCCCATGGGCTTTACCCTTCCCTGCGGGCTCTCTCCCCGACACATTTG >93149_93149_1_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000341197_EIF5A_chr17_7212934_ENST00000336452_length(amino acids)=298AA_BP=137 MDVRLYPSAPAVGARPGAEPAGLAHLDYYHGGKFDGDSAYVGMSDGNPELLSTSQTYNGQSENNEDYEIPPITPPNLPEPSLLHLGDHEA SYHSLCHGLTPNGLLPAYSYQAMDLPAIMVSNMLAQDSHLLSGQLPTLESKPLKMADDLDFETGDAGASATFPMQCSALRKNGFVVLKGR PCKIVEMSTSKTGKHGHAKVHLVGIDIFTGKKYEDICPSTHNMDVPNIKRNDFQLIGIQDGYLSLLQDSGEVREDLRLPEGDLGKEIEQK -------------------------------------------------------------- >93149_93149_2_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000358131_EIF5A_chr17_7212934_ENST00000336452_length(transcript)=1777nt_BP=646nt ACCAGGAGCTCCGAACACGCAGGCCCAAATTGGAATAGGAGATGGATTTTGATAAAAGAGGTTAAATAACAGGTTTTTCCATTTCACCTT ATACGAATGGGATGGGGGGTGGGGGTGCTGGGCCTCCAGCCAATAGAGGACCAAGAGTTGTCAGGAGGGCTTATAAAGCTTGCTATTTCT GGCATTTTTTCCTCTTTCTCTGCTGATTATGCAGCAGACTCGCACAGAGGCTGTCGCGGGCGCGTTCTCTCGCTGCCTGGGCTTCTGTGG AATGAGACTCGGGCTCCTTCTACTTGCAAGACACTGGTGCATTGCAGGTGTGTTTCCGCAGAAGTTTGATGGTGACAGTGCCTACGTGGG GATGAGTGACGGAAACCCAGAGCTCCTGTCAACCAGCCAGACCTACAACGGCCAGAGCGAGAACAACGAAGACTATGAGATCCCCCCGAT AACACCTCCCAACCTCCCGGAGCCATCCCTCCTGCACCTGGGGGACCACGAAGCCAGCTACCACTCGCTGTGCCACGGCCTCACCCCCAA CGGTCTGCTCCCTGCCTACTCCTATCAGGCCATGGACCTCCCAGCCATCATGGTGTCCAACATGCTAGCACAGGACAGCCACCTGCTGTC GGGCCAGCTGCCCACGTTGGAATCGAAGCCTCTTAAAATGGCAGATGACTTGGACTTCGAGACAGGAGATGCAGGGGCCTCAGCCACCTT CCCAATGCAGTGCTCAGCATTACGTAAGAATGGCTTTGTGGTGCTCAAAGGCCGGCCATGTAAGATCGTCGAGATGTCTACTTCGAAGAC TGGCAAGCACGGCCACGCCAAGGTCCATCTGGTTGGTATTGACATCTTTACTGGGAAGAAATATGAAGATATCTGCCCGTCAACTCATAA TATGGATGTCCCCAACATCAAAAGGAATGACTTCCAGCTGATTGGCATCCAGGATGGGTACCTATCACTGCTCCAGGACAGCGGGGAGGT ACGAGAGGACCTTCGTCTCCCTGAGGGAGACCTTGGCAAGGAGATTGAGCAGAAGTACGACTGTGGAGAAGAGATCCTGATCACGGTGCT GTCTGCCATGACAGAGGAGGCAGCTGTTGCAATCAAGGCCATGGCAAAATAACTGGCTCCCAGGGTGGCGGTGGTGGCAGCAGTGATCCT CTGAACCTGCAGAGGCCCCCTCCCCGAGCCTGGCCTGGCTCTGGCCCGGTCCTAAGCTGGACTCCTCCTACACAATTTATTTGACGTTTT ATTTTGGTTTTCCCCACCCCCTCAATCTGTCGGGGAGCCCCTGCCCTTCACCTAGCTCCCTTGGCCAGGAGCGAGCGAAGCTGTGGCCTT GGTGAAGCTGCCCTCCTCTTCTCCCCTCACACTACAGCCCTGGTGGGGGAGAAGGGGGTGGGTGCTGCTTGTGGTTTAGTCTTTTTTTTT TTTTTTTTTTTAATTCAATCTGGAATCAGAAAGCGGTGGATTCTGGCAAATGGTCCTTGTGCCCTCCCCACTCATCCCTGGTCTGGTCCC CTGTTGCCTATAGCCCTTTACCCTGAGCACCACCCCAACAGACTGGGGACCAGCCCCCTCGCCTGCCTGTGTCTCTCCCCAAACCCCTTT AGATGGGGAGGGAAGAGGAGGAGAGGGGAGGGGACCTGCCCCCTCCTCAGGCATCTGGGAGGGCCCTGCCCCCATGGGCTTTACCCTTCC >93149_93149_2_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000358131_EIF5A_chr17_7212934_ENST00000336452_length(amino acids)=320AA_BP=159 MLFLAFFPLSLLIMQQTRTEAVAGAFSRCLGFCGMRLGLLLLARHWCIAGVFPQKFDGDSAYVGMSDGNPELLSTSQTYNGQSENNEDYE IPPITPPNLPEPSLLHLGDHEASYHSLCHGLTPNGLLPAYSYQAMDLPAIMVSNMLAQDSHLLSGQLPTLESKPLKMADDLDFETGDAGA SATFPMQCSALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVGIDIFTGKKYEDICPSTHNMDVPNIKRNDFQLIGIQDGYLSLLQD -------------------------------------------------------------- >93149_93149_3_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000372999_EIF5A_chr17_7212934_ENST00000336452_length(transcript)=1610nt_BP=479nt CGCAGCCCTGGCGCAGACGCGTGGGCTCCGTGGCGATGCGGGGTGTTGCCTGAGGCTCCACTGAAGCTATGGCATAATTTGCAGAATTTG CACTTCATTACTTTTCTGAAATTCAAACAAATTCTGAAACTGCACGAGTTCTGGCTGAGAGCTGTGGATCTGTGCATTTTGATGGTGACA GTGCCTACGTGGGGATGAGTGACGGAAACCCAGAGCTCCTGTCAACCAGCCAGACCTACAACGGCCAGAGCGAGAACAACGAAGACTATG AGATCCCCCCGATAACACCTCCCAACCTCCCGGAGCCATCCCTCCTGCACCTGGGGGACCACGAAGCCAGCTACCACTCGCTGTGCCACG GCCTCACCCCCAACGGTCTGCTCCCTGCCTACTCCTATCAGGCCATGGACCTCCCAGCCATCATGGTGTCCAACATGCTAGCACAGGACA GCCACCTGCTGTCGGGCCAGCTGCCCACGTTGGAATCGAAGCCTCTTAAAATGGCAGATGACTTGGACTTCGAGACAGGAGATGCAGGGG CCTCAGCCACCTTCCCAATGCAGTGCTCAGCATTACGTAAGAATGGCTTTGTGGTGCTCAAAGGCCGGCCATGTAAGATCGTCGAGATGT CTACTTCGAAGACTGGCAAGCACGGCCACGCCAAGGTCCATCTGGTTGGTATTGACATCTTTACTGGGAAGAAATATGAAGATATCTGCC CGTCAACTCATAATATGGATGTCCCCAACATCAAAAGGAATGACTTCCAGCTGATTGGCATCCAGGATGGGTACCTATCACTGCTCCAGG ACAGCGGGGAGGTACGAGAGGACCTTCGTCTCCCTGAGGGAGACCTTGGCAAGGAGATTGAGCAGAAGTACGACTGTGGAGAAGAGATCC TGATCACGGTGCTGTCTGCCATGACAGAGGAGGCAGCTGTTGCAATCAAGGCCATGGCAAAATAACTGGCTCCCAGGGTGGCGGTGGTGG CAGCAGTGATCCTCTGAACCTGCAGAGGCCCCCTCCCCGAGCCTGGCCTGGCTCTGGCCCGGTCCTAAGCTGGACTCCTCCTACACAATT TATTTGACGTTTTATTTTGGTTTTCCCCACCCCCTCAATCTGTCGGGGAGCCCCTGCCCTTCACCTAGCTCCCTTGGCCAGGAGCGAGCG AAGCTGTGGCCTTGGTGAAGCTGCCCTCCTCTTCTCCCCTCACACTACAGCCCTGGTGGGGGAGAAGGGGGTGGGTGCTGCTTGTGGTTT AGTCTTTTTTTTTTTTTTTTTTTTAATTCAATCTGGAATCAGAAAGCGGTGGATTCTGGCAAATGGTCCTTGTGCCCTCCCCACTCATCC CTGGTCTGGTCCCCTGTTGCCTATAGCCCTTTACCCTGAGCACCACCCCAACAGACTGGGGACCAGCCCCCTCGCCTGCCTGTGTCTCTC CCCAAACCCCTTTAGATGGGGAGGGAAGAGGAGGAGAGGGGAGGGGACCTGCCCCCTCCTCAGGCATCTGGGAGGGCCCTGCCCCCATGG >93149_93149_3_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000372999_EIF5A_chr17_7212934_ENST00000336452_length(amino acids)=274AA_BP=113 MAESCGSVHFDGDSAYVGMSDGNPELLSTSQTYNGQSENNEDYEIPPITPPNLPEPSLLHLGDHEASYHSLCHGLTPNGLLPAYSYQAMD LPAIMVSNMLAQDSHLLSGQLPTLESKPLKMADDLDFETGDAGASATFPMQCSALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVG IDIFTGKKYEDICPSTHNMDVPNIKRNDFQLIGIQDGYLSLLQDSGEVREDLRLPEGDLGKEIEQKYDCGEEILITVLSAMTEEAAVAIK -------------------------------------------------------------- >93149_93149_4_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000423191_EIF5A_chr17_7212934_ENST00000336452_length(transcript)=1502nt_BP=371nt AGCGCGCGTGGGTTTCCCGCAGCCCTGGCGCAGACGCGTGGGCTCCGTGGCGATGCGGGTTTGATGGTGACAGTGCCTACGTGGGGATGA GTGACGGAAACCCAGAGCTCCTGTCAACCAGCCAGACCTACAACGGCCAGAGCGAGAACAACGAAGACTATGAGATCCCCCCGATAACAC CTCCCAACCTCCCGGAGCCATCCCTCCTGCACCTGGGGGACCACGAAGCCAGCTACCACTCGCTGTGCCACGGCCTCACCCCCAACGGTC TGCTCCCTGCCTACTCCTATCAGGCCATGGACCTCCCAGCCATCATGGTGTCCAACATGCTAGCACAGGACAGCCACCTGCTGTCGGGCC AGCTGCCCACGTTGGAATCGAAGCCTCTTAAAATGGCAGATGACTTGGACTTCGAGACAGGAGATGCAGGGGCCTCAGCCACCTTCCCAA TGCAGTGCTCAGCATTACGTAAGAATGGCTTTGTGGTGCTCAAAGGCCGGCCATGTAAGATCGTCGAGATGTCTACTTCGAAGACTGGCA AGCACGGCCACGCCAAGGTCCATCTGGTTGGTATTGACATCTTTACTGGGAAGAAATATGAAGATATCTGCCCGTCAACTCATAATATGG ATGTCCCCAACATCAAAAGGAATGACTTCCAGCTGATTGGCATCCAGGATGGGTACCTATCACTGCTCCAGGACAGCGGGGAGGTACGAG AGGACCTTCGTCTCCCTGAGGGAGACCTTGGCAAGGAGATTGAGCAGAAGTACGACTGTGGAGAAGAGATCCTGATCACGGTGCTGTCTG CCATGACAGAGGAGGCAGCTGTTGCAATCAAGGCCATGGCAAAATAACTGGCTCCCAGGGTGGCGGTGGTGGCAGCAGTGATCCTCTGAA CCTGCAGAGGCCCCCTCCCCGAGCCTGGCCTGGCTCTGGCCCGGTCCTAAGCTGGACTCCTCCTACACAATTTATTTGACGTTTTATTTT GGTTTTCCCCACCCCCTCAATCTGTCGGGGAGCCCCTGCCCTTCACCTAGCTCCCTTGGCCAGGAGCGAGCGAAGCTGTGGCCTTGGTGA AGCTGCCCTCCTCTTCTCCCCTCACACTACAGCCCTGGTGGGGGAGAAGGGGGTGGGTGCTGCTTGTGGTTTAGTCTTTTTTTTTTTTTT TTTTTTAATTCAATCTGGAATCAGAAAGCGGTGGATTCTGGCAAATGGTCCTTGTGCCCTCCCCACTCATCCCTGGTCTGGTCCCCTGTT GCCTATAGCCCTTTACCCTGAGCACCACCCCAACAGACTGGGGACCAGCCCCCTCGCCTGCCTGTGTCTCTCCCCAAACCCCTTTAGATG GGGAGGGAAGAGGAGGAGAGGGGAGGGGACCTGCCCCCTCCTCAGGCATCTGGGAGGGCCCTGCCCCCATGGGCTTTACCCTTCCCTGCG >93149_93149_4_TOX2-EIF5A_TOX2_chr20_42635432_ENST00000423191_EIF5A_chr17_7212934_ENST00000336452_length(amino acids)=256AA_BP=95 MSDGNPELLSTSQTYNGQSENNEDYEIPPITPPNLPEPSLLHLGDHEASYHSLCHGLTPNGLLPAYSYQAMDLPAIMVSNMLAQDSHLLS GQLPTLESKPLKMADDLDFETGDAGASATFPMQCSALRKNGFVVLKGRPCKIVEMSTSKTGKHGHAKVHLVGIDIFTGKKYEDICPSTHN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TOX2-EIF5A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TOX2-EIF5A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TOX2-EIF5A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C0007097 | Carcinoma | 1 | CTD_human | |
| Tgene | C0022548 | Keloid | 1 | CTD_human | |
| Tgene | C0024667 | Animal Mammary Neoplasms | 1 | CTD_human | |
| Tgene | C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human | |
| Tgene | C0205696 | Anaplastic carcinoma | 1 | CTD_human | |
| Tgene | C0205697 | Carcinoma, Spindle-Cell | 1 | CTD_human | |
| Tgene | C0205698 | Undifferentiated carcinoma | 1 | CTD_human | |
| Tgene | C0205699 | Carcinomatosis | 1 | CTD_human | |
| Tgene | C1257925 | Mammary Carcinoma, Animal | 1 | CTD_human |
