
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ASMTL-CLEC7A (FusionGDB2 ID:HG8623TG64581) |
Fusion Gene Summary for ASMTL-CLEC7A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ASMTL-CLEC7A | Fusion gene ID: hg8623tg64581 | Hgene | Tgene | Gene symbol | ASMTL | CLEC7A | Gene ID | 8623 | 64581 |
| Gene name | acetylserotonin O-methyltransferase like | C-type lectin domain containing 7A | |
| Synonyms | ASMTLX|ASMTLY|ASTML | BGR|CANDF4|CD369|CLECSF12|DECTIN1|SCARE2 | |
| Cytomap | ('ASMTL')('CLEC7A') Xp22.33 and Yp11.2 | 12p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | probable bifunctional dTTP/UTP pyrophosphatase/methyltransferase proteinN-acetylserotonin O-methyltransferase-like proteinacetylserotonin N-methyltransferase-like | C-type lectin domain family 7 member AC-type (calcium dependent, carbohydrate-recognition domain) lectin, superfamily member 12C-type lectin superfamily member 12DC-associated C-type lectin 1beta-glucan receptordectin-1dendritic cell-associated C-ty | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O95671 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000381317, ENST00000381333, ENST00000416733, ENST00000534940, ENST00000463763, | ||
| Fusion gene scores | * DoF score | 15 X 11 X 4=660 | 1 X 1 X 1=1 |
| # samples | 10 | 2 | |
| ** MAII score | log2(10/660*10)=-2.72246602447109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/1*10)=4.32192809488736 | |
| Context | PubMed: ASMTL [Title/Abstract] AND CLEC7A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ASMTL(1540550)-CLEC7A(10271189), # samples:1 ASMTL(1540551)-CLEC7A(10271189), # samples:1 ASMTL(1490551)-CLEC7A(10271189), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ASMTL-CLEC7A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ASMTL-CLEC7A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ASMTL-CLEC7A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ASMTL-CLEC7A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ASMTL-CLEC7A seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CLEC7A | GO:0006910 | phagocytosis, recognition | 11567029 |
| Tgene | CLEC7A | GO:0008037 | cell recognition | 11567029 |
| Tgene | CLEC7A | GO:0010628 | positive regulation of gene expression | 19358895 |
| Tgene | CLEC7A | GO:0016046 | detection of fungus | 19358895 |
| Tgene | CLEC7A | GO:0032491 | detection of molecule of fungal origin | 19358895 |
| Tgene | CLEC7A | GO:0032731 | positive regulation of interleukin-1 beta production | 22267217 |
| Tgene | CLEC7A | GO:0032930 | positive regulation of superoxide anion generation | 19358895 |
| Tgene | CLEC7A | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0045078 | positive regulation of interferon-gamma biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0045082 | positive regulation of interleukin-10 biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0045084 | positive regulation of interleukin-12 biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0045086 | positive regulation of interleukin-2 biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0045410 | positive regulation of interleukin-6 biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 19358895 |
| Tgene | CLEC7A | GO:0050766 | positive regulation of phagocytosis | 19358895 |
| Tgene | CLEC7A | GO:0051251 | positive regulation of lymphocyte activation | 19358895 |
| Tgene | CLEC7A | GO:0051712 | positive regulation of killing of cells of other organism | 19358895 |
| Tgene | CLEC7A | GO:0060267 | positive regulation of respiratory burst | 19358895 |
| Tgene | CLEC7A | GO:0071226 | cellular response to molecule of fungal origin | 22267217 |
| Tgene | CLEC7A | GO:1900017 | positive regulation of cytokine production involved in inflammatory response | 22267217 |
| Tgene | CLEC7A | GO:1903431 | positive regulation of cell maturation | 19358895 |
 Fusion gene breakpoints across ASMTL (5'-gene) Fusion gene breakpoints across ASMTL (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
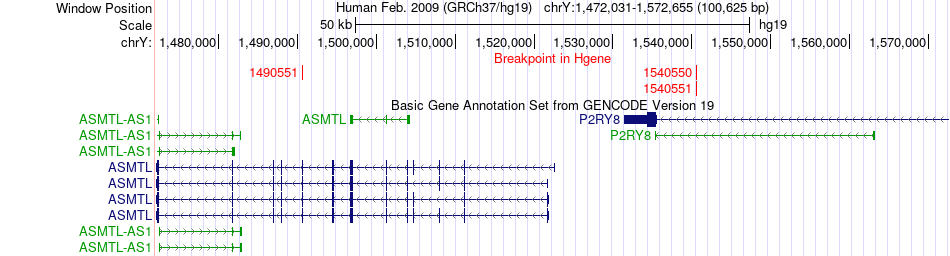 |
 Fusion gene breakpoints across CLEC7A (3'-gene) Fusion gene breakpoints across CLEC7A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
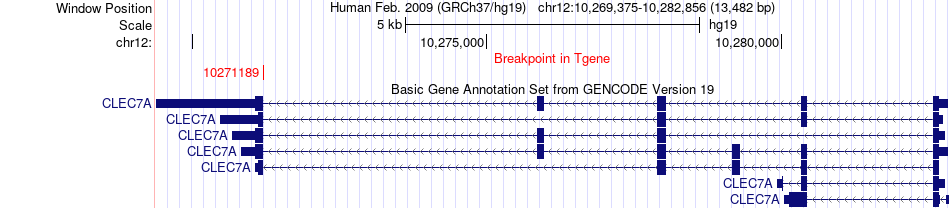 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-0908-01B | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| ChimerDB4 | OV | TCGA-13-0908-01B | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| ChimerDB4 | OV | TCGA-13-0908 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
Top |
Fusion Gene ORF analysis for ASMTL-CLEC7A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000381317 | ENST00000310002 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000381317 | ENST00000525605 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000381333 | ENST00000310002 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000381333 | ENST00000525605 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000416733 | ENST00000310002 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000416733 | ENST00000525605 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000534940 | ENST00000310002 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| 5CDS-intron | ENST00000534940 | ENST00000525605 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381317 | ENST00000298523 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381317 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381317 | ENST00000396484 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381317 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381333 | ENST00000298523 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381333 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381333 | ENST00000396484 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000381333 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000416733 | ENST00000298523 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000416733 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000416733 | ENST00000396484 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000534940 | ENST00000298523 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000534940 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| Frame-shift | ENST00000534940 | ENST00000396484 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| In-frame | ENST00000381317 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| In-frame | ENST00000381333 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| In-frame | ENST00000416733 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| In-frame | ENST00000416733 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| In-frame | ENST00000534940 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| In-frame | ENST00000534940 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000298523 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000298523 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000304084 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000304084 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000353231 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000353231 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000396484 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000396484 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000533022 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381317 | ENST00000533022 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000298523 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000298523 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000304084 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000304084 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000353231 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000353231 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000396484 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000396484 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000533022 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000381333 | ENST00000533022 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000298523 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000298523 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000304084 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000304084 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000353231 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000353231 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000396484 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000396484 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000533022 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000416733 | ENST00000533022 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000298523 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000298523 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000298523 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000304084 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000304084 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000353231 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000353231 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000396484 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000396484 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000396484 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000533022 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000463763 | ENST00000533022 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000298523 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000298523 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000304084 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000304084 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000353231 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000353231 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000396484 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000396484 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000533022 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-3CDS | ENST00000534940 | ENST00000533022 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381317 | ENST00000310002 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381317 | ENST00000310002 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381317 | ENST00000525605 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381317 | ENST00000525605 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381333 | ENST00000310002 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381333 | ENST00000310002 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381333 | ENST00000525605 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000381333 | ENST00000525605 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000416733 | ENST00000310002 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000416733 | ENST00000310002 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000416733 | ENST00000525605 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000416733 | ENST00000525605 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000463763 | ENST00000310002 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000463763 | ENST00000310002 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000463763 | ENST00000310002 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000463763 | ENST00000525605 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000463763 | ENST00000525605 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000463763 | ENST00000525605 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000534940 | ENST00000310002 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000534940 | ENST00000310002 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000534940 | ENST00000525605 | ASMTL | chrX | 1540551 | - | CLEC7A | chr12 | 10271189 | - |
| intron-intron | ENST00000534940 | ENST00000525605 | ASMTL | chrX | 1540550 | - | CLEC7A | chr12 | 10271189 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000416733 | ASMTL | chrY | 1490551 | - | ENST00000304084 | CLEC7A | chr12 | 10271189 | - | 1579 | 1215 | 54 | 1292 | 412 |
| ENST00000416733 | ASMTL | chrY | 1490551 | - | ENST00000533022 | CLEC7A | chr12 | 10271189 | - | 1348 | 1215 | 54 | 1292 | 412 |
| ENST00000534940 | ASMTL | chrY | 1490551 | - | ENST00000304084 | CLEC7A | chr12 | 10271189 | - | 1661 | 1297 | 187 | 1374 | 395 |
| ENST00000534940 | ASMTL | chrY | 1490551 | - | ENST00000533022 | CLEC7A | chr12 | 10271189 | - | 1430 | 1297 | 187 | 1374 | 395 |
| ENST00000381317 | ASMTL | chrY | 1490551 | - | ENST00000353231 | CLEC7A | chr12 | 10271189 | - | 3092 | 1278 | 33 | 1355 | 440 |
| ENST00000381333 | ASMTL | chrY | 1490551 | - | ENST00000353231 | CLEC7A | chr12 | 10271189 | - | 3100 | 1286 | 8 | 1363 | 451 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000416733 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - | 0.007565234 | 0.99243474 |
| ENST00000416733 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - | 0.010999179 | 0.9890008 |
| ENST00000534940 | ENST00000304084 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - | 0.004784044 | 0.995216 |
| ENST00000534940 | ENST00000533022 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - | 0.005895723 | 0.9941043 |
| ENST00000381317 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - | 0.000780121 | 0.9992199 |
| ENST00000381333 | ENST00000353231 | ASMTL | chrY | 1490551 | - | CLEC7A | chr12 | 10271189 | - | 0.001123858 | 0.99887615 |
Top |
Fusion Genomic Features for ASMTL-CLEC7A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
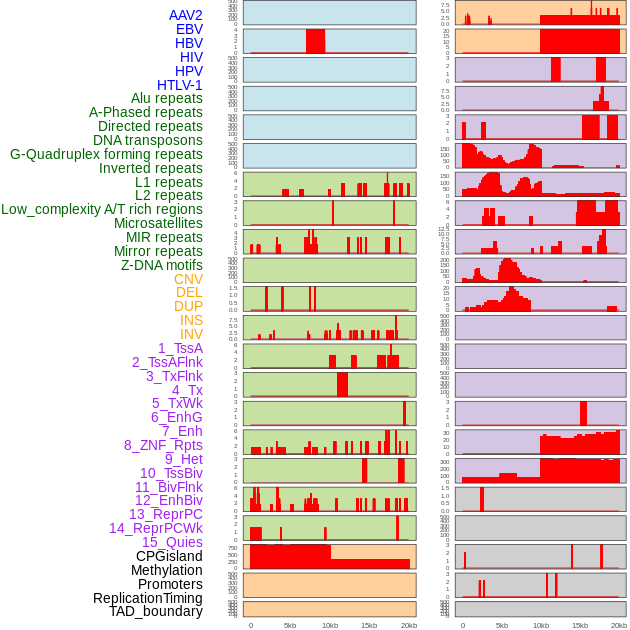 |
Top |
Fusion Protein Features for ASMTL-CLEC7A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrY:1540550/chr12:10271189) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ASMTL | . |
| FUNCTION: Nucleoside triphosphate pyrophosphatase that hydrolyzes dTTP and UTP. Can also hydrolyze CTP and the modified nucleotides pseudo-UTP, 5-methyl-UTP (m(5)UTP) and 5-methyl-CTP (m(5)CTP). Has weak activity with dCTP, 8-oxo-GTP and N(4)-methyl-dCTP (PubMed:24210219). May have a dual role in cell division arrest and in preventing the incorporation of modified nucleotides into cellular nucleic acids (PubMed:24210219). In addition, the presence of the putative catalytic domain of S-adenosyl-L-methionine binding in the C-terminal region argues for a methyltransferase activity (Probable). {ECO:0000269|PubMed:24210219, ECO:0000305}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000381317 | - | 9 | 13 | 11_223 | 415 | 622.0 | Region | Note=MAF-like |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000381333 | - | 8 | 12 | 11_223 | 399 | 606.0 | Region | Note=MAF-like |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000534940 | - | 9 | 13 | 11_223 | 357 | 564.0 | Region | Note=MAF-like |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000298523 | 2 | 4 | 127_242 | 118 | 144.0 | Domain | C-type lectin | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000310002 | 0 | 3 | 127_242 | 0 | 78.0 | Domain | C-type lectin | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000396484 | 2 | 4 | 127_242 | 124 | 169.0 | Domain | C-type lectin | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000310002 | 0 | 3 | 15_18 | 0 | 78.0 | Motif | Note=ITAM-like | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000310002 | 0 | 3 | 1_44 | 0 | 78.0 | Topological domain | Cytoplasmic | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000310002 | 0 | 3 | 66_247 | 0 | 78.0 | Topological domain | Extracellular | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000310002 | 0 | 3 | 45_65 | 0 | 78.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000381317 | - | 9 | 13 | 277_621 | 415 | 622.0 | Region | Note=ASMT-like |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000381317 | - | 9 | 13 | 508_510 | 415 | 622.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000381333 | - | 8 | 12 | 277_621 | 399 | 606.0 | Region | Note=ASMT-like |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000381333 | - | 8 | 12 | 508_510 | 399 | 606.0 | Region | S-adenosyl-L-methionine binding |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000534940 | - | 9 | 13 | 277_621 | 357 | 564.0 | Region | Note=ASMT-like |
| Hgene | ASMTL | chrY:1490551 | chr12:10271189 | ENST00000534940 | - | 9 | 13 | 508_510 | 357 | 564.0 | Region | S-adenosyl-L-methionine binding |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000304084 | 4 | 6 | 127_242 | 203 | 248.0 | Domain | C-type lectin | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000353231 | 3 | 5 | 127_242 | 157 | 202.0 | Domain | C-type lectin | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000533022 | 3 | 5 | 127_242 | 164 | 190.0 | Domain | C-type lectin | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000298523 | 2 | 4 | 15_18 | 118 | 144.0 | Motif | Note=ITAM-like | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000304084 | 4 | 6 | 15_18 | 203 | 248.0 | Motif | Note=ITAM-like | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000353231 | 3 | 5 | 15_18 | 157 | 202.0 | Motif | Note=ITAM-like | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000396484 | 2 | 4 | 15_18 | 124 | 169.0 | Motif | Note=ITAM-like | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000533022 | 3 | 5 | 15_18 | 164 | 190.0 | Motif | Note=ITAM-like | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000298523 | 2 | 4 | 1_44 | 118 | 144.0 | Topological domain | Cytoplasmic | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000298523 | 2 | 4 | 66_247 | 118 | 144.0 | Topological domain | Extracellular | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000304084 | 4 | 6 | 1_44 | 203 | 248.0 | Topological domain | Cytoplasmic | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000304084 | 4 | 6 | 66_247 | 203 | 248.0 | Topological domain | Extracellular | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000353231 | 3 | 5 | 1_44 | 157 | 202.0 | Topological domain | Cytoplasmic | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000353231 | 3 | 5 | 66_247 | 157 | 202.0 | Topological domain | Extracellular | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000396484 | 2 | 4 | 1_44 | 124 | 169.0 | Topological domain | Cytoplasmic | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000396484 | 2 | 4 | 66_247 | 124 | 169.0 | Topological domain | Extracellular | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000533022 | 3 | 5 | 1_44 | 164 | 190.0 | Topological domain | Cytoplasmic | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000533022 | 3 | 5 | 66_247 | 164 | 190.0 | Topological domain | Extracellular | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000298523 | 2 | 4 | 45_65 | 118 | 144.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000304084 | 4 | 6 | 45_65 | 203 | 248.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000353231 | 3 | 5 | 45_65 | 157 | 202.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000396484 | 2 | 4 | 45_65 | 124 | 169.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | CLEC7A | chrY:1490551 | chr12:10271189 | ENST00000533022 | 3 | 5 | 45_65 | 164 | 190.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for ASMTL-CLEC7A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >7168_7168_1_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000381317_CLEC7A_chr12_10271189_ENST00000353231_length(transcript)=3092nt_BP=1278nt AAGTGCGGACGCCCGGCTCCCGGCGTGGACGCCATGGTGCTGTGCCCGGTGATTGGGAAGCTGCTGCACAAGCGCGTGGTGCTGGCCAGC GCCTCCCCACGCCGTCAGGAGATCCTCAGCAACGCGGGTCTCAGGTTTGAGGTGGTCCCCTCCAAGTTTAAAGAGAAGCTGGACAAAGCC TCCTTCGCTACTCCGTATGGGTACGCCATGGAGACCGCCAAGCAGAAGGCCCTGGAGGTGGCCAACCGGCTGTACCAGAAAGACCTGCGG GCCCCCGACGTGGTCATTGGAGCGGACACGATCGTGACAGTCGGGGGGCTGATTCTGGAGAAGCCGGTGGACAAGCAGGACGCCTACAGG ATGCTGTCCCGGTTGAGTGGGAGAGAACACAGCGTGTTCACAGGTGTCGCGATCGTCCACTGCTCCAGCAAAGACCATCAGCTGGACACC AGGGTCTCGGAATTCTACGAGGAAACGAAGGTGAAGTTCTCGGAGCTGTCCGAGGAGCTGCTCTGGGAATACGTCCACAGCGGGGAGCCC ATGGACAAAGCTGGCGGCTACGGGATCCAGGCCCTGGGCGGCATGCTGGTGGAGTCCGTACACGGGGACTTTCTGAACGTGGTGGGATTC CCGCTGAACCACTTCTGCAAGCAGCTGGTGAAGCTCTACTACCCGCCCCGTCCGGAGGACCTGCGGCGGAGTGTCAAGCACGACTCCATC CCGGCCGCGGACACCTTCGAAGACCTCAGTGACGTGGAGGGGGGCGGCTCGGAGCCCACTCAGAGGGACGCGGGCAGCCGCGATGAGAAG GCCGAGGCGGGAGAGGCGGGACAGGCCACGGCAGAGGCTGAGTGTCACAGGACTCGGGAGACCCTGCCTCCGTTCCCGACACGCCTCCTG GAGCTGATTGAGGGCTTTATGCTATCCAAGGGCCTGCTCACCGCTTGCAAACTGAAGGTGTTCGATTTGTTAAAAGATGAAGCACCCCAG AAGGCTGCGGATATTGCCAGCAAAGTGGACGCCTCTGCGTGTGGAATGGAGAGGCTTCTGGACATCTGTGCTGCCATGGGGCTCCTGGAG AAGACAGAGCAAGGTTACAGTAACACAGAGACAGCGAACGTCTACCTGGCATCGGATGGCGAATACTCTCTGCACGGCTTCATCATGCAC AATAATGACCTCACATGGAACCTCTTTACATACCTGGAGTTTGCCATCCGAGAGGGAACAAACCAGCACCACAGGGCGTTGGGGAAGAAG GCGGAAGATCTGTTCCAGATTTCAGATCAGAACCACAGCTACCCAAGAAAACCCATCTCCAAATTGTGTATGGATTCACGTGTCAGTCAT TTATGACCAACTGTGTAGTGTGCCCTCATATAGTATTTGTGAGAAGAAGTTTTCAATGTAAGAGGAAGGGTGGAGAAGGAGAGAGAAATA TGTGAGGTAGTAAGGAGGACAGAAAACAGAACAGAAAAGAGTAACAGCTGAGGTCAAGATAAATGCAGAAAATGTTTAGAGAGCTTGGCC AACTGTAATCTTAACCAAGAAATTGAAGGGAGAGGCTGTGATTTCTGTATTTGTCGACCTACAGGTAGGCTAGTATTATTTTTCTAGTTA GTAGATCCCTAGACATGGAATCAGGGCAGCCAAGCTTGAGTTTTTATTTTTTATTTATTTATTTTTTTGAGATAGGGTCTCACTTTGTTA CCCAGGCTGGAGTGCAGTGGCACAATCTCGACTCACTGCAGCTATCTCTCGCCTCAGCCCCTCAAGTAGCTGGGACTACAGGTGCATGCC ACCATGCCAGGCTAATTTTTGGTGTTTTTTGTAGAGACTGGGTTTTGCCATGTTGACCAAGCTGGTCTCTAACTCCTGGGCTTAAGTGAT CTGCCCGCCTTGGCCTCCCAAAGTGCTGGGATTACAGATGTGAGCCACCACACCTGGCCCCAAGCTTGAATTTTCATTCTGCCATTGACT TGGCATTTACCTTGGGTAAGCCATAAGCGAATCTTAATTTCTGGCTCTATCAGAGTTGTTTCATGCTCAACAATGCCATTGAAGTGCACG GTGTGTTGCCACGATTTGACCCTCAACTTCTAGCAGTATATCAGTTATGAACTGAGGGTGAAATATATTTCTGAATAGCTAAATGAAGAA ATGGGAAAAAATCTTCACCACAGTCAGAGCAATTTTATTATTTTCATCAGTATGATCATAATTATGATTATCATCTTAGTAAAAAGCAGG AACTCCTACTTTTTCTTTATCAATTAAATAGCTCAGAGAGTACATCTGCCATATCTCTAATAGAATCTTTTTTTTTTTTTTTTTTTTTGA GACAGAGTTTCGCTCTTGTTGCCCAGGCTGGAGTGCAACGGCACGATCTCGGCTCACCGCAACCTCCGCCCCCTGGGTTCAAGCAATTCT CCTGCCTCAGCCTCCCAAGTAGCTGGGATTACAGTCAGGCACCACCACACCCGGCTAATTTTGTATTTTTTTAGTAGAGACAGGGTTTCT CCATGTCGGTCAGGGTAGTCCCGAACTCCTGACCTCAAGTGATCTGCCTGCCTCGGCCTCCCAAGTGCTGGGATTACAGGCGTGAGCCAC TGCACCCAGCCTAGAATCTTGTATAATATGTAATTGTAGGGAAACTGCTCTCATAGGAAAGTTTTCTGCTTTTTAAATACAAAAATACAT AAAAATACATAAAATCTGATGATGAATATAAAAAAGTAACCAACCTCATTGGAACAAGTATTAACATTTTGGAATATGTTTTATTAGTTT TGTGATGTACTGTTTTACAATTTTTACCATTTTTTTCAGTAATTACTGTAAAATGGTATTATTGGAATGAAACTATATTTCCTCATGTGC TGATTTGTCTTATTTTTTTCATACTTTCCCACTGGTGCTATTTTTATTTCCAATGGATATTTCTGTATTACTAGGGAGGCATTTACAGTC CTCTAATGTTGATTAATATGTGAAAAGAAATTGTACCAATTTTACTAAATTATGCAGTTTAAAATGGATGATTTTATGTTATGTGGATTT >7168_7168_1_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000381317_CLEC7A_chr12_10271189_ENST00000353231_length(amino acids)=440AA_BP=414 MVLCPVIGKLLHKRVVLASASPRRQEILSNAGLRFEVVPSKFKEKLDKASFATPYGYAMETAKQKALEVANRLYQKDLRAPDVVIGADTI VTVGGLILEKPVDKQDAYRMLSRLSGREHSVFTGVAIVHCSSKDHQLDTRVSEFYEETKVKFSELSEELLWEYVHSGEPMDKAGGYGIQA LGGMLVESVHGDFLNVVGFPLNHFCKQLVKLYYPPRPEDLRRSVKHDSIPAADTFEDLSDVEGGGSEPTQRDAGSRDEKAEAGEAGQATA EAECHRTRETLPPFPTRLLELIEGFMLSKGLLTACKLKVFDLLKDEAPQKAADIASKVDASACGMERLLDICAAMGLLEKTEQGYSNTET -------------------------------------------------------------- >7168_7168_2_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000381333_CLEC7A_chr12_10271189_ENST00000353231_length(transcript)=3100nt_BP=1286nt CTGGCCGCCTGTTTTTTTTGCAGCCGCGCTGCGCGCACCGCGGGCTCCGGGCTCAGAAGTGCGGACGCCCGGCTCCCGGCGTGGACGCCA TGGTGCTGTGCCCGGTGATTGGGAAGCTGCTGCACAAGCGCGTGGTGCTGGCCAGCGCCTCCCCACGCCGTCAGGAGATCCTCAGCAACG CGGGTCTCAGGTTTGAGGTGGTCCCCTCCAAGTTTAAAGAGAAGCTGGACAAAGCCTCCTTCGCTACTCCGTATGGGTACGCCATGGAGA CCGCCAAGCAGAAGGCCCTGGAGGTGGCCAACCGGCTGTACCAGACAGTCGGGGGGCTGATTCTGGAGAAGCCGGTGGACAAGCAGGACG CCTACAGGATGCTGTCCCGGTTGAGTGGGAGAGAACACAGCGTGTTCACAGGTGTCGCGATCGTCCACTGCTCCAGCAAAGACCATCAGC TGGACACCAGGGTCTCGGAATTCTACGAGGAAACGAAGGTGAAGTTCTCGGAGCTGTCCGAGGAGCTGCTCTGGGAATACGTCCACAGCG GGGAGCCCATGGACAAAGCTGGCGGCTACGGGATCCAGGCCCTGGGCGGCATGCTGGTGGAGTCCGTACACGGGGACTTTCTGAACGTGG TGGGATTCCCGCTGAACCACTTCTGCAAGCAGCTGGTGAAGCTCTACTACCCGCCCCGTCCGGAGGACCTGCGGCGGAGTGTCAAGCACG ACTCCATCCCGGCCGCGGACACCTTCGAAGACCTCAGTGACGTGGAGGGGGGCGGCTCGGAGCCCACTCAGAGGGACGCGGGCAGCCGCG ATGAGAAGGCCGAGGCGGGAGAGGCGGGACAGGCCACGGCAGAGGCTGAGTGTCACAGGACTCGGGAGACCCTGCCTCCGTTCCCGACAC GCCTCCTGGAGCTGATTGAGGGCTTTATGCTATCCAAGGGCCTGCTCACCGCTTGCAAACTGAAGGTGTTCGATTTGTTAAAAGATGAAG CACCCCAGAAGGCTGCGGATATTGCCAGCAAAGTGGACGCCTCTGCGTGTGGAATGGAGAGGCTTCTGGACATCTGTGCTGCCATGGGGC TCCTGGAGAAGACAGAGCAAGGTTACAGTAACACAGAGACAGCGAACGTCTACCTGGCATCGGATGGCGAATACTCTCTGCACGGCTTCA TCATGCACAATAATGACCTCACATGGAACCTCTTTACATACCTGGAGTTTGCCATCCGAGAGGGAACAAACCAGCACCACAGGGCGTTGG GGAAGAAGGCGGAAGATCTGTTCCAGATTTCAGATCAGAACCACAGCTACCCAAGAAAACCCATCTCCAAATTGTGTATGGATTCACGTG TCAGTCATTTATGACCAACTGTGTAGTGTGCCCTCATATAGTATTTGTGAGAAGAAGTTTTCAATGTAAGAGGAAGGGTGGAGAAGGAGA GAGAAATATGTGAGGTAGTAAGGAGGACAGAAAACAGAACAGAAAAGAGTAACAGCTGAGGTCAAGATAAATGCAGAAAATGTTTAGAGA GCTTGGCCAACTGTAATCTTAACCAAGAAATTGAAGGGAGAGGCTGTGATTTCTGTATTTGTCGACCTACAGGTAGGCTAGTATTATTTT TCTAGTTAGTAGATCCCTAGACATGGAATCAGGGCAGCCAAGCTTGAGTTTTTATTTTTTATTTATTTATTTTTTTGAGATAGGGTCTCA CTTTGTTACCCAGGCTGGAGTGCAGTGGCACAATCTCGACTCACTGCAGCTATCTCTCGCCTCAGCCCCTCAAGTAGCTGGGACTACAGG TGCATGCCACCATGCCAGGCTAATTTTTGGTGTTTTTTGTAGAGACTGGGTTTTGCCATGTTGACCAAGCTGGTCTCTAACTCCTGGGCT TAAGTGATCTGCCCGCCTTGGCCTCCCAAAGTGCTGGGATTACAGATGTGAGCCACCACACCTGGCCCCAAGCTTGAATTTTCATTCTGC CATTGACTTGGCATTTACCTTGGGTAAGCCATAAGCGAATCTTAATTTCTGGCTCTATCAGAGTTGTTTCATGCTCAACAATGCCATTGA AGTGCACGGTGTGTTGCCACGATTTGACCCTCAACTTCTAGCAGTATATCAGTTATGAACTGAGGGTGAAATATATTTCTGAATAGCTAA ATGAAGAAATGGGAAAAAATCTTCACCACAGTCAGAGCAATTTTATTATTTTCATCAGTATGATCATAATTATGATTATCATCTTAGTAA AAAGCAGGAACTCCTACTTTTTCTTTATCAATTAAATAGCTCAGAGAGTACATCTGCCATATCTCTAATAGAATCTTTTTTTTTTTTTTT TTTTTTGAGACAGAGTTTCGCTCTTGTTGCCCAGGCTGGAGTGCAACGGCACGATCTCGGCTCACCGCAACCTCCGCCCCCTGGGTTCAA GCAATTCTCCTGCCTCAGCCTCCCAAGTAGCTGGGATTACAGTCAGGCACCACCACACCCGGCTAATTTTGTATTTTTTTAGTAGAGACA GGGTTTCTCCATGTCGGTCAGGGTAGTCCCGAACTCCTGACCTCAAGTGATCTGCCTGCCTCGGCCTCCCAAGTGCTGGGATTACAGGCG TGAGCCACTGCACCCAGCCTAGAATCTTGTATAATATGTAATTGTAGGGAAACTGCTCTCATAGGAAAGTTTTCTGCTTTTTAAATACAA AAATACATAAAAATACATAAAATCTGATGATGAATATAAAAAAGTAACCAACCTCATTGGAACAAGTATTAACATTTTGGAATATGTTTT ATTAGTTTTGTGATGTACTGTTTTACAATTTTTACCATTTTTTTCAGTAATTACTGTAAAATGGTATTATTGGAATGAAACTATATTTCC TCATGTGCTGATTTGTCTTATTTTTTTCATACTTTCCCACTGGTGCTATTTTTATTTCCAATGGATATTTCTGTATTACTAGGGAGGCAT TTACAGTCCTCTAATGTTGATTAATATGTGAAAAGAAATTGTACCAATTTTACTAAATTATGCAGTTTAAAATGGATGATTTTATGTTAT >7168_7168_2_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000381333_CLEC7A_chr12_10271189_ENST00000353231_length(amino acids)=451AA_BP=425 MFFLQPRCAHRGLRAQKCGRPAPGVDAMVLCPVIGKLLHKRVVLASASPRRQEILSNAGLRFEVVPSKFKEKLDKASFATPYGYAMETAK QKALEVANRLYQTVGGLILEKPVDKQDAYRMLSRLSGREHSVFTGVAIVHCSSKDHQLDTRVSEFYEETKVKFSELSEELLWEYVHSGEP MDKAGGYGIQALGGMLVESVHGDFLNVVGFPLNHFCKQLVKLYYPPRPEDLRRSVKHDSIPAADTFEDLSDVEGGGSEPTQRDAGSRDEK AEAGEAGQATAEAECHRTRETLPPFPTRLLELIEGFMLSKGLLTACKLKVFDLLKDEAPQKAADIASKVDASACGMERLLDICAAMGLLE KTEQGYSNTETANVYLASDGEYSLHGFIMHNNDLTWNLFTYLEFAIREGTNQHHRALGKKAEDLFQISDQNHSYPRKPISKLCMDSRVSH -------------------------------------------------------------- >7168_7168_3_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000416733_CLEC7A_chr12_10271189_ENST00000304084_length(transcript)=1579nt_BP=1215nt AGAAGTGCGGACGCCCGGCTCCCGGCGTGGACGCCATGGTGCTGTGCCCGGTGATTGGGAAGCTGCTGCACAAGCGCGTGGTGCTGGCCA GCGCCTCCCCACGCCGTCAGGAGATCCTCAGCAACGCGGGTCTCAGGTTTGAGGTGGTCCCCTCCAAGTTTAAAGAGAAGCTGGACAAAG CCTCCTTCGCTACTCCGTATGGGTACGCCATGGAGACCGCCAAGCAGAAGGCCCTGGAGGTGGCCAACCGGCTGTACCAGAAAGACCTGC GGGCCCCCGACGTGGTCATTGGAGCGGACACGATCGTGGTTGAGTGGGAGAGAACACAGCGTGTTCACAGGTGTCGCGATCGTCCACTGC TCCAGCAAAGACCATCAGCTGGACACCAGGGTCTCGGAATTCTACGAGGAAACGAAGGTGAAGTTCTCGGAGCTGTCCGAGGAGCTGCTC TGGGAATACGTCCACAGCGGGGAGCCCATGGACAAAGCTGGCGGCTACGGGATCCAGGCCCTGGGCGGCATGCTGGTGGAGTCCGTACAC GGGGACTTTCTGAACGTGGTGGGATTCCCGCTGAACCACTTCTGCAAGCAGCTGGTGAAGCTCTACTACCCGCCCCGTCCGGAGGACCTG CGGCGGAGTGTCAAGCACGACTCCATCCCGGCCGCGGACACCTTCGAAGACCTCAGTGACGTGGAGGGGGGCGGCTCGGAGCCCACTCAG AGGGACGCGGGCAGCCGCGATGAGAAGGCCGAGGCGGGAGAGGCGGGACAGGCCACGGCAGAGGCTGAGTGTCACAGGACTCGGGAGACC CTGCCTCCGTTCCCGACACGCCTCCTGGAGCTGATTGAGGGCTTTATGCTATCCAAGGGCCTGCTCACCGCTTGCAAACTGAAGGTGTTC GATTTGTTAAAAGATGAAGCACCCCAGAAGGCTGCGGATATTGCCAGCAAAGTGGACGCCTCTGCGTGTGGAATGGAGAGGCTTCTGGAC ATCTGTGCTGCCATGGGGCTCCTGGAGAAGACAGAGCAAGGTTACAGTAACACAGAGACAGCGAACGTCTACCTGGCATCGGATGGCGAA TACTCTCTGCACGGCTTCATCATGCACAATAATGACCTCACATGGAACCTCTTTACATACCTGGAGTTTGCCATCCGAGAGGGAACAAAC CAGCACCACAGGGCGTTGGGGAAGAAGGCGGAAGATCTGTTCCAGATTTCAGATCAGAACCACAGCTACCCAAGAAAACCCATCTCCAAA TTGTGTATGGATTCACGTGTCAGTCATTTATGACCAACTGTGTAGTGTGCCCTCATATAGTATTTGTGAGAAGAAGTTTTCAATGTAAGA GGAAGGGTGGAGAAGGAGAGAGAAATATGTGAGGTAGTAAGGAGGACAGAAAACAGAACAGAAAAGAGTAACAGCTGAGGTCAAGATAAA TGCAGAAAATGTTTAGAGAGCTTGGCCAACTGTAATCTTAACCAAGAAATTGAAGGGAGAGGCTGTGATTTCTGTATTTGTCGACCTACA >7168_7168_3_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000416733_CLEC7A_chr12_10271189_ENST00000304084_length(amino acids)=412AA_BP=386 MGSCCTSAWCWPAPPHAVRRSSATRVSGLRWSPPSLKRSWTKPPSLLRMGTPWRPPSRRPWRWPTGCTRKTCGPPTWSLERTRSWLSGRE HSVFTGVAIVHCSSKDHQLDTRVSEFYEETKVKFSELSEELLWEYVHSGEPMDKAGGYGIQALGGMLVESVHGDFLNVVGFPLNHFCKQL VKLYYPPRPEDLRRSVKHDSIPAADTFEDLSDVEGGGSEPTQRDAGSRDEKAEAGEAGQATAEAECHRTRETLPPFPTRLLELIEGFMLS KGLLTACKLKVFDLLKDEAPQKAADIASKVDASACGMERLLDICAAMGLLEKTEQGYSNTETANVYLASDGEYSLHGFIMHNNDLTWNLF -------------------------------------------------------------- >7168_7168_4_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000416733_CLEC7A_chr12_10271189_ENST00000533022_length(transcript)=1348nt_BP=1215nt AGAAGTGCGGACGCCCGGCTCCCGGCGTGGACGCCATGGTGCTGTGCCCGGTGATTGGGAAGCTGCTGCACAAGCGCGTGGTGCTGGCCA GCGCCTCCCCACGCCGTCAGGAGATCCTCAGCAACGCGGGTCTCAGGTTTGAGGTGGTCCCCTCCAAGTTTAAAGAGAAGCTGGACAAAG CCTCCTTCGCTACTCCGTATGGGTACGCCATGGAGACCGCCAAGCAGAAGGCCCTGGAGGTGGCCAACCGGCTGTACCAGAAAGACCTGC GGGCCCCCGACGTGGTCATTGGAGCGGACACGATCGTGGTTGAGTGGGAGAGAACACAGCGTGTTCACAGGTGTCGCGATCGTCCACTGC TCCAGCAAAGACCATCAGCTGGACACCAGGGTCTCGGAATTCTACGAGGAAACGAAGGTGAAGTTCTCGGAGCTGTCCGAGGAGCTGCTC TGGGAATACGTCCACAGCGGGGAGCCCATGGACAAAGCTGGCGGCTACGGGATCCAGGCCCTGGGCGGCATGCTGGTGGAGTCCGTACAC GGGGACTTTCTGAACGTGGTGGGATTCCCGCTGAACCACTTCTGCAAGCAGCTGGTGAAGCTCTACTACCCGCCCCGTCCGGAGGACCTG CGGCGGAGTGTCAAGCACGACTCCATCCCGGCCGCGGACACCTTCGAAGACCTCAGTGACGTGGAGGGGGGCGGCTCGGAGCCCACTCAG AGGGACGCGGGCAGCCGCGATGAGAAGGCCGAGGCGGGAGAGGCGGGACAGGCCACGGCAGAGGCTGAGTGTCACAGGACTCGGGAGACC CTGCCTCCGTTCCCGACACGCCTCCTGGAGCTGATTGAGGGCTTTATGCTATCCAAGGGCCTGCTCACCGCTTGCAAACTGAAGGTGTTC GATTTGTTAAAAGATGAAGCACCCCAGAAGGCTGCGGATATTGCCAGCAAAGTGGACGCCTCTGCGTGTGGAATGGAGAGGCTTCTGGAC ATCTGTGCTGCCATGGGGCTCCTGGAGAAGACAGAGCAAGGTTACAGTAACACAGAGACAGCGAACGTCTACCTGGCATCGGATGGCGAA TACTCTCTGCACGGCTTCATCATGCACAATAATGACCTCACATGGAACCTCTTTACATACCTGGAGTTTGCCATCCGAGAGGGAACAAAC CAGCACCACAGGGCGTTGGGGAAGAAGGCGGAAGATCTGTTCCAGATTTCAGATCAGAACCACAGCTACCCAAGAAAACCCATCTCCAAA >7168_7168_4_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000416733_CLEC7A_chr12_10271189_ENST00000533022_length(amino acids)=412AA_BP=386 MGSCCTSAWCWPAPPHAVRRSSATRVSGLRWSPPSLKRSWTKPPSLLRMGTPWRPPSRRPWRWPTGCTRKTCGPPTWSLERTRSWLSGRE HSVFTGVAIVHCSSKDHQLDTRVSEFYEETKVKFSELSEELLWEYVHSGEPMDKAGGYGIQALGGMLVESVHGDFLNVVGFPLNHFCKQL VKLYYPPRPEDLRRSVKHDSIPAADTFEDLSDVEGGGSEPTQRDAGSRDEKAEAGEAGQATAEAECHRTRETLPPFPTRLLELIEGFMLS KGLLTACKLKVFDLLKDEAPQKAADIASKVDASACGMERLLDICAAMGLLEKTEQGYSNTETANVYLASDGEYSLHGFIMHNNDLTWNLF -------------------------------------------------------------- >7168_7168_5_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000534940_CLEC7A_chr12_10271189_ENST00000304084_length(transcript)=1661nt_BP=1297nt ATCTCCACTCCCCTCCCACCAGGCACGAGGACATCCCCATCCCCACGCCACTCGCACCGGGTCCGCGGGGAACCCCCTATCTCTACCTTC CCCGACGCGGCTCCCACAAGACCCCCATCTCTGCGCCGGGCCCCTCCACGTGCGGGGTCTCAGGTTTGAGGTGGTCCCCTCCAAGTTTAA AGAGAAGCTGGACAAAGCCTCCTTCGCTACTCCGTATGGGTACGCCATGGAGACCGCCAAGCAGAAGGCCCTGGAGGTGGCCAACCGGCT GTACCAGAAAGACCTGCGGGCCCCCGACGTGGTCATTGGAGCGGACACGATCGTGACAGTCGGGGGGCTGATTCTGGAGAAGCCGGTGGA CAAGCAGGACGCCTACAGGATGCTGTCCCGGTTGAGTGGGAGAGAACACAGCGTGTTCACAGGTGTCGCGATCGTCCACTGCTCCAGCAA AGACCATCAGCTGGACACCAGGGTCTCGGAATTCTACGAGGAAACGAAGGTGAAGTTCTCGGAGCTGTCCGAGGAGCTGCTCTGGGAATA CGTCCACAGCGGGGAGCCCATGGACAAAGCTGGCGGCTACGGGATCCAGGCCCTGGGCGGCATGCTGGTGGAGTCCGTACACGGGGACTT TCTGAACGTGGTGGGATTCCCGCTGAACCACTTCTGCAAGCAGCTGGTGAAGCTCTACTACCCGCCCCGTCCGGAGGACCTGCGGCGGAG TGTCAAGCACGACTCCATCCCGGCCGCGGACACCTTCGAAGACCTCAGTGACGTGGAGGGGGGCGGCTCGGAGCCCACTCAGAGGGACGC GGGCAGCCGCGATGAGAAGGCCGAGGCGGGAGAGGCGGGACAGGCCACGGCAGAGGCTGAGTGTCACAGGACTCGGGAGACCCTGCCTCC GTTCCCGACACGCCTCCTGGAGCTGATTGAGGGCTTTATGCTATCCAAGGGCCTGCTCACCGCTTGCAAACTGAAGGTGTTCGATTTGTT AAAAGATGAAGCACCCCAGAAGGCTGCGGATATTGCCAGCAAAGTGGACGCCTCTGCGTGTGGAATGGAGAGGCTTCTGGACATCTGTGC TGCCATGGGGCTCCTGGAGAAGACAGAGCAAGGTTACAGTAACACAGAGACAGCGAACGTCTACCTGGCATCGGATGGCGAATACTCTCT GCACGGCTTCATCATGCACAATAATGACCTCACATGGAACCTCTTTACATACCTGGAGTTTGCCATCCGAGAGGGAACAAACCAGCACCA CAGGGCGTTGGGGAAGAAGGCGGAAGATCTGTTCCAGATTTCAGATCAGAACCACAGCTACCCAAGAAAACCCATCTCCAAATTGTGTAT GGATTCACGTGTCAGTCATTTATGACCAACTGTGTAGTGTGCCCTCATATAGTATTTGTGAGAAGAAGTTTTCAATGTAAGAGGAAGGGT GGAGAAGGAGAGAGAAATATGTGAGGTAGTAAGGAGGACAGAAAACAGAACAGAAAAGAGTAACAGCTGAGGTCAAGATAAATGCAGAAA ATGTTTAGAGAGCTTGGCCAACTGTAATCTTAACCAAGAAATTGAAGGGAGAGGCTGTGATTTCTGTATTTGTCGACCTACAGGTAGGCT >7168_7168_5_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000534940_CLEC7A_chr12_10271189_ENST00000304084_length(amino acids)=395AA_BP=369 MDKASFATPYGYAMETAKQKALEVANRLYQKDLRAPDVVIGADTIVTVGGLILEKPVDKQDAYRMLSRLSGREHSVFTGVAIVHCSSKDH QLDTRVSEFYEETKVKFSELSEELLWEYVHSGEPMDKAGGYGIQALGGMLVESVHGDFLNVVGFPLNHFCKQLVKLYYPPRPEDLRRSVK HDSIPAADTFEDLSDVEGGGSEPTQRDAGSRDEKAEAGEAGQATAEAECHRTRETLPPFPTRLLELIEGFMLSKGLLTACKLKVFDLLKD EAPQKAADIASKVDASACGMERLLDICAAMGLLEKTEQGYSNTETANVYLASDGEYSLHGFIMHNNDLTWNLFTYLEFAIREGTNQHHRA -------------------------------------------------------------- >7168_7168_6_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000534940_CLEC7A_chr12_10271189_ENST00000533022_length(transcript)=1430nt_BP=1297nt ATCTCCACTCCCCTCCCACCAGGCACGAGGACATCCCCATCCCCACGCCACTCGCACCGGGTCCGCGGGGAACCCCCTATCTCTACCTTC CCCGACGCGGCTCCCACAAGACCCCCATCTCTGCGCCGGGCCCCTCCACGTGCGGGGTCTCAGGTTTGAGGTGGTCCCCTCCAAGTTTAA AGAGAAGCTGGACAAAGCCTCCTTCGCTACTCCGTATGGGTACGCCATGGAGACCGCCAAGCAGAAGGCCCTGGAGGTGGCCAACCGGCT GTACCAGAAAGACCTGCGGGCCCCCGACGTGGTCATTGGAGCGGACACGATCGTGACAGTCGGGGGGCTGATTCTGGAGAAGCCGGTGGA CAAGCAGGACGCCTACAGGATGCTGTCCCGGTTGAGTGGGAGAGAACACAGCGTGTTCACAGGTGTCGCGATCGTCCACTGCTCCAGCAA AGACCATCAGCTGGACACCAGGGTCTCGGAATTCTACGAGGAAACGAAGGTGAAGTTCTCGGAGCTGTCCGAGGAGCTGCTCTGGGAATA CGTCCACAGCGGGGAGCCCATGGACAAAGCTGGCGGCTACGGGATCCAGGCCCTGGGCGGCATGCTGGTGGAGTCCGTACACGGGGACTT TCTGAACGTGGTGGGATTCCCGCTGAACCACTTCTGCAAGCAGCTGGTGAAGCTCTACTACCCGCCCCGTCCGGAGGACCTGCGGCGGAG TGTCAAGCACGACTCCATCCCGGCCGCGGACACCTTCGAAGACCTCAGTGACGTGGAGGGGGGCGGCTCGGAGCCCACTCAGAGGGACGC GGGCAGCCGCGATGAGAAGGCCGAGGCGGGAGAGGCGGGACAGGCCACGGCAGAGGCTGAGTGTCACAGGACTCGGGAGACCCTGCCTCC GTTCCCGACACGCCTCCTGGAGCTGATTGAGGGCTTTATGCTATCCAAGGGCCTGCTCACCGCTTGCAAACTGAAGGTGTTCGATTTGTT AAAAGATGAAGCACCCCAGAAGGCTGCGGATATTGCCAGCAAAGTGGACGCCTCTGCGTGTGGAATGGAGAGGCTTCTGGACATCTGTGC TGCCATGGGGCTCCTGGAGAAGACAGAGCAAGGTTACAGTAACACAGAGACAGCGAACGTCTACCTGGCATCGGATGGCGAATACTCTCT GCACGGCTTCATCATGCACAATAATGACCTCACATGGAACCTCTTTACATACCTGGAGTTTGCCATCCGAGAGGGAACAAACCAGCACCA CAGGGCGTTGGGGAAGAAGGCGGAAGATCTGTTCCAGATTTCAGATCAGAACCACAGCTACCCAAGAAAACCCATCTCCAAATTGTGTAT >7168_7168_6_ASMTL-CLEC7A_ASMTL_chrY_1490551_ENST00000534940_CLEC7A_chr12_10271189_ENST00000533022_length(amino acids)=395AA_BP=369 MDKASFATPYGYAMETAKQKALEVANRLYQKDLRAPDVVIGADTIVTVGGLILEKPVDKQDAYRMLSRLSGREHSVFTGVAIVHCSSKDH QLDTRVSEFYEETKVKFSELSEELLWEYVHSGEPMDKAGGYGIQALGGMLVESVHGDFLNVVGFPLNHFCKQLVKLYYPPRPEDLRRSVK HDSIPAADTFEDLSDVEGGGSEPTQRDAGSRDEKAEAGEAGQATAEAECHRTRETLPPFPTRLLELIEGFMLSKGLLTACKLKVFDLLKD EAPQKAADIASKVDASACGMERLLDICAAMGLLEKTEQGYSNTETANVYLASDGEYSLHGFIMHNNDLTWNLFTYLEFAIREGTNQHHRA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ASMTL-CLEC7A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ASMTL-CLEC7A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ASMTL-CLEC7A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ASMTL | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | ASMTL | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | ASMTL | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Tgene | C0006845 | Candidiasis, Chronic Mucocutaneous | 1 | ORPHANET |
