
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:IFT20-GLS (FusionGDB2 ID:HG90410TG2744) |
Fusion Gene Summary for IFT20-GLS |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: IFT20-GLS | Fusion gene ID: hg90410tg2744 | Hgene | Tgene | Gene symbol | IFT20 | GLS | Gene ID | 90410 | 2744 |
| Gene name | intraflagellar transport 20 | glutaminase | |
| Synonyms | - | AAD20|CASGID|EIEE71|GAC|GAM|GDPAG|GLS1|KGA | |
| Cytomap | ('IFT20')('GLS') 17q11.2 | 2q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | intraflagellar transport protein 20 homologintraflagellar transport 20 homologintraflagellar transport protein IFT20 | glutaminase kidney isoform, mitochondrialK-glutaminaseL-glutamine amidohydrolaseglutaminase Cglutaminase, phosphate-activated | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q8IY31 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000357896, ENST00000395418, ENST00000578122, ENST00000578985, ENST00000579419, ENST00000585089, ENST00000585313, | ||
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 12 X 9 X 5=540 |
| # samples | 3 | 12 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(12/540*10)=-2.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: IFT20 [Title/Abstract] AND GLS [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | IFT20(26658882)-GLS(191827556), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | IFT20-GLS seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IFT20-GLS seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IFT20-GLS seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IFT20-GLS seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IFT20-GLS seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. IFT20-GLS seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. IFT20-GLS seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | GLS | GO:0006537 | glutamate biosynthetic process | 16899818|22049910 |
| Tgene | GLS | GO:0006543 | glutamine catabolic process | 22049910 |
| Tgene | GLS | GO:0051289 | protein homotetramerization | 22049910 |
 Fusion gene breakpoints across IFT20 (5'-gene) Fusion gene breakpoints across IFT20 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
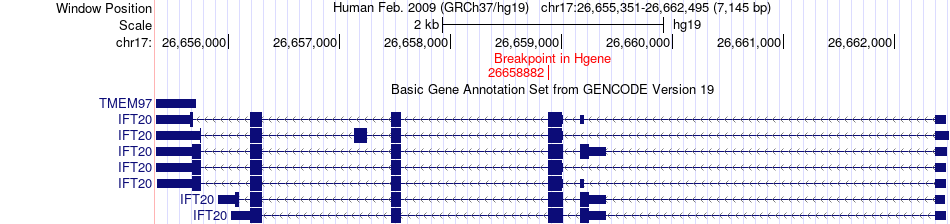 |
 Fusion gene breakpoints across GLS (3'-gene) Fusion gene breakpoints across GLS (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
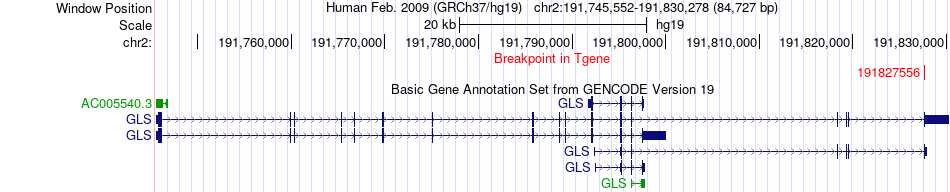 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | DC316034 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
Top |
Fusion Gene ORF analysis for IFT20-GLS |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000357896 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000357896 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000357896 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000357896 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000357896 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000395418 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000395418 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000395418 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000395418 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000395418 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578122 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578122 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578122 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578122 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578122 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578985 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578985 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578985 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578985 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000578985 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000579419 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000579419 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000579419 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000579419 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000579419 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585089 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585089 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585089 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585089 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585089 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585313 | ENST00000338435 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585313 | ENST00000409215 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585313 | ENST00000409428 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585313 | ENST00000409626 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| 5CDS-intron | ENST00000585313 | ENST00000471443 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| Frame-shift | ENST00000357896 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| Frame-shift | ENST00000395418 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| Frame-shift | ENST00000579419 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| Frame-shift | ENST00000585313 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| In-frame | ENST00000578122 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| In-frame | ENST00000578985 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
| In-frame | ENST00000585089 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000585089 | IFT20 | chr17 | 26658882 | - | ENST00000320717 | GLS | chr2 | 191827556 | + | 3199 | 476 | 256 | 672 | 138 |
| ENST00000578122 | IFT20 | chr17 | 26658882 | - | ENST00000320717 | GLS | chr2 | 191827556 | + | 3185 | 462 | 242 | 658 | 138 |
| ENST00000578985 | IFT20 | chr17 | 26658882 | - | ENST00000320717 | GLS | chr2 | 191827556 | + | 3190 | 467 | 247 | 663 | 138 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000585089 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + | 0.017680187 | 0.9823198 |
| ENST00000578122 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + | 0.017822243 | 0.98217773 |
| ENST00000578985 | ENST00000320717 | IFT20 | chr17 | 26658882 | - | GLS | chr2 | 191827556 | + | 0.017833116 | 0.9821669 |
Top |
Fusion Genomic Features for IFT20-GLS |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| IFT20 | chr17 | 26658884 | - | GLS | chr2 | 191827555 | + | 0.000121266 | 0.99987876 |
| IFT20 | chr17 | 26658884 | - | GLS | chr2 | 191827555 | + | 0.000121266 | 0.99987876 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for IFT20-GLS |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:26658882/chr2:191827556) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IFT20 | . |
| FUNCTION: Part of intraflagellar transport (IFT) particles involved in ciliary process assembly. May play a role in the trafficking of ciliary membrane proteins from the Golgi complex to the cilium (PubMed:16775004). Regulates the platelet-derived growth factor receptor-alpha (PDGFRA) signaling pathway. Required for protein stability of E3 ubiquitin ligases CBL and CBLB that mediate ubiquitination and internalization of PDGFRA for proper feedback inhibition of PDGFRA signaling (PubMed:29237719). Essential for male fertility. Plays an important role in spermatogenesis, particularly spermiogenesis, when germ cells form flagella. May play a role in the transport of flagellar proteins ODF2 and SPAG16 to build sperm flagella and in the removal of redundant sperm cytoplasm (By similarity). Also involved in autophagy since it is required for trafficking of ATG16L and the expansion of the autophagic compartment (By similarity). {ECO:0000250|UniProtKB:Q61025, ECO:0000269|PubMed:16775004, ECO:0000269|PubMed:29237719}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GLS | chr17:26658882 | chr2:191827556 | ENST00000338435 | 0 | 15 | 315_322 | 0 | 599.0 | Region | Highly mobile activation loop | |
| Tgene | GLS | chr17:26658882 | chr2:191827556 | ENST00000320717 | 16 | 18 | 619_648 | 617 | 670.0 | Repeat | Note=ANK 2 | |
| Tgene | GLS | chr17:26658882 | chr2:191827556 | ENST00000338435 | 0 | 15 | 585_614 | 0 | 599.0 | Repeat | Note=ANK 1 | |
| Tgene | GLS | chr17:26658882 | chr2:191827556 | ENST00000338435 | 0 | 15 | 619_648 | 0 | 599.0 | Repeat | Note=ANK 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000357896 | - | 2 | 6 | 74_114 | 42 | 149.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000395418 | - | 2 | 5 | 74_114 | 42 | 133.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000579419 | - | 3 | 6 | 74_114 | 42 | 115.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000585089 | - | 3 | 6 | 74_114 | 68 | 159.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000585313 | - | 3 | 6 | 74_114 | 42 | 133.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000357896 | - | 2 | 6 | 70_132 | 42 | 149.0 | Region | IFT57-binding |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000395418 | - | 2 | 5 | 70_132 | 42 | 133.0 | Region | IFT57-binding |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000579419 | - | 3 | 6 | 70_132 | 42 | 115.0 | Region | IFT57-binding |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000585089 | - | 3 | 6 | 70_132 | 68 | 159.0 | Region | IFT57-binding |
| Hgene | IFT20 | chr17:26658882 | chr2:191827556 | ENST00000585313 | - | 3 | 6 | 70_132 | 42 | 133.0 | Region | IFT57-binding |
| Tgene | GLS | chr17:26658882 | chr2:191827556 | ENST00000320717 | 16 | 18 | 315_322 | 617 | 670.0 | Region | Highly mobile activation loop | |
| Tgene | GLS | chr17:26658882 | chr2:191827556 | ENST00000320717 | 16 | 18 | 585_614 | 617 | 670.0 | Repeat | Note=ANK 1 |
Top |
Fusion Gene Sequence for IFT20-GLS |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >38416_38416_1_IFT20-GLS_IFT20_chr17_26658882_ENST00000578122_GLS_chr2_191827556_ENST00000320717_length(transcript)=3185nt_BP=462nt GGCAGGCTGGCTGGGTACCGGCTGTCGCTGACCCAGGAGAAGCTGCCTGTCTACATCAGCCTGGGCTGCAGCGCGCTGCCGCCGCGGGGC CGGCAGCTGAACTATGTGCTCTTCAGGGCGGGCACCGTGTTGCATTCATCTTTGTACCCCCAGCATCTAGCAGTGTTGGCATGTAGTAGG CACTCAAGAAATGTGTGTTGAATGAACGATGCCTGTGACAAGCAAGCGGACTTTATTCTTTCCTGACCCTTGCTCCTATGACACACCTCC TCCTGACTGCCACTGTCACTCCTTCAGAGCAGAACTCCTCTAGGGAACCTGGATGGGAAACAGCCATGGCCAAGGACATCCTGGGTGAAG CAGGGCTACACTTTGATGAACTGAACAAGCTGAGGGTGTTGGACCCAGAGGTTACCCAGCAGACCATAGAGCTGAAGGAAGAGTGCAAAG ACTTTGTGGACAGTGGAATAACACTCCCATGGATGAAGCACTGCACTTTGGACACCATGATGTATTTAAAATTCTCCAAGAATACCAAGT CCAGTACACACCTCAAGGAGATTCTGACAACGGGAAGGAAAATCAAACCGTCCATAAGAATCTTGATGGATTGTTGTAATGGTCTCAAAT CCCAAGATTTAAATCACTTACCTATTTAATTGTGGAAAATGATTATGAAGAACATGTGTATTTCTATCTGGTAGTGATGTATATTTTACA TTTGTCATTTCAGTGTTACTGGAGTTTTCTTCATTGTGCACACAGGACAAATCTGATCTCTTTGGGAAAAAATAGAAATAAAACAATCTC CCTCCATAATGTGAGCAATATTACCTCGTGCATTGTATAATTTGATGTAAAAGAAATAGTTACCAATGCTAGCTTGTGTGGTCTTCCATG ATTTATTTGTGTTTTGTGAATTTTCAATTTATGGTGATGATCTGCTGATATGCATTTATAAAGTAAGCTCTGTTGTACAGTCTGTCCAAA TGGGTCAAGGTTGCCTTTAGAAGCAAATAGTGTGATTTTCAAGACTTCAAATACAAATTTAGTTTAAGTGTTTGAACAACTATATGCACT TACGGTTGTGTGTTTAAAATGTCTCTCTCACCCCCTAGCTTCATGATGTGACTCTTAAAAAACTATAATAGTTAACAACTGTTAGTAAGA TAGACCAATTCTGATTAGACTTTATCAGGGAATCTGTTTAAGATATGTTTGGTGACCAAAACGTATGTGTGAATGTAGTTATAATGCTTT TGAAAAATTTTCCTTTTTCTATATCCCCTTAGTCCAGCCTCTCTTCTCAGACATTTAGCTATCTGCCTCTTTCCTTTAGCTGGGAAAGTG AGAGCTGGCATACTATGCAGTTTTTATGTTTTCCATAGTAAGTCAGAAAATGCCTCCTATTTCTGGCATCAGAACTTTGCCATTTGTCTA CAGAAGACGAACCAGAGACAAAATTACTAAGTATAAATTAGTCAAGTTTATCAGTCTAAAAAACGAAGGGATGTGCAACTGCAGCTCTTT AAGAAGTTTTTTTTTTTTAGCTTCTAGGGTAAAGATAAATTCAGAAATGCTCTAAGCTACCAAAGTTATTCTGAAAGTATGGGAACTGCT ACAACTAACAAACATTTGTTTCCAAGCCTGTCATTAAGAGTCTGCATCAAGAGATTTGTCCTCCTTGGGGGACCACTGGATCATTCCAGA TTTCTTGTGATTTTTCTATTGTGTAATTCTTGGTGGGCTCTGTAGTTTAATAATAAGAAAAAGGCCATTTCATTTTAAATTGTGACCTAT AATTCTTTGTCTTGGGTTGGTAATTCAGGATTCATTTGGAAAGTGGGTAAAAGGGGCTTCAAAAAACGGATAGAACAGGATTTTCTAGGA GTTACACATACATTTTATCCTGTCATACCTCGAGATAAAGTGGCATGTTAGTGAGGAGTTCTGATATTAAGCACACACACACATGCACAC AAATGGACTTCTCTGAAGCTGTGTTTAGTGAAATGAGCTCAAGTACATGAATGTTAGTTGTTATCACATACAGCAAATTCCTTTTTTTTT CTTTTTCTATGAGCACACTCTGCTGCTTCTAAACTTTACATGCCTGATGGCACCTTACTCCAGCAGCCTCCAGGTGCTTTCATTTTCACT TCCAGTCTAAGCCAGTGGCTCCTGCCACTGCCCTCCCATTACCTAGATGGCACCTCCTTTGGTGAAACCACGGCCAATGTTCCTTAGCTG CACCAGGCCCGAAGCTGTTCCCATGCTTGAGCTTCCATGGGGAGGATGCTGAGTGAGCAGTTTCCTACCCCGTGGATCTAGCAAGCCATG GAGACAGGTAGCATTTGTAAGATGCTGCACAGGAGCAGCATTATCCCCAAAGATATTACAGGGTAGACACGTTTTAACTGAAATCAATCA AGATAACTTTATTCAAAGAGCAGCCCGCTTTGTGTGACTAAAATGAAACAAGACAGTTGAATTGTGTGACTTGAAGATTACCAATGATTT TGAGGCTTTTCTATAATAAAAAGAGGTTCTAACCATTATTTGGGAACAAAGAGAGTTTTCATCTTTTTTCAGATCAAAACCATTCTGTAA AATCTTTGTTGTTTAATTAAATGTGCCGTTATTTACCCCTGATGTTATTTATGACTATGTGCCGATTCCTGCTCGGGCTGTTTGCTGTTG GCTGGTAATAATATATTTGATTTAAATGCTGTTGACTGTGCTATTAACTGCTGCCGTCAGTAAACTCCAAAGATCTTTTTGTTTTGGCTT TAGTATCATATGTGCTTTTTCTGTATCCTGAGCGCTCTATATGATCATGTTAATTTAAAGCTTTATACACATTGTTGTTTTTGCTGGTCT CATCTTTGGTAATATGCTATACCCCACTGCTGCCCGACACTGCCCTTTAGCTGCAGAGCTGGATTAGCTGTTGACCATTTGATGCTGTTG TCTGTCTGGCAGGGACTGAATGACCTGATGTCAGATTTAGATTCTTCCTGGGGATTACACAGCTATGAATGTATTTGCTTCTAAAACCTC CCAAAGTGAATCTAATCTTAAAACTACAAGTTGTAAGTATTCTGAAATTGGGAAACATTTATTTTAAATGCAATCAGGTAGTGTTGCTTT >38416_38416_1_IFT20-GLS_IFT20_chr17_26658882_ENST00000578122_GLS_chr2_191827556_ENST00000320717_length(amino acids)=138AA_BP=74 MTLAPMTHLLLTATVTPSEQNSSREPGWETAMAKDILGEAGLHFDELNKLRVLDPEVTQQTIELKEECKDFVDSGITLPWMKHCTLDTMM -------------------------------------------------------------- >38416_38416_2_IFT20-GLS_IFT20_chr17_26658882_ENST00000578985_GLS_chr2_191827556_ENST00000320717_length(transcript)=3190nt_BP=467nt GCCGTGGCAGGCTGGCTGGGTACCGGCTGTCGCTGACCCAGGAGAAGCTGCCTGTCTACATCAGCCTGGGCTGCAGCGCGCTGCCGCCGC GGGGCCGGCAGCTGAACTATGTGCTCTTCAGGGCGGGCACCGTGTTGCATTCATCTTTGTACCCCCAGCATCTAGCAGTGTTGGCATGTA GTAGGCACTCAAGAAATGTGTGTTGAATGAACGATGCCTGTGACAAGCAAGCGGACTTTATTCTTTCCTGACCCTTGCTCCTATGACACA CCTCCTCCTGACTGCCACTGTCACTCCTTCAGAGCAGAACTCCTCTAGGGAACCTGGATGGGAAACAGCCATGGCCAAGGACATCCTGGG TGAAGCAGGGCTACACTTTGATGAACTGAACAAGCTGAGGGTGTTGGACCCAGAGGTTACCCAGCAGACCATAGAGCTGAAGGAAGAGTG CAAAGACTTTGTGGACAGTGGAATAACACTCCCATGGATGAAGCACTGCACTTTGGACACCATGATGTATTTAAAATTCTCCAAGAATAC CAAGTCCAGTACACACCTCAAGGAGATTCTGACAACGGGAAGGAAAATCAAACCGTCCATAAGAATCTTGATGGATTGTTGTAATGGTCT CAAATCCCAAGATTTAAATCACTTACCTATTTAATTGTGGAAAATGATTATGAAGAACATGTGTATTTCTATCTGGTAGTGATGTATATT TTACATTTGTCATTTCAGTGTTACTGGAGTTTTCTTCATTGTGCACACAGGACAAATCTGATCTCTTTGGGAAAAAATAGAAATAAAACA ATCTCCCTCCATAATGTGAGCAATATTACCTCGTGCATTGTATAATTTGATGTAAAAGAAATAGTTACCAATGCTAGCTTGTGTGGTCTT CCATGATTTATTTGTGTTTTGTGAATTTTCAATTTATGGTGATGATCTGCTGATATGCATTTATAAAGTAAGCTCTGTTGTACAGTCTGT CCAAATGGGTCAAGGTTGCCTTTAGAAGCAAATAGTGTGATTTTCAAGACTTCAAATACAAATTTAGTTTAAGTGTTTGAACAACTATAT GCACTTACGGTTGTGTGTTTAAAATGTCTCTCTCACCCCCTAGCTTCATGATGTGACTCTTAAAAAACTATAATAGTTAACAACTGTTAG TAAGATAGACCAATTCTGATTAGACTTTATCAGGGAATCTGTTTAAGATATGTTTGGTGACCAAAACGTATGTGTGAATGTAGTTATAAT GCTTTTGAAAAATTTTCCTTTTTCTATATCCCCTTAGTCCAGCCTCTCTTCTCAGACATTTAGCTATCTGCCTCTTTCCTTTAGCTGGGA AAGTGAGAGCTGGCATACTATGCAGTTTTTATGTTTTCCATAGTAAGTCAGAAAATGCCTCCTATTTCTGGCATCAGAACTTTGCCATTT GTCTACAGAAGACGAACCAGAGACAAAATTACTAAGTATAAATTAGTCAAGTTTATCAGTCTAAAAAACGAAGGGATGTGCAACTGCAGC TCTTTAAGAAGTTTTTTTTTTTTAGCTTCTAGGGTAAAGATAAATTCAGAAATGCTCTAAGCTACCAAAGTTATTCTGAAAGTATGGGAA CTGCTACAACTAACAAACATTTGTTTCCAAGCCTGTCATTAAGAGTCTGCATCAAGAGATTTGTCCTCCTTGGGGGACCACTGGATCATT CCAGATTTCTTGTGATTTTTCTATTGTGTAATTCTTGGTGGGCTCTGTAGTTTAATAATAAGAAAAAGGCCATTTCATTTTAAATTGTGA CCTATAATTCTTTGTCTTGGGTTGGTAATTCAGGATTCATTTGGAAAGTGGGTAAAAGGGGCTTCAAAAAACGGATAGAACAGGATTTTC TAGGAGTTACACATACATTTTATCCTGTCATACCTCGAGATAAAGTGGCATGTTAGTGAGGAGTTCTGATATTAAGCACACACACACATG CACACAAATGGACTTCTCTGAAGCTGTGTTTAGTGAAATGAGCTCAAGTACATGAATGTTAGTTGTTATCACATACAGCAAATTCCTTTT TTTTTCTTTTTCTATGAGCACACTCTGCTGCTTCTAAACTTTACATGCCTGATGGCACCTTACTCCAGCAGCCTCCAGGTGCTTTCATTT TCACTTCCAGTCTAAGCCAGTGGCTCCTGCCACTGCCCTCCCATTACCTAGATGGCACCTCCTTTGGTGAAACCACGGCCAATGTTCCTT AGCTGCACCAGGCCCGAAGCTGTTCCCATGCTTGAGCTTCCATGGGGAGGATGCTGAGTGAGCAGTTTCCTACCCCGTGGATCTAGCAAG CCATGGAGACAGGTAGCATTTGTAAGATGCTGCACAGGAGCAGCATTATCCCCAAAGATATTACAGGGTAGACACGTTTTAACTGAAATC AATCAAGATAACTTTATTCAAAGAGCAGCCCGCTTTGTGTGACTAAAATGAAACAAGACAGTTGAATTGTGTGACTTGAAGATTACCAAT GATTTTGAGGCTTTTCTATAATAAAAAGAGGTTCTAACCATTATTTGGGAACAAAGAGAGTTTTCATCTTTTTTCAGATCAAAACCATTC TGTAAAATCTTTGTTGTTTAATTAAATGTGCCGTTATTTACCCCTGATGTTATTTATGACTATGTGCCGATTCCTGCTCGGGCTGTTTGC TGTTGGCTGGTAATAATATATTTGATTTAAATGCTGTTGACTGTGCTATTAACTGCTGCCGTCAGTAAACTCCAAAGATCTTTTTGTTTT GGCTTTAGTATCATATGTGCTTTTTCTGTATCCTGAGCGCTCTATATGATCATGTTAATTTAAAGCTTTATACACATTGTTGTTTTTGCT GGTCTCATCTTTGGTAATATGCTATACCCCACTGCTGCCCGACACTGCCCTTTAGCTGCAGAGCTGGATTAGCTGTTGACCATTTGATGC TGTTGTCTGTCTGGCAGGGACTGAATGACCTGATGTCAGATTTAGATTCTTCCTGGGGATTACACAGCTATGAATGTATTTGCTTCTAAA ACCTCCCAAAGTGAATCTAATCTTAAAACTACAAGTTGTAAGTATTCTGAAATTGGGAAACATTTATTTTAAATGCAATCAGGTAGTGTT >38416_38416_2_IFT20-GLS_IFT20_chr17_26658882_ENST00000578985_GLS_chr2_191827556_ENST00000320717_length(amino acids)=138AA_BP=74 MTLAPMTHLLLTATVTPSEQNSSREPGWETAMAKDILGEAGLHFDELNKLRVLDPEVTQQTIELKEECKDFVDSGITLPWMKHCTLDTMM -------------------------------------------------------------- >38416_38416_3_IFT20-GLS_IFT20_chr17_26658882_ENST00000585089_GLS_chr2_191827556_ENST00000320717_length(transcript)=3199nt_BP=476nt GTGGCAGCGGCCGTGGCAGGCTGGCTGGGTACCGGCTGTCGCTGACCCAGGAGAAGCTGCCTGTCTACATCAGCCTGGGCTGCAGCGCGC TGCCGCCGCGGGGCCGGCAGCTGAACTATGTGCTCTTCAGGGCGGGCACCGTGTTGCATTCATCTTTGTACCCCCAGCATCTAGCAGTGT TGGCATGTAGTAGGCACTCAAGAAATGTGTGTTGAATGAACGATGCCTGTGACAAGCAAGCGGACTTTATTCTTTCCTGACCCTTGCTCC TATGACACACCTCCTCCTGACTGCCACTGTCACTCCTTCAGAGCAGAACTCCTCTAGGGAACCTGGATGGGAAACAGCCATGGCCAAGGA CATCCTGGGTGAAGCAGGGCTACACTTTGATGAACTGAACAAGCTGAGGGTGTTGGACCCAGAGGTTACCCAGCAGACCATAGAGCTGAA GGAAGAGTGCAAAGACTTTGTGGACAGTGGAATAACACTCCCATGGATGAAGCACTGCACTTTGGACACCATGATGTATTTAAAATTCTC CAAGAATACCAAGTCCAGTACACACCTCAAGGAGATTCTGACAACGGGAAGGAAAATCAAACCGTCCATAAGAATCTTGATGGATTGTTG TAATGGTCTCAAATCCCAAGATTTAAATCACTTACCTATTTAATTGTGGAAAATGATTATGAAGAACATGTGTATTTCTATCTGGTAGTG ATGTATATTTTACATTTGTCATTTCAGTGTTACTGGAGTTTTCTTCATTGTGCACACAGGACAAATCTGATCTCTTTGGGAAAAAATAGA AATAAAACAATCTCCCTCCATAATGTGAGCAATATTACCTCGTGCATTGTATAATTTGATGTAAAAGAAATAGTTACCAATGCTAGCTTG TGTGGTCTTCCATGATTTATTTGTGTTTTGTGAATTTTCAATTTATGGTGATGATCTGCTGATATGCATTTATAAAGTAAGCTCTGTTGT ACAGTCTGTCCAAATGGGTCAAGGTTGCCTTTAGAAGCAAATAGTGTGATTTTCAAGACTTCAAATACAAATTTAGTTTAAGTGTTTGAA CAACTATATGCACTTACGGTTGTGTGTTTAAAATGTCTCTCTCACCCCCTAGCTTCATGATGTGACTCTTAAAAAACTATAATAGTTAAC AACTGTTAGTAAGATAGACCAATTCTGATTAGACTTTATCAGGGAATCTGTTTAAGATATGTTTGGTGACCAAAACGTATGTGTGAATGT AGTTATAATGCTTTTGAAAAATTTTCCTTTTTCTATATCCCCTTAGTCCAGCCTCTCTTCTCAGACATTTAGCTATCTGCCTCTTTCCTT TAGCTGGGAAAGTGAGAGCTGGCATACTATGCAGTTTTTATGTTTTCCATAGTAAGTCAGAAAATGCCTCCTATTTCTGGCATCAGAACT TTGCCATTTGTCTACAGAAGACGAACCAGAGACAAAATTACTAAGTATAAATTAGTCAAGTTTATCAGTCTAAAAAACGAAGGGATGTGC AACTGCAGCTCTTTAAGAAGTTTTTTTTTTTTAGCTTCTAGGGTAAAGATAAATTCAGAAATGCTCTAAGCTACCAAAGTTATTCTGAAA GTATGGGAACTGCTACAACTAACAAACATTTGTTTCCAAGCCTGTCATTAAGAGTCTGCATCAAGAGATTTGTCCTCCTTGGGGGACCAC TGGATCATTCCAGATTTCTTGTGATTTTTCTATTGTGTAATTCTTGGTGGGCTCTGTAGTTTAATAATAAGAAAAAGGCCATTTCATTTT AAATTGTGACCTATAATTCTTTGTCTTGGGTTGGTAATTCAGGATTCATTTGGAAAGTGGGTAAAAGGGGCTTCAAAAAACGGATAGAAC AGGATTTTCTAGGAGTTACACATACATTTTATCCTGTCATACCTCGAGATAAAGTGGCATGTTAGTGAGGAGTTCTGATATTAAGCACAC ACACACATGCACACAAATGGACTTCTCTGAAGCTGTGTTTAGTGAAATGAGCTCAAGTACATGAATGTTAGTTGTTATCACATACAGCAA ATTCCTTTTTTTTTCTTTTTCTATGAGCACACTCTGCTGCTTCTAAACTTTACATGCCTGATGGCACCTTACTCCAGCAGCCTCCAGGTG CTTTCATTTTCACTTCCAGTCTAAGCCAGTGGCTCCTGCCACTGCCCTCCCATTACCTAGATGGCACCTCCTTTGGTGAAACCACGGCCA ATGTTCCTTAGCTGCACCAGGCCCGAAGCTGTTCCCATGCTTGAGCTTCCATGGGGAGGATGCTGAGTGAGCAGTTTCCTACCCCGTGGA TCTAGCAAGCCATGGAGACAGGTAGCATTTGTAAGATGCTGCACAGGAGCAGCATTATCCCCAAAGATATTACAGGGTAGACACGTTTTA ACTGAAATCAATCAAGATAACTTTATTCAAAGAGCAGCCCGCTTTGTGTGACTAAAATGAAACAAGACAGTTGAATTGTGTGACTTGAAG ATTACCAATGATTTTGAGGCTTTTCTATAATAAAAAGAGGTTCTAACCATTATTTGGGAACAAAGAGAGTTTTCATCTTTTTTCAGATCA AAACCATTCTGTAAAATCTTTGTTGTTTAATTAAATGTGCCGTTATTTACCCCTGATGTTATTTATGACTATGTGCCGATTCCTGCTCGG GCTGTTTGCTGTTGGCTGGTAATAATATATTTGATTTAAATGCTGTTGACTGTGCTATTAACTGCTGCCGTCAGTAAACTCCAAAGATCT TTTTGTTTTGGCTTTAGTATCATATGTGCTTTTTCTGTATCCTGAGCGCTCTATATGATCATGTTAATTTAAAGCTTTATACACATTGTT GTTTTTGCTGGTCTCATCTTTGGTAATATGCTATACCCCACTGCTGCCCGACACTGCCCTTTAGCTGCAGAGCTGGATTAGCTGTTGACC ATTTGATGCTGTTGTCTGTCTGGCAGGGACTGAATGACCTGATGTCAGATTTAGATTCTTCCTGGGGATTACACAGCTATGAATGTATTT GCTTCTAAAACCTCCCAAAGTGAATCTAATCTTAAAACTACAAGTTGTAAGTATTCTGAAATTGGGAAACATTTATTTTAAATGCAATCA >38416_38416_3_IFT20-GLS_IFT20_chr17_26658882_ENST00000585089_GLS_chr2_191827556_ENST00000320717_length(amino acids)=138AA_BP=74 MTLAPMTHLLLTATVTPSEQNSSREPGWETAMAKDILGEAGLHFDELNKLRVLDPEVTQQTIELKEECKDFVDSGITLPWMKHCTLDTMM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for IFT20-GLS |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for IFT20-GLS |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for IFT20-GLS |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C0000786 | Spontaneous abortion | 1 | CTD_human | |
| Tgene | C0000822 | Abortion, Tubal | 1 | CTD_human | |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human | |
| Tgene | C0029408 | Degenerative polyarthritis | 1 | CTD_human | |
| Tgene | C0086743 | Osteoarthrosis Deformans | 1 | CTD_human | |
| Tgene | C3830362 | Early Pregnancy Loss | 1 | CTD_human | |
| Tgene | C4552766 | Miscarriage | 1 | CTD_human |
