
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DDX21-BEX2 (FusionGDB2 ID:HG9188TG84707) |
Fusion Gene Summary for DDX21-BEX2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DDX21-BEX2 | Fusion gene ID: hg9188tg84707 | Hgene | Tgene | Gene symbol | DDX21 | BEX2 | Gene ID | 9188 | 84707 |
| Gene name | DExD-box helicase 21 | brain expressed X-linked 2 | |
| Synonyms | GUA|GURDB|RH-II/GU|RH-II/GuA | BEX1|DJ79P11.1 | |
| Cytomap | ('DDX21')('BEX2') 10q22.1 | Xq22.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nucleolar RNA helicase 2DEAD (Asp-Glu-Ala-Asp) box helicase 21DEAD (Asp-Glu-Ala-Asp) box polypeptide 21DEAD box protein 21DEAD-box helicase 21DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 21Gu proteinRH II/GuRNA helicase II/Gu alphagu-alphanucleo | protein BEX2brain-expressed X-linked protein 2hBex2 | |
| Modification date | 20200313 | 20200315 | |
| UniProtAcc | Q9NR30 | Q9BXY8 | |
| Ensembl transtripts involved in fusion gene | ENST00000354185, | ||
| Fusion gene scores | * DoF score | 11 X 11 X 2=242 | 3 X 2 X 1=6 |
| # samples | 12 | 3 | |
| ** MAII score | log2(12/242*10)=-1.01197264166608 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/6*10)=2.32192809488736 | |
| Context | PubMed: DDX21 [Title/Abstract] AND BEX2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DDX21(70742402)-BEX2(102564533), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | DDX21-BEX2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. DDX21-BEX2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. DDX21-BEX2 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. DDX21-BEX2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. DDX21-BEX2 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BEX2 | GO:0042981 | regulation of apoptotic process | 19711341 |
| Tgene | BEX2 | GO:0051726 | regulation of cell cycle | 19711341 |
 Fusion gene breakpoints across DDX21 (5'-gene) Fusion gene breakpoints across DDX21 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
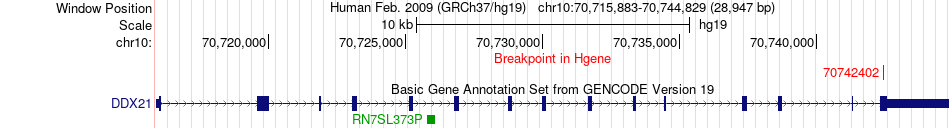 |
 Fusion gene breakpoints across BEX2 (3'-gene) Fusion gene breakpoints across BEX2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
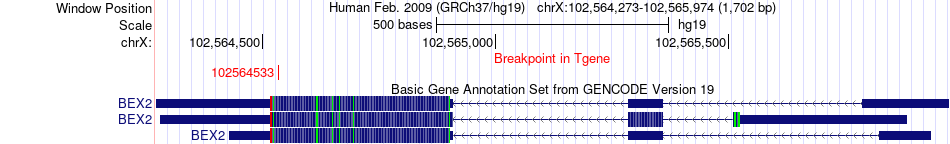 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | AW379411 | DDX21 | chr10 | 70742402 | - | BEX2 | chrX | 102564533 | - |
Top |
Fusion Gene ORF analysis for DDX21-BEX2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000354185 | ENST00000372674 | DDX21 | chr10 | 70742402 | - | BEX2 | chrX | 102564533 | - |
| Frame-shift | ENST00000354185 | ENST00000372677 | DDX21 | chr10 | 70742402 | - | BEX2 | chrX | 102564533 | - |
| In-frame | ENST00000354185 | ENST00000536889 | DDX21 | chr10 | 70742402 | - | BEX2 | chrX | 102564533 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000354185 | DDX21 | chr10 | 70742402 | - | ENST00000536889 | BEX2 | chrX | 102564533 | - | 4859 | 4607 | 92 | 2248 | 718 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000354185 | ENST00000536889 | DDX21 | chr10 | 70742402 | - | BEX2 | chrX | 102564533 | - | 0.000162236 | 0.99983776 |
Top |
Fusion Genomic Features for DDX21-BEX2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
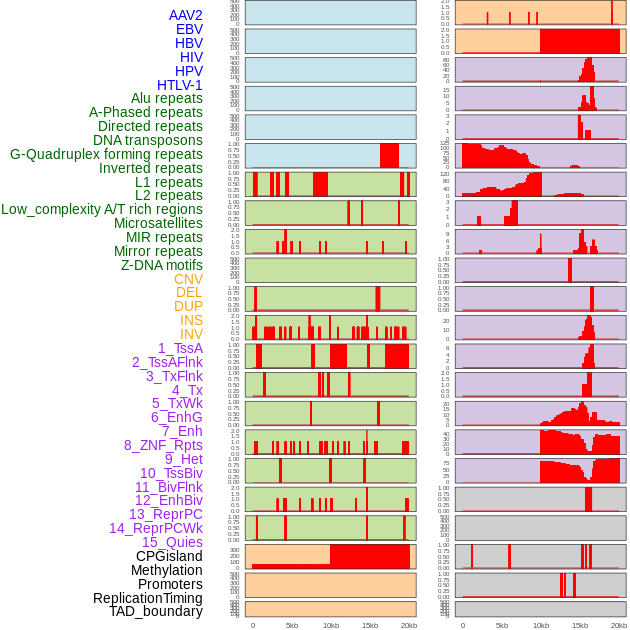 |
Top |
Fusion Protein Features for DDX21-BEX2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:70742402/chrX:102564533) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| DDX21 | BEX2 |
| FUNCTION: RNA helicase that acts as a sensor of the transcriptional status of both RNA polymerase (Pol) I and II: promotes ribosomal RNA (rRNA) processing and transcription from polymerase II (Pol II) (PubMed:25470060, PubMed:28790157). Binds various RNAs, such as rRNAs, snoRNAs, 7SK and, at lower extent, mRNAs (PubMed:25470060). In the nucleolus, localizes to rDNA locus, where it directly binds rRNAs and snoRNAs, and promotes rRNA transcription, processing and modification. Required for rRNA 2'-O-methylation, possibly by promoting the recruitment of late-acting snoRNAs SNORD56 and SNORD58 with pre-ribosomal complexes (PubMed:25470060, PubMed:25477391). In the nucleoplasm, binds 7SK RNA and is recruited to the promoters of Pol II-transcribed genes: acts by facilitating the release of P-TEFb from inhibitory 7SK snRNP in a manner that is dependent on its helicase activity, thereby promoting transcription of its target genes (PubMed:25470060). Functions as cofactor for JUN-activated transcription: required for phosphorylation of JUN at 'Ser-77' (PubMed:11823437, PubMed:25260534). Can unwind double-stranded RNA (helicase) and can fold or introduce a secondary structure to a single-stranded RNA (foldase) (PubMed:9461305). Together with SIRT7, required to prevent R-loop-associated DNA damage and transcription-associated genomic instability: deacetylation by SIRT7 activates the helicase activity, thereby overcoming R-loop-mediated stalling of RNA polymerases (PubMed:28790157). Involved in rRNA processing (PubMed:14559904, PubMed:18180292). May bind to specific miRNA hairpins (PubMed:28431233). Component of a multi-helicase-TICAM1 complex that acts as a cytoplasmic sensor of viral double-stranded RNA (dsRNA) and plays a role in the activation of a cascade of antiviral responses including the induction of proinflammatory cytokines via the adapter molecule TICAM1 (By similarity). {ECO:0000250|UniProtKB:Q9JIK5, ECO:0000269|PubMed:11823437, ECO:0000269|PubMed:14559904, ECO:0000269|PubMed:18180292, ECO:0000269|PubMed:25260534, ECO:0000269|PubMed:25470060, ECO:0000269|PubMed:25477391, ECO:0000269|PubMed:28431233, ECO:0000269|PubMed:28790157, ECO:0000269|PubMed:9461305}. | FUNCTION: Regulator of mitochondrial apoptosis and G1 cell cycle in breast cancer. Protects the breast cancer cells against mitochondrial apoptosis and this effect is mediated through the modulation of BCL2 protein family, which involves the positive regulation of anti-apoptotic member BCL2 and the negative regulation of pro-apoptotic members BAD, BAK1 and PUMA. Required for the normal cell cycle progression during G1 in breast cancer cells through the regulation of CCND1 and CDKN1A. Regulates the level of PP2A regulatory subunit B and PP2A phosphatase activity. {ECO:0000269|PubMed:19711341}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 217_396 | 0 | 784.0 | Domain | Helicase ATP-binding |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 429_573 | 0 | 784.0 | Domain | Helicase C-terminal |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 186_214 | 0 | 784.0 | Motif | Q motif |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 339_342 | 0 | 784.0 | Motif | DEAD box |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 230_237 | 0 | 784.0 | Nucleotide binding | ATP |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 724_748 | 0 | 784.0 | Region | Note=3 X 5 AA repeats |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 724_728 | 0 | 784.0 | Repeat | Note=1 |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 734_738 | 0 | 784.0 | Repeat | Note=2 |
| Hgene | DDX21 | chr10:70742402 | chrX:102564533 | ENST00000354185 | - | 1 | 15 | 744_748 | 0 | 784.0 | Repeat | Note=3 |
Top |
Fusion Gene Sequence for DDX21-BEX2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >21927_21927_1_DDX21-BEX2_DDX21_chr10_70742402_ENST00000354185_BEX2_chrX_102564533_ENST00000536889_length(transcript)=4859nt_BP=4607nt GGGAAGTGACGAGAGCCGGGTGGACCCAGGGTGGGGAACTACCTCTTCCTCTCCACGCGGTTGAGAAGACCGGTCGGCCTGGGCAACCTG CGCTGAAGATGCCGGGAAAACTCCGTAGTGACGCTGGTTTGGAATCAGACACCGCAATGAAAAAAGGGGAGACACTGCGAAAGCAAACCG AGGAGAAAGAGAAAAAAGAGAAGCCAAAATCTGATAAGACTGAAGAGATAGCAGAAGAGGAAGAAACTGTTTTCCCCAAAGCTAAACAAG TTAAAAAGAAAGCAGAGCCTTCTGAAGTTGACATGAATTCTCCTAAATCCAAAAAGGCAAAAAAGAAAGAGGAGCCATCTCAAAATGACA TTTCTCCTAAAACCAAAAGTTTGAGAAAGAAAAAGGAGCCCATTGAAAAGAAAGTGGTTTCTTCTAAAACCAAAAAAGTGACAAAAAATG AGGAGCCTTCTGAGGAAGAAATAGATGCTCCTAAGCCCAAGAAGATGAAGAAAGAAAAGGAAATGAATGGAGAAACTAGAGAGAAAAGCC CCAAACTGAAGAATGGATTTCCTCATCCTGAACCGGACTGTAACCCCAGTGAAGCTGCCAGTGAAGAAAGTAACAGTGAGATAGAGCAGG AAATACCTGTGGAACAAAAAGAAGGCGCTTTCTCTAATTTTCCCATATCTGAAGAAACTATTAAACTTCTCAAAGGCCGAGGAGTGACCT TCCTATTTCCTATACAAGCAAAGACATTCCATCATGTTTACAGCGGGAAGGACTTAATTGCACAGGCACGGACAGGAACTGGGAAGACAT TCTCCTTTGCCATCCCTTTGATTGAGAAACTTCATGGGGAACTGCAAGACAGGAAGAGAGGCCGTGCCCCTCAGGTACTGGTTCTTGCAC CTACAAGAGAGTTGGCAAATCAAGTAAGCAAAGACTTCAGTGACATCACAAAAAAGCTGTCAGTGGCTTGTTTTTATGGTGGAACTCCCT ATGGAGGTCAATTTGAACGCATGAGGAATGGGATTGATATCCTGGTTGGAACACCAGGTCGTATCAAAGACCACATACAGAATGGCAAAC TAGATCTCACCAAACTTAAGCATGTTGTCCTGGATGAAGTGGACCAGATGTTGGATATGGGATTTGCTGATCAAGTGGAAGAGATTTTAA GTGTGGCATACAAGAAAGATTCTGAAGACAATCCCCAAACATTGCTTTTTTCTGCAACTTGCCCTCATTGGGTATTTAATGTTGCCAAGA AATACATGAAATCTACATATGAACAGGTGGACCTGATTGGTAAAAAGACTCAGAAAACGGCAATAACTGTGGAGCATCTGGCTATTAAGT GCCACTGGACTCAGAGGGCAGCAGTTATTGGGGATGTCATCCGAGTATATAGTGGTCATCAAGGACGCACTATCATCTTTTGTGAAACCA AGAAAGAAGCCCAGGAGCTGTCCCAGAATTCAGCTATAAAGCAGGATGCTCAGTCCTTGCATGGAGACATTCCACAGAAGCAAAGGGAAA TCACCCTGAAAGGTTTTAGAAATGGTAGTTTTGGAGTTTTGGTGGCAACCAATGTTGCTGCACGTGGGTTAGACATCCCTGAGGTTGATT TGGTTATACAAAGCTCTCCACCAAAGGATGTAGAGTCCTACATTCATCGATCCGGGCGGACAGGCAGAGCTGGAAGGACGGGGGTGTGCA TCTGCTTTTATCAGCACAAGGAAGAATATCAGTTAGTACAAGTGGAGCAAAAAGCGGGAATTAAGTTCAAACGAATAGGTGTTCCTTCTG CAACAGAAATAATAAAAGCTTCCAGCAAAGATGCCATCAGGCTTTTGGATTCCGTGCCTCCCACTGCCATTAGTCACTTCAAACAATCAG CTGAGAAGCTGATAGAGGAGAAGGGAGCTGTGGAAGCTCTGGCAGCAGCACTGGCCCATATTTCAGGTGCCACGTCCGTAGACCAGCGCT CCTTGATCAACTCAAATGTGGGTTTTGTGACCATGATCTTGCAGTGCTCAATTGAAATGCCAAATATTAGTTATGCTTGGAAAGAACTTA AAGAGCAGCTGGGCGAGGAGATTGATTCCAAAGTGAAGGGAATGGTTTTTCTCAAAGGAAAGCTGGGTGTTTGCTTTGATGTACCTACCG CATCAGTAACAGAAATACAGAGGCAGTCGAGGCTTCAGGGGACAGCGGGACGGAAACAGAAGATTCAGAGGACAGCGGGAAGGCAGTAGA GGCCCGAGAGGACAGCGATCAGGAGGTGGCAACAAAAGTAACAGATCCCAAAACAAAGGCCAGAAGCGGAGTTTCAGTAAAGCATTTGGT CAATAATTAGAAATAGAAGATTTATATAGCAAAAAGAGAATGATGTTTGGCAATATAGAACTGAACATTATTTTTCATGCAAAGTTAAAA GCACATTGTGCCTCCTTTTGACCACTTGCCAAGTCCCTGTCTCTTTCAGACACAGACAAGCTTCATTTAAATTATTTCATCTGATCATTA TCATTTATAACTTTATTGTTACTTCTTCATCAGTTTTTCCTTTTGAAAGGTGTATGAATTCATTACATTTTTATTCTAATGTATTATCTG TAGATTAGAAGATAAAATCAAGCATGTATCTGCCTATACTTTGTGAGTTCACCTGTCTTTATACTCAAAAGTGTCCCTTAATAGTGTCCT TCCCTGAAATAAATACCTAAGGGAGTGTAACAGTCTCTGGAGGACCACTTTGAGCCTTTGGAAGTTAAGGTTTCCTCAGCCACCTGCCGA ACAGTTTCTCATGTGGTCCTATTATTTGTCTACTGAGACTTAATACTGAGCAATGTTTTGAAACAAGATTTCAAACTAATCTGGGTTGTA ATACAGTTTATACCAGTGTATGCTCTAGACTTGGAAGATGTAGTATGTTTGATGTGGATTACCTATACTTATGTTCGTTTTGATACATTT TTAGCTTCTCATTATAAGGTGATTCATGCTTTAGTGAATTCTTCATAGATAGTATATATAAAAGTACATTTTAATAGAAAGCCAGGGTTT TAAGGAATTTCACATGTATAAGGTGGCTCCATAGCTTTATTTGTAAGTAGGCTGGATAAATGGTGCTTAAATGGTAATGTACTCCACTTC TTCCTATTGGAAGATTAACATTATTTACCAAGAAGGACTTAAGGGAGTAGGGGGCGCAGATTAGCATTGCTCAAGAGTATGTAAAAAAAA AAAAAAAAAAAAAGAACCAAACCACTGGAAATAATCAAATGCAAAAAGGTAACAAATTCATAACTGGAAAGCAAAGAGAAGAACAAGTAT GATTTGGATGATAAAGCATTGTTTTAATGGTGAAAACTTCACAGATCACTAATGTTTCTAGAGGTTAACTTCAAGTGGGCAAGCTGGGGT TTTTAGGTAGTCAGTGGCCTAGTTCCTAAAGCCACAGTATAGGATCTGTTAAACTGAATGTCTGTTGAAAGTTTGTTTTAGCTGCTTGGA GGCTTCCTTTTAAGACAAACTGTATGTGATTAAGTTGTTTTGAGGGAACTGAAGAACCTGATGTAGCCCCTGGCCAGATAACTGCCTGAT TTCTCAGATATTATTTCTCTGGGAAACATTCTACATAGCACAGGAGCTTAAGAGTGGCATTATCTTCTCGCCTTAATTTCCAGAGATTAT TTCTGTACTGAGAATCCTGGAACTACTATGCTAGGAAATTTAAAGCTGCATGGTCTGTCTTGTTTTCATTTAATTATTGTGAATACCTAG AATCTTTCTTGGTCCTGATTTCTCTTGCTTAATCCAGTCTTTATCTCTAACTGCCCCTTATTTGATCACCATGTACTAGGAGCTCTGATA GCCAGCTCAGCTCCTAATCCTTGAGGCAACATTCTTTTTCTATTTGAACTTCAGTTCTGTCCTTGAATCCCGACTAGATATTTCTTGCCC TCTGGTCTCAGAATTCTCTTGGCTTTTATTCCTTGATCCACTTGCCAGTTTTATCACTTTACCCTTGTTCCTCATGGCTTCCCATCAAGC CATGGGTATTAGGTGACAGTGTAATTTATTAGATTCTGGTTTTGCCCAAATACTGGGCATGCTTTAATAATAACTGAACCATTTCATTAT TTGGATAGGCATGGGTACCTTATCAAGCAGATTAAAAGGATATGGTACCCGTCCTTTAGAAAAGAACAGCTAAAACCTTGTTGTGGATTA TGGATTTAGCCTAAAGAAAAATAATCTGGCATAAATTAAGAGTAAGAGAGAAGATTAATAGAAATTTCACTTCACATAACTTAAAACATG GCTATTTCAATAAAGGACTAAAGTTTCTCCTGGATCCCAGAATTCAACCTGTATTTATAAATGTATAATGTATTTAGCTACTTTTTGGTT TAAATGAACTTGTTGGGTTAGCTTGGTAAATGTTATAATTTTTACTATTTTCTACAAAGAAAATATTTTCTAATTTAAGTTGGAGCTATC TGTGCAGCAGTTTCTCTACAGTTGTGCATAAATGTTTTTACTATAAAATGAGCTAATGTATAAAATACTGCTGTATACCATAATAAAGAT AGTAATACTTGATTTTATTGCCTTATGCCCTGAATCCTGATGGTTTCCCTGAAGTTAATAGGGAGACCCCTGCTTCCTAAACTTACACAT TTGTGGTGTACCTTTGTCGTAAACGTTTTGATGTTACCTATTTCTTGTGGGTCTCCTATTACCAGCTTCTAAATGAATGTTGTTTTTGAC >21927_21927_1_DDX21-BEX2_DDX21_chr10_70742402_ENST00000354185_BEX2_chrX_102564533_ENST00000536889_length(amino acids)=718AA_BP= MKMPGKLRSDAGLESDTAMKKGETLRKQTEEKEKKEKPKSDKTEEIAEEEETVFPKAKQVKKKAEPSEVDMNSPKSKKAKKKEEPSQNDI SPKTKSLRKKKEPIEKKVVSSKTKKVTKNEEPSEEEIDAPKPKKMKKEKEMNGETREKSPKLKNGFPHPEPDCNPSEAASEESNSEIEQE IPVEQKEGAFSNFPISEETIKLLKGRGVTFLFPIQAKTFHHVYSGKDLIAQARTGTGKTFSFAIPLIEKLHGELQDRKRGRAPQVLVLAP TRELANQVSKDFSDITKKLSVACFYGGTPYGGQFERMRNGIDILVGTPGRIKDHIQNGKLDLTKLKHVVLDEVDQMLDMGFADQVEEILS VAYKKDSEDNPQTLLFSATCPHWVFNVAKKYMKSTYEQVDLIGKKTQKTAITVEHLAIKCHWTQRAAVIGDVIRVYSGHQGRTIIFCETK KEAQELSQNSAIKQDAQSLHGDIPQKQREITLKGFRNGSFGVLVATNVAARGLDIPEVDLVIQSSPPKDVESYIHRSGRTGRAGRTGVCI CFYQHKEEYQLVQVEQKAGIKFKRIGVPSATEIIKASSKDAIRLLDSVPPTAISHFKQSAEKLIEEKGAVEALAAALAHISGATSVDQRS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DDX21-BEX2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DDX21-BEX2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DDX21-BEX2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
