
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACVR2A-CASP8 (FusionGDB2 ID:HG92TG841) |
Fusion Gene Summary for ACVR2A-CASP8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACVR2A-CASP8 | Fusion gene ID: hg92tg841 | Hgene | Tgene | Gene symbol | ACVR2A | CASP8 | Gene ID | 92 | 841 |
| Gene name | activin A receptor type 2A | caspase 8 | |
| Synonyms | ACTRII|ACVR2 | ALPS2B|CAP4|Casp-8|FLICE|MACH|MCH5 | |
| Cytomap | ('ACVR2A')('CASP8') 2q22.3-q23.1 | 2q33.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | activin receptor type-2Aactivin A receptor, type IIA | caspase-8FADD-homologous ICE/CED-3-like proteaseFADD-like ICEICE-like apoptotic protease 5MACH-alpha-1/2/3 proteinMACH-beta-1/2/3/4 proteinMORT1-associated ced-3 homologapoptotic cysteine proteaseapoptotic protease Mch-5caspase 8, apoptosis-relat | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000241416, ENST00000404590, ENST00000535787, ENST00000495775, | ||
| Fusion gene scores | * DoF score | 8 X 7 X 7=392 | 4 X 4 X 3=48 |
| # samples | 10 | 5 | |
| ** MAII score | log2(10/392*10)=-1.97085365434048 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/48*10)=0.0588936890535686 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: ACVR2A [Title/Abstract] AND CASP8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACVR2A(148674995)-CASP8(202131184), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. ACVR2A-CASP8 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACVR2A | GO:0030509 | BMP signaling pathway | 18436533 |
| Hgene | ACVR2A | GO:0032927 | positive regulation of activin receptor signaling pathway | 12665502 |
| Hgene | ACVR2A | GO:0045648 | positive regulation of erythrocyte differentiation | 9032295 |
| Tgene | CASP8 | GO:0006508 | proteolysis | 12888622 |
| Tgene | CASP8 | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 21785459 |
| Tgene | CASP8 | GO:0045862 | positive regulation of proteolysis | 18387192 |
| Tgene | CASP8 | GO:0097191 | extrinsic apoptotic signaling pathway | 21785459 |
| Tgene | CASP8 | GO:0097202 | activation of cysteine-type endopeptidase activity | 18387192 |
 Fusion gene breakpoints across ACVR2A (5'-gene) Fusion gene breakpoints across ACVR2A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
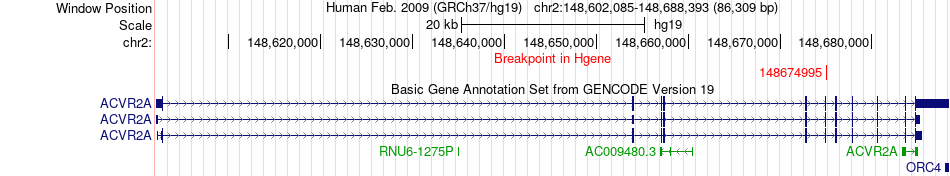 |
 Fusion gene breakpoints across CASP8 (3'-gene) Fusion gene breakpoints across CASP8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
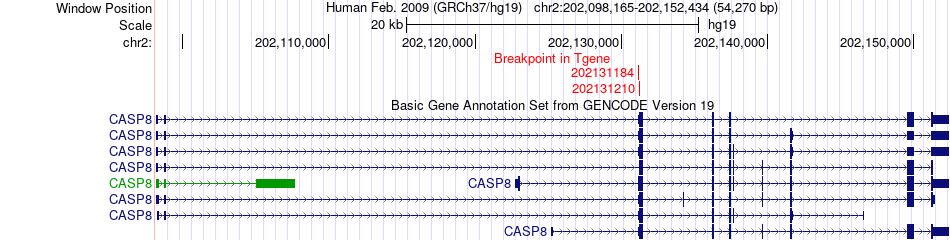 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-21-1082-01A | ACVR2A | chr2 | 148674995 | - | CASP8 | chr2 | 202131184 | + |
| ChimerDB4 | LUSC | TCGA-21-1082-01A | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| ChimerDB4 | LUSC | TCGA-21-1082 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| ChimerDB4 | LUSC | TCGA-21-1082 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
Top |
Fusion Gene ORF analysis for ACVR2A-CASP8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000241416 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000241416 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000241416 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000241416 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000241416 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000241416 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000404590 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000404590 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000404590 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000404590 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000404590 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000404590 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000535787 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000535787 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000535787 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000535787 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000535787 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-5UTR | ENST00000535787 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000241416 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000241416 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000241416 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000241416 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000241416 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000241416 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000404590 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000404590 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000404590 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000404590 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000404590 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000404590 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000535787 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000535787 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000535787 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| 5CDS-intron | ENST00000535787 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000535787 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| 5CDS-intron | ENST00000535787 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000241416 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000241416 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000241416 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000241416 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000241416 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000404590 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000404590 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000404590 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000404590 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000404590 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000535787 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000535787 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000535787 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000535787 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| Frame-shift | ENST00000535787 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| In-frame | ENST00000241416 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| In-frame | ENST00000404590 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| In-frame | ENST00000535787 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-3CDS | ENST00000495775 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-3CDS | ENST00000495775 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-3CDS | ENST00000495775 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-3CDS | ENST00000495775 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-3CDS | ENST00000495775 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-3CDS | ENST00000495775 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-5UTR | ENST00000495775 | ENST00000264274 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-5UTR | ENST00000495775 | ENST00000264275 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-5UTR | ENST00000495775 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-5UTR | ENST00000495775 | ENST00000392258 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-5UTR | ENST00000495775 | ENST00000392259 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-5UTR | ENST00000495775 | ENST00000392266 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-intron | ENST00000495775 | ENST00000323492 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-intron | ENST00000495775 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-intron | ENST00000495775 | ENST00000358485 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
| intron-intron | ENST00000495775 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-intron | ENST00000495775 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131184 | + |
| intron-intron | ENST00000495775 | ENST00000490682 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535787 | ACVR2A | chr2 | 148674995 | + | ENST00000432109 | CASP8 | chr2 | 202131210 | + | 2328 | 888 | 90 | 2327 | 745 |
| ENST00000241416 | ACVR2A | chr2 | 148674995 | + | ENST00000432109 | CASP8 | chr2 | 202131210 | + | 2892 | 1452 | 636 | 2891 | 751 |
| ENST00000404590 | ACVR2A | chr2 | 148674995 | + | ENST00000432109 | CASP8 | chr2 | 202131210 | + | 2426 | 986 | 170 | 2425 | 752 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535787 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + | 0.002276216 | 0.99772376 |
| ENST00000241416 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + | 0.000623458 | 0.9993766 |
| ENST00000404590 | ENST00000432109 | ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131210 | + | 0.001343028 | 0.998657 |
Top |
Fusion Genomic Features for ACVR2A-CASP8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131183 | + | 1.99E-06 | 0.999998 |
| ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131183 | + | 1.99E-06 | 0.999998 |
| ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131209 | + | 4.96E-07 | 0.9999995 |
| ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131183 | + | 1.99E-06 | 0.999998 |
| ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131183 | + | 1.99E-06 | 0.999998 |
| ACVR2A | chr2 | 148674995 | + | CASP8 | chr2 | 202131209 | + | 4.96E-07 | 0.9999995 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
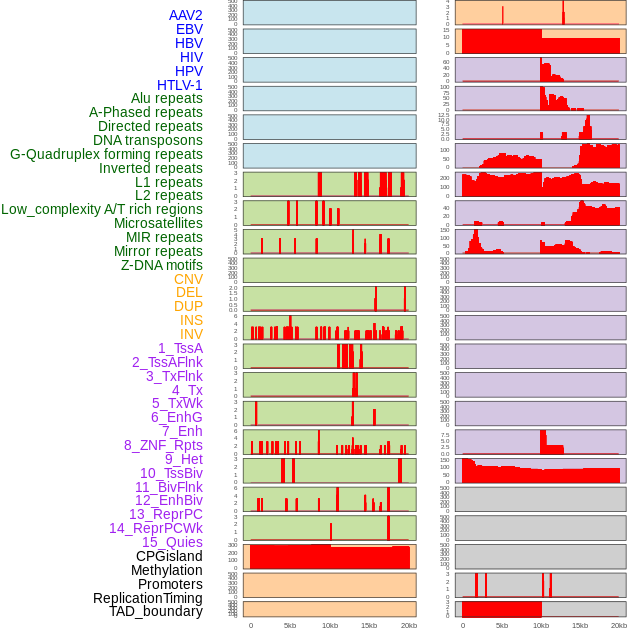 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
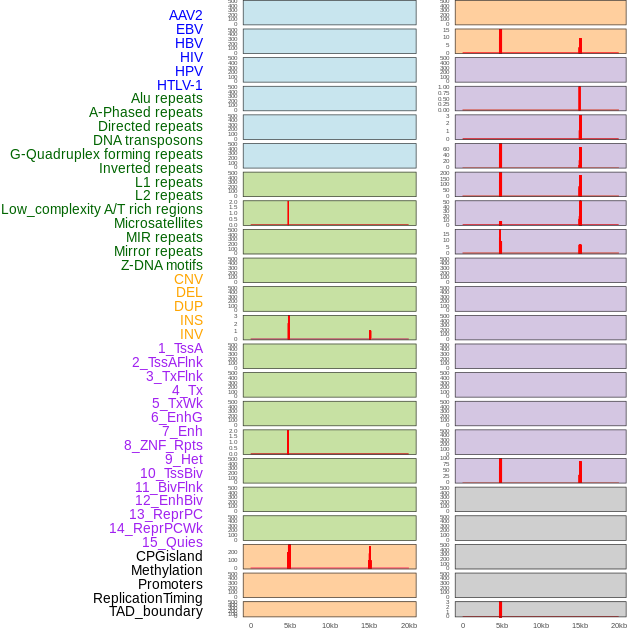 |
Top |
Fusion Protein Features for ACVR2A-CASP8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:148674995/chr2:202131184) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000241416 | + | 6 | 11 | 198_206 | 272 | 514.0 | Nucleotide binding | ATP |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000404590 | + | 7 | 12 | 198_206 | 272 | 514.0 | Nucleotide binding | ATP |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000241416 | + | 6 | 11 | 20_135 | 272 | 514.0 | Topological domain | Extracellular |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000404590 | + | 7 | 12 | 20_135 | 272 | 514.0 | Topological domain | Extracellular |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000241416 | + | 6 | 11 | 136_161 | 272 | 514.0 | Transmembrane | Helical |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000404590 | + | 7 | 12 | 136_161 | 272 | 514.0 | Transmembrane | Helical |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000264274 | 0 | 7 | 100_177 | 0 | 396.0 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000264274 | 0 | 7 | 2_80 | 0 | 396.0 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000264275 | 0 | 10 | 100_177 | 0 | 497.0 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000264275 | 0 | 10 | 2_80 | 0 | 497.0 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000323492 | 0 | 8 | 100_177 | 0 | 465.0 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000323492 | 0 | 8 | 2_80 | 0 | 465.0 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000358485 | 0 | 9 | 100_177 | 0 | 539.0 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000358485 | 0 | 9 | 2_80 | 0 | 539.0 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000392258 | 0 | 8 | 100_177 | 0 | 224.66666666666666 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000392258 | 0 | 8 | 2_80 | 0 | 224.66666666666666 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000392259 | 0 | 9 | 100_177 | 0 | 751.3333333333334 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000392259 | 0 | 9 | 2_80 | 0 | 751.3333333333334 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000392266 | 0 | 8 | 100_177 | 0 | 736.3333333333334 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000392266 | 0 | 8 | 2_80 | 0 | 736.3333333333334 | Domain | DED 1 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000432109 | 1 | 10 | 100_177 | 0 | 480.0 | Domain | DED 2 | |
| Tgene | CASP8 | chr2:148674995 | chr2:202131210 | ENST00000432109 | 1 | 10 | 2_80 | 0 | 480.0 | Domain | DED 1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000241416 | + | 6 | 11 | 192_485 | 272 | 514.0 | Domain | Protein kinase |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000404590 | + | 7 | 12 | 192_485 | 272 | 514.0 | Domain | Protein kinase |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000241416 | + | 6 | 11 | 162_513 | 272 | 514.0 | Topological domain | Cytoplasmic |
| Hgene | ACVR2A | chr2:148674995 | chr2:202131210 | ENST00000404590 | + | 7 | 12 | 162_513 | 272 | 514.0 | Topological domain | Cytoplasmic |
Top |
Fusion Gene Sequence for ACVR2A-CASP8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1905_1905_1_ACVR2A-CASP8_ACVR2A_chr2_148674995_ENST00000241416_CASP8_chr2_202131210_ENST00000432109_length(transcript)=2892nt_BP=1452nt AGGAGACCGAAAACGCGGCCGAGCCCGGAGCCCGGAGCTGGAGCCAGAGCCTGGACCAGAACTTGGCCGCCGCCTGCACCGCCGCCGCCG CTGCCGCCCGCCGCCCCTTCCCCGCGCCGCAGCCGCCTCGCCGCCACCGCCGCGAGCTCGGCCGCCAGTGGTCCTCGGACTTTAGGTGTC TGGGTTGAAGGTCGGTCCGGACATCGGACCCAGTCAGTTCCCAGACTGTCCGGGCGGCAGCGGACGCCGCTGCCGCCGCCGCCTCTTCGT CGCCTCAGCCTGGCGTTTTGTTCCGAGAGACGGGAGAGGCGAGCGGAGCTGACAGTGATTTTGACAGTGATTTAAACCCGCTTTTGTTGT TGTTGGCTTTTCGTTGTTTGGTTTTGTGTGTTGTGCGTGTGTGTGGCGGTTTTTCCCCCGTGGTGCATTTTATTTTTTATCGTTGTGCTC TTTTTTTTCTTTTTTTTTTTTAACCCAGTGAGCGTTTTTTTTTTTTTTTTTTTTTTTTTTTGGTCTGGGCTTCCGAATATGTTTTATGAC GGTTGATTTTACACCAGGAGGTTTGTCTCCGAGGAAGACCCAGGGAACTGGATATCTAGCGAGAACTTCCTCCGGATTCCCCGGCGCCTC GGGAAAATGGGAGCTGCTGCAAAGTTGGCGTTTGCCGTCTTTCTTATCTCCTGTTCTTCAGGTGCTATACTTGGTAGATCAGAAACTCAG GAGTGTCTTTTCTTTAATGCTAATTGGGAAAAAGACAGAACCAATCAAACTGGTGTTGAACCGTGTTATGGTGACAAAGATAAACGGCGG CATTGTTTTGCTACCTGGAAGAATATTTCTGGTTCCATTGAAATAGTGAAACAAGGTTGTTGGCTGGATGATATCAACTGCTATGACAGG ACTGATTGTGTAGAAAAAAAAGACAGCCCTGAAGTATATTTTTGTTGCTGTGAGGGCAATATGTGTAATGAAAAGTTTTCTTATTTTCCG GAGATGGAAGTCACACAGCCCACTTCAAATCCAGTTACACCTAAGCCACCCTATTACAACATCCTGCTCTATTCCTTGGTGCCACTTATG TTAATTGCGGGGATTGTCATTTGTGCATTTTGGGTGTACAGGCATCACAAGATGGCCTACCCTCCTGTACTTGTTCCAACTCAAGACCCA GGACCACCCCCACCTTCTCCATTACTAGGTTTGAAACCACTGCAGTTATTAGAAGTGAAAGCAAGGGGAAGATTTGGTTGTGTCTGGAAA GCCCAGTTGCTTAACGAATATGTGGCTGTCAAAATATTTCCAATACAGGACAAACAGTCATGGCAAAATGAATACGAAGTCTACAGTTTG CCTGGAATGAAGCATGAGAACATATTACAGTTCATTGGTGCAGAAAAACGAGGCACCAGTGTTGATGTGGATCTTTGGCTGATCACAGCA TTTCATGAAAAGATGGACTTCAGCAGAAATCTTTATGATATTGGGGAACAACTGGACAGTGAAGATCTGGCCTCCCTCAAGTTCCTGAGC CTGGACTACATTCCGCAAAGGAAGCAAGAACCCATCAAGGATGCCTTGATGTTATTCCAGAGACTCCAGGAAAAGAGAATGTTGGAGGAA AGCAATCTGTCCTTCCTGAAGGAGCTGCTCTTCCGAATTAATAGACTGGATTTGCTGATTACCTACCTAAACACTAGAAAGGAGGAGATG GAAAGGGAACTTCAGACACCAGGCAGGGCTCAAATTTCTGCCTACAGGGTCATGCTCTATCAGATTTCAGAAGAAGTGAGCAGATCAGAA TTGAGGTCTTTTAAGTTTCTTTTGCAAGAGGAAATCTCCAAATGCAAACTGGATGATGACATGAACCTGCTGGATATTTTCATAGAGATG GAGAAGAGGGTCATCCTGGGAGAAGGAAAGTTGGACATCCTGAAAAGAGTCTGTGCCCAAATCAACAAGAGCCTGCTGAAGATAATCAAC GACTATGAAGAATTCAGCAAAGAGAGAAGCAGCAGCCTTGAAGGAAGTCCTGATGAATTTTCAAATGGGGAGGAGTTGTGTGGGGTAATG ACAATCTCGGACTCTCCAAGAGAACAGGATAGTGAATCACAGACTTTGGACAAAGTTTACCAAATGAAAAGCAAACCTCGGGGATACTGT CTGATCATCAACAATCACAATTTTGCAAAAGCACGGGAGAAAGTGCCCAAACTTCACAGCATTAGGGACAGGAATGGAACACACTTGGAT GCAGGGGCTTTGACCACGACCTTTGAAGAGCTTCATTTTGAGATCAAGCCCCACGATGACTGCACAGTAGAGCAAATCTATGAGATTTTG AAAATCTACCAACTCATGGACCACAGTAACATGGACTGCTTCATCTGCTGTATCCTCTCCCATGGAGACAAGGGCATCATCTATGGCACT GATGGACAGGAGGCCCCCATCTATGAGCTGACATCTCAGTTCACTGGTTTGAAGTGCCCTTCCCTTGCTGGAAAACCCAAAGTGTTTTTT ATTCAGGCTTGTCAGGGGGATAACTACCAGAAAGGTATACCTGTTGAGACTGATTCAGAGGAGCAACCCTATTTAGAAATGGATTTATCA TCACCTCAAACGAGATATATCCCGGATGAGGCTGACTTTCTGCTGGGGATGGCCACTGTGAATAACTGTGTTTCCTACCGAAACCCTGCA GAGGGAACCTGGTACATCCAGTCACTTTGCCAGAGCCTGAGAGAGCGATGTCCTCGAGGCGATGATATTCTCACCATCCTGACTGAAGTG AACTATGAAGTAAGCAACAAGGATGACAAGAAAAACATGGGGAAACAGATGCCTCAGCCTACTTTCACACTAAGAAAAAAACTTGTCTTC >1905_1905_1_ACVR2A-CASP8_ACVR2A_chr2_148674995_ENST00000241416_CASP8_chr2_202131210_ENST00000432109_length(amino acids)=751AA_BP=271 MGAAAKLAFAVFLISCSSGAILGRSETQECLFFNANWEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGSIEIVKQGCWLDDINCYDRTD CVEKKDSPEVYFCCCEGNMCNEKFSYFPEMEVTQPTSNPVTPKPPYYNILLYSLVPLMLIAGIVICAFWVYRHHKMAYPPVLVPTQDPGP PPPSPLLGLKPLQLLEVKARGRFGCVWKAQLLNEYVAVKIFPIQDKQSWQNEYEVYSLPGMKHENILQFIGAEKRGTSVDVDLWLITAFH EKMDFSRNLYDIGEQLDSEDLASLKFLSLDYIPQRKQEPIKDALMLFQRLQEKRMLEESNLSFLKELLFRINRLDLLITYLNTRKEEMER ELQTPGRAQISAYRVMLYQISEEVSRSELRSFKFLLQEEISKCKLDDDMNLLDIFIEMEKRVILGEGKLDILKRVCAQINKSLLKIINDY EEFSKERSSSLEGSPDEFSNGEELCGVMTISDSPREQDSESQTLDKVYQMKSKPRGYCLIINNHNFAKAREKVPKLHSIRDRNGTHLDAG ALTTTFEELHFEIKPHDDCTVEQIYEILKIYQLMDHSNMDCFICCILSHGDKGIIYGTDGQEAPIYELTSQFTGLKCPSLAGKPKVFFIQ ACQGDNYQKGIPVETDSEEQPYLEMDLSSPQTRYIPDEADFLLGMATVNNCVSYRNPAEGTWYIQSLCQSLRERCPRGDDILTILTEVNY -------------------------------------------------------------- >1905_1905_2_ACVR2A-CASP8_ACVR2A_chr2_148674995_ENST00000404590_CASP8_chr2_202131210_ENST00000432109_length(transcript)=2426nt_BP=986nt GCCGCCCCTTCCCCGCGCCGCAGCCGCCTCGCCGCCACCGCCGCGAGCTCGGCCGCCAGTGGTCCTCGGACTTTAGGTGTCTGGGTTGAA GGAGGTTTGTCTCCGAGGAAGACCCAGGGAACTGGATATCTAGCGAGAACTTCCTCCGGATTCCCCGGCGCCTCGGGAAAATGGGAGCTG CTGCAAAGTTGGCGTTTGCCGTCTTTCTTATCTCCTGTTCTTCAGGTGCTATACTTGGTAGATCAGAAACTCAGGAGTGTCTTTTCTTTA ATGCTAATTGGGAAAAAGACAGAACCAATCAAACTGGTGTTGAACCGTGTTATGGTGACAAAGATAAACGGCGGCATTGTTTTGCTACCT GGAAGAATATTTCTGGTTCCATTGAAATAGTGAAACAAGGTTGTTGGCTGGATGATATCAACTGCTATGACAGGACTGATTGTGTAGAAA AAAAAGACAGCCCTGAAGTATATTTTTGTTGCTGTGAGGGCAATATGTGTAATGAAAAGTTTTCTTATTTTCCGGAGATGGAAGTCACAC AGCCCACTTCAAATCCAGTTACACCTAAGCCACCCTATTACAACATCCTGCTCTATTCCTTGGTGCCACTTATGTTAATTGCGGGGATTG TCATTTGTGCATTTTGGGTGTACAGGCATCACAAGATGGCCTACCCTCCTGTACTTGTTCCAACTCAAGACCCAGGACCACCCCCACCTT CTCCATTACTAGGTTTGAAACCACTGCAGTTATTAGAAGTGAAAGCAAGGGGAAGATTTGGTTGTGTCTGGAAAGCCCAGTTGCTTAACG AATATGTGGCTGTCAAAATATTTCCAATACAGGACAAACAGTCATGGCAAAATGAATACGAAGTCTACAGTTTGCCTGGAATGAAGCATG AGAACATATTACAGTTCATTGGTGCAGAAAAACGAGGCACCAGTGTTGATGTGGATCTTTGGCTGATCACAGCATTTCATGAAAAGATGG ACTTCAGCAGAAATCTTTATGATATTGGGGAACAACTGGACAGTGAAGATCTGGCCTCCCTCAAGTTCCTGAGCCTGGACTACATTCCGC AAAGGAAGCAAGAACCCATCAAGGATGCCTTGATGTTATTCCAGAGACTCCAGGAAAAGAGAATGTTGGAGGAAAGCAATCTGTCCTTCC TGAAGGAGCTGCTCTTCCGAATTAATAGACTGGATTTGCTGATTACCTACCTAAACACTAGAAAGGAGGAGATGGAAAGGGAACTTCAGA CACCAGGCAGGGCTCAAATTTCTGCCTACAGGGTCATGCTCTATCAGATTTCAGAAGAAGTGAGCAGATCAGAATTGAGGTCTTTTAAGT TTCTTTTGCAAGAGGAAATCTCCAAATGCAAACTGGATGATGACATGAACCTGCTGGATATTTTCATAGAGATGGAGAAGAGGGTCATCC TGGGAGAAGGAAAGTTGGACATCCTGAAAAGAGTCTGTGCCCAAATCAACAAGAGCCTGCTGAAGATAATCAACGACTATGAAGAATTCA GCAAAGAGAGAAGCAGCAGCCTTGAAGGAAGTCCTGATGAATTTTCAAATGGGGAGGAGTTGTGTGGGGTAATGACAATCTCGGACTCTC CAAGAGAACAGGATAGTGAATCACAGACTTTGGACAAAGTTTACCAAATGAAAAGCAAACCTCGGGGATACTGTCTGATCATCAACAATC ACAATTTTGCAAAAGCACGGGAGAAAGTGCCCAAACTTCACAGCATTAGGGACAGGAATGGAACACACTTGGATGCAGGGGCTTTGACCA CGACCTTTGAAGAGCTTCATTTTGAGATCAAGCCCCACGATGACTGCACAGTAGAGCAAATCTATGAGATTTTGAAAATCTACCAACTCA TGGACCACAGTAACATGGACTGCTTCATCTGCTGTATCCTCTCCCATGGAGACAAGGGCATCATCTATGGCACTGATGGACAGGAGGCCC CCATCTATGAGCTGACATCTCAGTTCACTGGTTTGAAGTGCCCTTCCCTTGCTGGAAAACCCAAAGTGTTTTTTATTCAGGCTTGTCAGG GGGATAACTACCAGAAAGGTATACCTGTTGAGACTGATTCAGAGGAGCAACCCTATTTAGAAATGGATTTATCATCACCTCAAACGAGAT ATATCCCGGATGAGGCTGACTTTCTGCTGGGGATGGCCACTGTGAATAACTGTGTTTCCTACCGAAACCCTGCAGAGGGAACCTGGTACA TCCAGTCACTTTGCCAGAGCCTGAGAGAGCGATGTCCTCGAGGCGATGATATTCTCACCATCCTGACTGAAGTGAACTATGAAGTAAGCA >1905_1905_2_ACVR2A-CASP8_ACVR2A_chr2_148674995_ENST00000404590_CASP8_chr2_202131210_ENST00000432109_length(amino acids)=752AA_BP=271 MGAAAKLAFAVFLISCSSGAILGRSETQECLFFNANWEKDRTNQTGVEPCYGDKDKRRHCFATWKNISGSIEIVKQGCWLDDINCYDRTD CVEKKDSPEVYFCCCEGNMCNEKFSYFPEMEVTQPTSNPVTPKPPYYNILLYSLVPLMLIAGIVICAFWVYRHHKMAYPPVLVPTQDPGP PPPSPLLGLKPLQLLEVKARGRFGCVWKAQLLNEYVAVKIFPIQDKQSWQNEYEVYSLPGMKHENILQFIGAEKRGTSVDVDLWLITAFH EKMDFSRNLYDIGEQLDSEDLASLKFLSLDYIPQRKQEPIKDALMLFQRLQEKRMLEESNLSFLKELLFRINRLDLLITYLNTRKEEMER ELQTPGRAQISAYRVMLYQISEEVSRSELRSFKFLLQEEISKCKLDDDMNLLDIFIEMEKRVILGEGKLDILKRVCAQINKSLLKIINDY EEFSKERSSSLEGSPDEFSNGEELCGVMTISDSPREQDSESQTLDKVYQMKSKPRGYCLIINNHNFAKAREKVPKLHSIRDRNGTHLDAG ALTTTFEELHFEIKPHDDCTVEQIYEILKIYQLMDHSNMDCFICCILSHGDKGIIYGTDGQEAPIYELTSQFTGLKCPSLAGKPKVFFIQ ACQGDNYQKGIPVETDSEEQPYLEMDLSSPQTRYIPDEADFLLGMATVNNCVSYRNPAEGTWYIQSLCQSLRERCPRGDDILTILTEVNY -------------------------------------------------------------- >1905_1905_3_ACVR2A-CASP8_ACVR2A_chr2_148674995_ENST00000535787_CASP8_chr2_202131210_ENST00000432109_length(transcript)=2328nt_BP=888nt AGGAGACCGAAAACGCGGCCGAGCCCGGAGCCCGGAGCTGGAGCCAGAGCCTGGACCAGAACTTGGCCGCCGCCTGCACCGCCGCCGCCG CTGCCGCCCGCCGCCCCTTCCCCGCGCCGCAGCCGCCTCGCCGCCACCGCCGCGAGCTCGGCCGCCAGTGGTCCTCGGACTTTAGGTGTC TGGGTTGAAGACAGAACCAATCAAACTGGTGTTGAACCGTGTTATGGTGACAAAGATAAACGGCGGCATTGTTTTGCTACCTGGAAGAAT ATTTCTGGTTCCATTGAAATAGTGAAACAAGGTTGTTGGCTGGATGATATCAACTGCTATGACAGGACTGATTGTGTAGAAAAAAAAGAC AGCCCTGAAGTATATTTTTGTTGCTGTGAGGGCAATATGTGTAATGAAAAGTTTTCTTATTTTCCGGAGATGGAAGTCACACAGCCCACT TCAAATCCAGTTACACCTAAGCCACCCTATTACAACATCCTGCTCTATTCCTTGGTGCCACTTATGTTAATTGCGGGGATTGTCATTTGT GCATTTTGGGTGTACAGGCATCACAAGATGGCCTACCCTCCTGTACTTGTTCCAACTCAAGACCCAGGACCACCCCCACCTTCTCCATTA CTAGGTTTGAAACCACTGCAGTTATTAGAAGTGAAAGCAAGGGGAAGATTTGGTTGTGTCTGGAAAGCCCAGTTGCTTAACGAATATGTG GCTGTCAAAATATTTCCAATACAGGACAAACAGTCATGGCAAAATGAATACGAAGTCTACAGTTTGCCTGGAATGAAGCATGAGAACATA TTACAGTTCATTGGTGCAGAAAAACGAGGCACCAGTGTTGATGTGGATCTTTGGCTGATCACAGCATTTCATGAAAAGATGGACTTCAGC AGAAATCTTTATGATATTGGGGAACAACTGGACAGTGAAGATCTGGCCTCCCTCAAGTTCCTGAGCCTGGACTACATTCCGCAAAGGAAG CAAGAACCCATCAAGGATGCCTTGATGTTATTCCAGAGACTCCAGGAAAAGAGAATGTTGGAGGAAAGCAATCTGTCCTTCCTGAAGGAG CTGCTCTTCCGAATTAATAGACTGGATTTGCTGATTACCTACCTAAACACTAGAAAGGAGGAGATGGAAAGGGAACTTCAGACACCAGGC AGGGCTCAAATTTCTGCCTACAGGGTCATGCTCTATCAGATTTCAGAAGAAGTGAGCAGATCAGAATTGAGGTCTTTTAAGTTTCTTTTG CAAGAGGAAATCTCCAAATGCAAACTGGATGATGACATGAACCTGCTGGATATTTTCATAGAGATGGAGAAGAGGGTCATCCTGGGAGAA GGAAAGTTGGACATCCTGAAAAGAGTCTGTGCCCAAATCAACAAGAGCCTGCTGAAGATAATCAACGACTATGAAGAATTCAGCAAAGAG AGAAGCAGCAGCCTTGAAGGAAGTCCTGATGAATTTTCAAATGGGGAGGAGTTGTGTGGGGTAATGACAATCTCGGACTCTCCAAGAGAA CAGGATAGTGAATCACAGACTTTGGACAAAGTTTACCAAATGAAAAGCAAACCTCGGGGATACTGTCTGATCATCAACAATCACAATTTT GCAAAAGCACGGGAGAAAGTGCCCAAACTTCACAGCATTAGGGACAGGAATGGAACACACTTGGATGCAGGGGCTTTGACCACGACCTTT GAAGAGCTTCATTTTGAGATCAAGCCCCACGATGACTGCACAGTAGAGCAAATCTATGAGATTTTGAAAATCTACCAACTCATGGACCAC AGTAACATGGACTGCTTCATCTGCTGTATCCTCTCCCATGGAGACAAGGGCATCATCTATGGCACTGATGGACAGGAGGCCCCCATCTAT GAGCTGACATCTCAGTTCACTGGTTTGAAGTGCCCTTCCCTTGCTGGAAAACCCAAAGTGTTTTTTATTCAGGCTTGTCAGGGGGATAAC TACCAGAAAGGTATACCTGTTGAGACTGATTCAGAGGAGCAACCCTATTTAGAAATGGATTTATCATCACCTCAAACGAGATATATCCCG GATGAGGCTGACTTTCTGCTGGGGATGGCCACTGTGAATAACTGTGTTTCCTACCGAAACCCTGCAGAGGGAACCTGGTACATCCAGTCA CTTTGCCAGAGCCTGAGAGAGCGATGTCCTCGAGGCGATGATATTCTCACCATCCTGACTGAAGTGAACTATGAAGTAAGCAACAAGGAT >1905_1905_3_ACVR2A-CASP8_ACVR2A_chr2_148674995_ENST00000535787_CASP8_chr2_202131210_ENST00000432109_length(amino acids)=745AA_BP=265 MPPAAPSPRRSRLAATAASSAASGPRTLGVWVEDRTNQTGVEPCYGDKDKRRHCFATWKNISGSIEIVKQGCWLDDINCYDRTDCVEKKD SPEVYFCCCEGNMCNEKFSYFPEMEVTQPTSNPVTPKPPYYNILLYSLVPLMLIAGIVICAFWVYRHHKMAYPPVLVPTQDPGPPPPSPL LGLKPLQLLEVKARGRFGCVWKAQLLNEYVAVKIFPIQDKQSWQNEYEVYSLPGMKHENILQFIGAEKRGTSVDVDLWLITAFHEKMDFS RNLYDIGEQLDSEDLASLKFLSLDYIPQRKQEPIKDALMLFQRLQEKRMLEESNLSFLKELLFRINRLDLLITYLNTRKEEMERELQTPG RAQISAYRVMLYQISEEVSRSELRSFKFLLQEEISKCKLDDDMNLLDIFIEMEKRVILGEGKLDILKRVCAQINKSLLKIINDYEEFSKE RSSSLEGSPDEFSNGEELCGVMTISDSPREQDSESQTLDKVYQMKSKPRGYCLIINNHNFAKAREKVPKLHSIRDRNGTHLDAGALTTTF EELHFEIKPHDDCTVEQIYEILKIYQLMDHSNMDCFICCILSHGDKGIIYGTDGQEAPIYELTSQFTGLKCPSLAGKPKVFFIQACQGDN YQKGIPVETDSEEQPYLEMDLSSPQTRYIPDEADFLLGMATVNNCVSYRNPAEGTWYIQSLCQSLRERCPRGDDILTILTEVNYEVSNKD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACVR2A-CASP8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACVR2A-CASP8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ACVR2A-CASP8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ACVR2A | C0920269 | Microsatellite Instability | 1 | CTD_human |
| Hgene | ACVR2A | C1721098 | Replication Error Phenotype | 1 | CTD_human |
| Tgene | C0006142 | Malignant neoplasm of breast | 4 | CGI;CTD_human | |
| Tgene | C0678222 | Breast Carcinoma | 4 | CGI;CTD_human | |
| Tgene | C1257931 | Mammary Neoplasms, Human | 4 | CTD_human | |
| Tgene | C1458155 | Mammary Neoplasms | 4 | CTD_human | |
| Tgene | C4704874 | Mammary Carcinoma, Human | 4 | CTD_human | |
| Tgene | C1846545 | Autoimmune Lymphoproliferative Syndrome Type 2B | 3 | CTD_human;ORPHANET;UNIPROT | |
| Tgene | C0014859 | Esophageal Neoplasms | 2 | CTD_human | |
| Tgene | C0025202 | melanoma | 2 | CTD_human | |
| Tgene | C0546837 | Malignant neoplasm of esophagus | 2 | CTD_human | |
| Tgene | C0001418 | Adenocarcinoma | 1 | CTD_human | |
| Tgene | C0005586 | Bipolar Disorder | 1 | PSYGENET | |
| Tgene | C0007114 | Malignant neoplasm of skin | 1 | CTD_human | |
| Tgene | C0007131 | Non-Small Cell Lung Carcinoma | 1 | CTD_human | |
| Tgene | C0007873 | Uterine Cervical Neoplasm | 1 | CTD_human | |
| Tgene | C0009402 | Colorectal Carcinoma | 1 | CTD_human | |
| Tgene | C0009404 | Colorectal Neoplasms | 1 | CTD_human | |
| Tgene | C0011616 | Contact Dermatitis | 1 | CTD_human | |
| Tgene | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human | |
| Tgene | C0013604 | Edema | 1 | CTD_human | |
| Tgene | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human | |
| Tgene | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human | |
| Tgene | C0024121 | Lung Neoplasms | 1 | CTD_human | |
| Tgene | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human | |
| Tgene | C0027055 | Myocardial Reperfusion Injury | 1 | CTD_human | |
| Tgene | C0035126 | Reperfusion Injury | 1 | CTD_human | |
| Tgene | C0037286 | Skin Neoplasms | 1 | CTD_human | |
| Tgene | C0038220 | Status Epilepticus | 1 | CTD_human | |
| Tgene | C0038356 | Stomach Neoplasms | 1 | CTD_human | |
| Tgene | C0151603 | Anasarca | 1 | CTD_human | |
| Tgene | C0162351 | Contact hypersensitivity | 1 | CTD_human | |
| Tgene | C0162820 | Dermatitis, Allergic Contact | 1 | CTD_human | |
| Tgene | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human | |
| Tgene | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human | |
| Tgene | C0205643 | Carcinoma, Cribriform | 1 | CTD_human | |
| Tgene | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human | |
| Tgene | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human | |
| Tgene | C0242379 | Malignant neoplasm of lung | 1 | CTD_human | |
| Tgene | C0270823 | Petit mal status | 1 | CTD_human | |
| Tgene | C0279607 | Adult Hepatocellular Carcinoma | 1 | ORPHANET | |
| Tgene | C0311335 | Grand Mal Status Epilepticus | 1 | CTD_human | |
| Tgene | C0343641 | Human papilloma virus infection | 1 | CTD_human | |
| Tgene | C0393734 | Complex Partial Status Epilepticus | 1 | CTD_human | |
| Tgene | C0751522 | Status Epilepticus, Subclinical | 1 | CTD_human | |
| Tgene | C0751523 | Non-Convulsive Status Epilepticus | 1 | CTD_human | |
| Tgene | C0751524 | Simple Partial Status Epilepticus | 1 | CTD_human | |
| Tgene | C1168401 | Squamous cell carcinoma of the head and neck | 1 | CTD_human | |
| Tgene | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human | |
| Tgene | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human | |
| Tgene | C2931456 | Prostate cancer, familial | 1 | CTD_human | |
| Tgene | C2937358 | Cerebral Hemorrhage | 1 | CTD_human | |
| Tgene | C4048328 | cervical cancer | 1 | CTD_human | |
| Tgene | C4722327 | PROSTATE CANCER, HEREDITARY, 1 | 1 | CTD_human |
