
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GTF3C3-COL5A2 (FusionGDB2 ID:HG9330TG1290) |
Fusion Gene Summary for GTF3C3-COL5A2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GTF3C3-COL5A2 | Fusion gene ID: hg9330tg1290 | Hgene | Tgene | Gene symbol | GTF3C3 | COL5A2 | Gene ID | 9330 | 1290 |
| Gene name | general transcription factor IIIC subunit 3 | collagen type V alpha 2 chain | |
| Synonyms | TFIIIC102|TFIIICgamma|TFiiiC2-102 | EDSC|EDSCL2 | |
| Cytomap | ('GTF3C3')('COL5A2') 2q33.1 | 2q32.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | general transcription factor 3C polypeptide 3TF3C-gammaTFIIIC 102 kDa subunitgeneral transcription factor IIIC, polypeptide 3, 102kDatranscription factor IIIC subunit gamma | collagen alpha-2(V) chainAB collagencollagen, fetal membrane, A polypeptidetype V preprocollagen alpha 2 chain | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9Y5Q9 | P05997 | |
| Ensembl transtripts involved in fusion gene | ENST00000263956, ENST00000409364, ENST00000470386, | ||
| Fusion gene scores | * DoF score | 2 X 3 X 2=12 | 8 X 9 X 4=288 |
| # samples | 3 | 9 | |
| ** MAII score | log2(3/12*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(9/288*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GTF3C3 [Title/Abstract] AND COL5A2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GTF3C3(197643759)-COL5A2(189917694), # samples:2 GTF3C3(197645282)-COL5A2(189917528), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | GTF3C3-COL5A2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GTF3C3-COL5A2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GTF3C3-COL5A2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. GTF3C3-COL5A2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | COL5A2 | GO:1903225 | negative regulation of endodermal cell differentiation | 23154389 |
 Fusion gene breakpoints across GTF3C3 (5'-gene) Fusion gene breakpoints across GTF3C3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
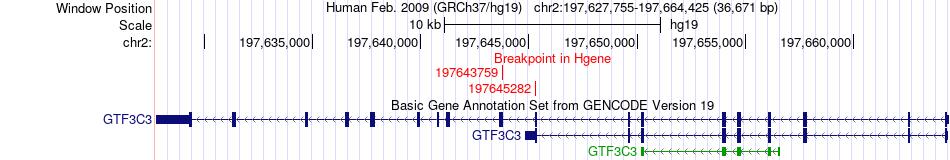 |
 Fusion gene breakpoints across COL5A2 (3'-gene) Fusion gene breakpoints across COL5A2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
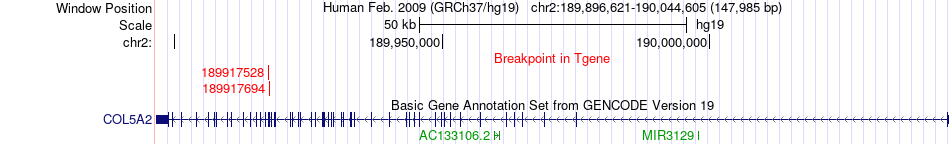 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-VR-A8EW-01A | GTF3C3 | chr2 | 197643759 | - | COL5A2 | chr2 | 189917694 | - |
| ChimerDB4 | ESCA | TCGA-VR-A8EW | GTF3C3 | chr2 | 197643759 | - | COL5A2 | chr2 | 189917694 | - |
| ChimerDB4 | ESCA | TCGA-VR-A8EW | GTF3C3 | chr2 | 197645282 | - | COL5A2 | chr2 | 189917528 | - |
Top |
Fusion Gene ORF analysis for GTF3C3-COL5A2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000263956 | ENST00000374866 | GTF3C3 | chr2 | 197645282 | - | COL5A2 | chr2 | 189917528 | - |
| intron-3CDS | ENST00000263956 | ENST00000374866 | GTF3C3 | chr2 | 197643759 | - | COL5A2 | chr2 | 189917694 | - |
| intron-3CDS | ENST00000409364 | ENST00000374866 | GTF3C3 | chr2 | 197643759 | - | COL5A2 | chr2 | 189917694 | - |
| intron-3CDS | ENST00000409364 | ENST00000374866 | GTF3C3 | chr2 | 197645282 | - | COL5A2 | chr2 | 189917528 | - |
| intron-3CDS | ENST00000470386 | ENST00000374866 | GTF3C3 | chr2 | 197643759 | - | COL5A2 | chr2 | 189917694 | - |
| intron-3CDS | ENST00000470386 | ENST00000374866 | GTF3C3 | chr2 | 197645282 | - | COL5A2 | chr2 | 189917528 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000263956 | GTF3C3 | chr2 | 197645282 | - | ENST00000374866 | COL5A2 | chr2 | 189917528 | - | 5321 | 1308 | 33 | 3146 | 1037 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000263956 | ENST00000374866 | GTF3C3 | chr2 | 197645282 | - | COL5A2 | chr2 | 189917528 | - | 0.000108975 | 0.99989104 |
Top |
Fusion Genomic Features for GTF3C3-COL5A2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
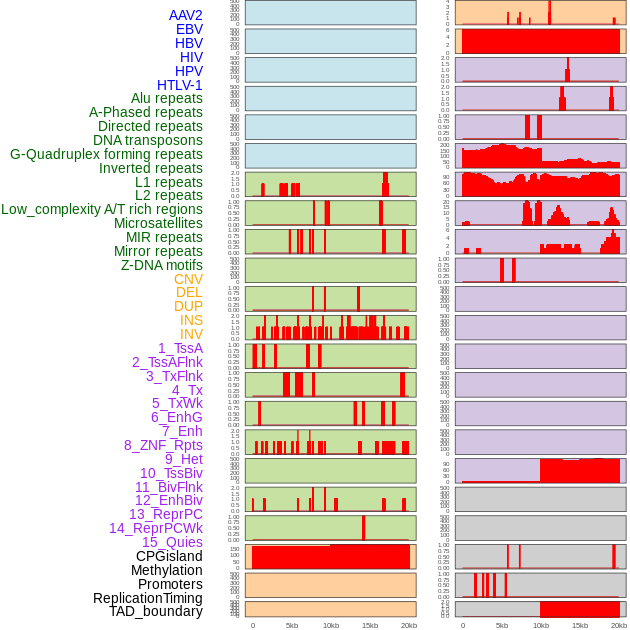 |
Top |
Fusion Protein Features for GTF3C3-COL5A2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:197643759/chr2:189917694) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GTF3C3 | COL5A2 |
| FUNCTION: Involved in RNA polymerase III-mediated transcription. Integral, tightly associated component of the DNA-binding TFIIIC2 subcomplex that directly binds tRNA and virus-associated RNA promoters. | FUNCTION: Type V collagen is a member of group I collagen (fibrillar forming collagen). It is a minor connective tissue component of nearly ubiquitous distribution. Type V collagen binds to DNA, heparan sulfate, thrombospondin, heparin, and insulin. Type V collagen is a key determinant in the assembly of tissue-specific matrices (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 92_114 | 406 | 887.0 | Compositional bias | Note=Glu-rich |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 149_182 | 406 | 887.0 | Repeat | Note=TPR 1 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 183_216 | 406 | 887.0 | Repeat | Note=TPR 2 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 217_250 | 406 | 887.0 | Repeat | Note=TPR 3 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 252_284 | 406 | 887.0 | Repeat | Note=TPR 4 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 290_323 | 406 | 887.0 | Repeat | Note=TPR 5 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 326_361 | 406 | 887.0 | Repeat | Note=TPR 6 |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 1266_1499 | 887 | 1500.0 | Domain | Fibrillar collagen NC1 | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 1067_1069 | 887 | 1500.0 | Motif | Cell attachment site | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 1070_1072 | 887 | 1500.0 | Motif | Cell attachment site | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 1100_1102 | 887 | 1500.0 | Motif | Cell attachment site | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 1127_1129 | 887 | 1500.0 | Motif | Cell attachment site | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 1136_1138 | 887 | 1500.0 | Motif | Cell attachment site | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 944_946 | 887 | 1500.0 | Motif | Cell attachment site |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 92_114 | 0 | 414.0 | Compositional bias | Note=Glu-rich |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 421_454 | 406 | 887.0 | Repeat | Note=TPR 7 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 456_489 | 406 | 887.0 | Repeat | Note=TPR 8 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 491_523 | 406 | 887.0 | Repeat | Note=TPR 9 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 733_766 | 406 | 887.0 | Repeat | Note=TPR 10 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000263956 | - | 9 | 18 | 811_844 | 406 | 887.0 | Repeat | Note=TPR 11 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 149_182 | 0 | 414.0 | Repeat | Note=TPR 1 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 183_216 | 0 | 414.0 | Repeat | Note=TPR 2 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 217_250 | 0 | 414.0 | Repeat | Note=TPR 3 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 252_284 | 0 | 414.0 | Repeat | Note=TPR 4 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 290_323 | 0 | 414.0 | Repeat | Note=TPR 5 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 326_361 | 0 | 414.0 | Repeat | Note=TPR 6 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 421_454 | 0 | 414.0 | Repeat | Note=TPR 7 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 456_489 | 0 | 414.0 | Repeat | Note=TPR 8 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 491_523 | 0 | 414.0 | Repeat | Note=TPR 9 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 733_766 | 0 | 414.0 | Repeat | Note=TPR 10 |
| Hgene | GTF3C3 | chr2:197645282 | chr2:189917528 | ENST00000409364 | - | 1 | 9 | 811_844 | 0 | 414.0 | Repeat | Note=TPR 11 |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 39_97 | 887 | 1500.0 | Domain | VWFC | |
| Tgene | COL5A2 | chr2:197645282 | chr2:189917528 | ENST00000374866 | 38 | 54 | 506_508 | 887 | 1500.0 | Motif | Cell attachment site |
Top |
Fusion Gene Sequence for GTF3C3-COL5A2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35282_35282_1_GTF3C3-COL5A2_GTF3C3_chr2_197645282_ENST00000263956_COL5A2_chr2_189917528_ENST00000374866_length(transcript)=5321nt_BP=1308nt TGCTGGCCCCGGCGAGGTAGCTTCTGGAAGGCGCTGCTCTTCCGGTTCTCTGTCCCGGTTCCTGGGGTTGCACAGACAGACCCTGTAAAC ATGTCAGGGTTCAGTCCGGAACTCATCGACTACTTGGAAGGGAAAATCTCCTTTGAGGAGTTCGAACGGCGGAGAGAAGAGAGAAAAACC CGCGAGAAGAAAAGTCTTCAGGAAAAAGGCAAGTTATCAGCTGAAGAAAATCCCGATGACTCTGAAGTTCCATCATCATCAGGAATTAAC TCTACCAAATCCCAAGACAAAGATGTCAATGAAGGAGAAACATCAGATGGAGTGAGGAAGTCAGTTCACAAGGTCTTTGCTTCCATGCTT GGAGAGAATGAAGATGATGAGGAGGAAGAGGAAGAAGAGGAGGAGGAGGAGGAGGAGGAAGAAACACCTGAGCAACCCACTGCGGGCGAT GTATTTGTATTGGAGATGGTTCTCAATCGTGAAACCAAGAAAATGATGAAAGAGAAAAGGCCTCGGAGTAAACTTCCCAGAGCTCTGAGA GGTCTCATGGGTGAAGCCAACATTCGTTTTGCTCGAGGAGAACGTGAAGAGGCGATATTGATGTGCATGGAAATCATAAGACAAGCTCCT CTGGCTTATGAGCCATTCTCTACTCTAGCCATGATATATGAGGACCAAGGTGACATGGAAAAATCATTGCAGTTTGAGTTGATTGCTGCG CATTTAAATCCCAGTGACACAGAAGAATGGGTTAGACTGGCAGAAATGTCTCTGGAACAAGACAATATTAAGCAGGCTATTTTTTGCTAT ACAAAAGCTCTTAAATATGAACCTACTAATGTCCGTTATCTGTGGGAGCGATCAAGCCTTTATGAACAGATGGGTGATCATAAAATGGCC ATGGATGGTTATAGGCGTATTTTAAACCTTTTGTCTCCATCTGATGGCGAACGTTTTATGCAGCTGGCTAGAGATATGGCAAAGAGTTAC TATGAAGCCAATGATGTTACTTCTGCTATTAACATAATTGATGAAGCTTTCTCAAAACACCAGGGCCTAGTCTCCATGGAAGATGTTAAC ATAGCAGCTGAACTATATATTTCTAACAAACAGTATGACAAAGCTTTGGAGATAATTACAGATTTTTCTGGAATTGTGCTGGAAAAAAAA ACTTCAGAAGAAGGCACCTCAGAAGAGAATAAAGCTCCTGAGAATGTTACCTGCACTATACCTGATGGCGTGCCAATAGATATCACAGTG AAGTTGATGGTCTGCCTTGTACATCTCAACATTCTTGAACCACTTAATGGTCCTAATGGTGTTCCTGGACTAAAAGGTGGTCGAGGAACC CAAGGTCCGCCTGGTGCTACAGGATTTCCTGGTTCTGCGGGCAGAGTTGGACCTCCAGGCCCTGCTGGAGCTCCAGGACCTGCGGGACCC CTAGGGGAACCCGGGAAGGAGGGACCTCCAGGTCTTCGTGGGGACCCTGGCTCTCATGGGCGTGTGGGAGATCGAGGACCAGCTGGCCCC CCTGGTGGCCCAGGAGACAAAGGGGACCCAGGAGAAGATGGGCAACCTGGTCCAGATGGCCCCCCTGGTCCAGCTGGAACGACCGGGCAG AGAGGAATTGTTGGCATGCCTGGGCAACGTGGAGAGAGAGGCATGCCCGGCCTACCAGGCCCAGCGGGAACACCAGGAAAAGTAGGACCA ACTGGTGCAACAGGAGATAAAGGTCCACCTGGACCTGTGGGGCCCCCAGGCTCCAATGGTCCTGTAGGGGAACCTGGACCAGAAGGTCCA GCTGGCAATGATGGTACCCCAGGACGGGATGGTGCTGTTGGAGAACGTGGTGATCGTGGAGACCCTGGGCCTGCAGGTCTGCCAGGCTCT CAGGGTGCCCCTGGAACTCCTGGCCCTGTGGGTGCTCCAGGAGATGCAGGACAAAGAGGAGATCCGGGTTCTCGGGGTCCTATAGGACCA CCTGGTCGAGCTGGGAAACGTGGATTACCTGGACCCCAAGGACCTCGTGGTGACAAAGGTGATCATGGAGACCGAGGTGACAGAGGTCAG AAGGGCCACAGAGGCTTTACTGGTCTTCAGGGTCTTCCTGGCCCTCCTGGTCCAAATGGTGAACAAGGAAGTGCTGGAATCCCTGGACCA TTTGGCCCAAGAGGTCCTCCAGGCCCAGTTGGTCCTTCAGGTAAAGAAGGAAACCCTGGGCCACTTGGGCCAATTGGACCTCCAGGTGTA CGAGGCAGTGTAGGAGAAGCAGGACCTGAGGGCCCTCCTGGTGAGCCTGGCCCACCTGGCCCTCCGGGTCCCCCTGGCCACCTTACAGCT GCTCTTGGGGATATCATGGGGCACTATGATGAAAGCATGCCAGATCCACTTCCTGAGTTTACTGAAGATCAGGCGGCTCCTGATGACAAA AACAAAACGGACCCAGGGGTTCATGCTACCCTGAAGTCACTCAGTAGTCAGATTGAAACCATGCGCAGCCCCGATGGCTCGAAAAAGCAC CCAGCCCGCACGTGTGATGACCTAAAGCTTTGCCATTCCGCAAAGCAGAGTGGTGAATACTGGATTGATCCTAACCAAGGATCTGTTGAA GATGCAATCAAAGTTTACTGCAACATGGAAACAGGAGAAACATGTATTTCAGCAAACCCATCCAGTGTACCACGTAAAACCTGGTGGGCC AGTAAATCTCCTGACAATAAACCTGTTTGGTATGGTCTTGATATGAACAGAGGGTCTCAGTTCGCTTATGGAGACCACCAATCACCTAAT ACAGCCATTACTCAGATGACTTTTTTGCGCCTTTTATCAAAAGAAGCCTCCCAGAACATCACTTACATCTGTAAAAACAGTGTAGGATAC ATGGACGATCAAGCTAAGAACCTCAAAAAAGCTGTGGTTCTCAAAGGGGCAAATGACTTAGATATCAAAGCAGAGGGAAATATTAGATTC CGGTATATCGTTCTTCAAGACACTTGCTCTAAGCGGAATGGAAATGTGGGCAAGACTGTCTTTGAATATAGAACACAGAATGTGGCACGC TTGCCCATCATAGATCTTGCTCCTGTGGATGTTGGCGGCACAGACCAGGAATTCGGCGTTGAAATTGGGCCAGTTTGTTTTGTGTAAAGT AAGCCAAGACACATCGACAATGAGCACCACCATCAATGACCACCGCCATTCACAAGAACTTTGACTGTTTGAAGTTGATCCTGAGACTCT TGAAGTAATGGCTGATCCTGCATCAGCATTGTATATATGGTCTTAAGTGCCTGGCCTCCTTATCCTTCAGAATATTTATTTTACTTACAA TCCTCAAGTTTTAATTGATTTTAAATATTTTTCAATACAACAGTTTAGGTTTAAGATGACCAATGACAATGACCACCTTTGCAGAAAGTA AACTGATTGAATAAATAAATCTCCGTTTTCTTCAATTTATTTCAGTGTAATGAAAAAGTTGCTTAGTATTTATGAGGAAATTCTTCTTCC TGGCAGGTAGCTTAAAGAGTGGGGTATATAGAGCCACAACACATGTTTATTTTGCTTGGCTGCAGTTGAAAAATAGAAATTAGTGCCCTT TTGTGACCTCTCATTCCAAGATTGTCAATTAAAAATGAGTTTAAAATGTTTAACTTGTGATCGAGACCTACATGCATGTCTTGATATTGT GTAACTATAATAGAGACTCTTTAAGGAGAATCTTAAAAAAAAAAAAACGTTTCTCACTGTCTTAAATAGAATTTTTAAATAGTATATATT CAGTGGCATTTTGGAGAACAAAGTGAATTTACTTCGACTTCTTAAATTTTTGTAAAAGACTATAAGTTTAGACATCTTTCTCATTCAAAT TTAAAGATATCTTTCTCCTCTTGATCAATCTATCAATATTGATAGAAGTCACACTAGTATATACCATTTAATACATTTACACTTTCTTAT TTAAGAAGATATTGAATGCAAAATAATTGACATATAGAACTTTACAAACATATGTCCAAGGACTCTAAATTGAGACTCTTCCACATGTAC AATCTCATCATCCTGAAGCCTATAATGAAGAAAAAGATCTAGAAACTGAGTTGTGGAGCTGACTCTAATCAAATGTGATGATTGGAATTA GACCATTTGGCCTTTGAACTTTCATAGGAAAAATGACCCAACATTTCTTAGCATGAGCTACCTCATCTCTAGAAGCTGGGATGGACTTAC TATTCTTGTTTATATTTTAGATACTGAAAGGTGCTATGCTTCTGTTATTATTCCAAGACTGGAGATAGGCAGGGCTAAAAAGGTATTATT ATTTTTCCTTTAATGATGGTGCTAAAATTCTTCCTATAAAATTCCTTAAAAATAAAGATGGTTTAATCACTACCATTGTGAAAACATAAC TGTTAGACTTCCCGTTTCTGAAAGAAAGAGCATCGTTCCAATGCTTGTTCACTGTTCCTCTGTCATACTGTATCTGGAATGCTTTGTAAT ACTTGCATGCTTCTTAGACCAGAACATGTAGGTCCCCTTGTGTCTCAATACTTTTTTTTTCTTAATTGCATTTGTTGGCTCTATTTTAAT TTTTTTCTTTTAAAATAAACAGCTGGGACCATCCCAAAAGACAAGCCATGCATACAACTTTGGTCATGTATCTCTGCAAAGCATCAAATT AAATGCACGCTTTTGTCATGTCAGTGGTTTTTGTTTTGTGAAATTCCTTTGACCATATTAGATCTATTTCATTTCCAATAGTGAAAAGGA GATGTGGTGGTATACTTTGTTTGCCATTTGTTTAAAAGATACAACGGATACCTTCTATCATGTATGTACTGGCTTATAAATGAAAATCTA TCTACAACATTACCCACAAAGGCAACATGACACCAATTATCACTGCCTCTGCCCTTAAAAATGTCAGAGTAGTATTATTGATAAAAAGGG CAAGCAATAGATTTTTCATGACTGAATAAACTGTAATAATAAAACATATGTCTCAAAGTGTATCACATATGAATTTAGCCTAATTGTTTT CAGTTTCATTCTCAATATTTAGTTTACAACATCATTTTCCCCTAAACTGGTTATATTTTGACCTGTATATCTTAAATTTGAGTATTTATA TGCCTAAATACATGTGTGAGTTTTGTTTGACTTCCAAGTCCAAACTATAAGATTATATAAGTTCATATAGATGAATCAGAAATATGTGGT AATACTATTAAGTCACAAACACTAACAATTTCCAACTATAGAAATAACAGTTCTTATTTGGATTTTGGGAATGCTACCAATAAAAGCCTG >35282_35282_1_GTF3C3-COL5A2_GTF3C3_chr2_197645282_ENST00000263956_COL5A2_chr2_189917528_ENST00000374866_length(amino acids)=1037AA_BP=455 MLFRFSVPVPGVAQTDPVNMSGFSPELIDYLEGKISFEEFERRREERKTREKKSLQEKGKLSAEENPDDSEVPSSSGINSTKSQDKDVNE GETSDGVRKSVHKVFASMLGENEDDEEEEEEEEEEEEEEETPEQPTAGDVFVLEMVLNRETKKMMKEKRPRSKLPRALRGLMGEANIRFA RGEREEAILMCMEIIRQAPLAYEPFSTLAMIYEDQGDMEKSLQFELIAAHLNPSDTEEWVRLAEMSLEQDNIKQAIFCYTKALKYEPTNV RYLWERSSLYEQMGDHKMAMDGYRRILNLLSPSDGERFMQLARDMAKSYYEANDVTSAINIIDEAFSKHQGLVSMEDVNIAAELYISNKQ YDKALEIITDFSGIVLEKKTSEEGTSEENKAPENVTCTIPDGVPIDITVKLMVCLVHLNILEPLNGPNGVPGLKGGRGTQGPPGATGFPG SAGRVGPPGPAGAPGPAGPLGEPGKEGPPGLRGDPGSHGRVGDRGPAGPPGGPGDKGDPGEDGQPGPDGPPGPAGTTGQRGIVGMPGQRG ERGMPGLPGPAGTPGKVGPTGATGDKGPPGPVGPPGSNGPVGEPGPEGPAGNDGTPGRDGAVGERGDRGDPGPAGLPGSQGAPGTPGPVG APGDAGQRGDPGSRGPIGPPGRAGKRGLPGPQGPRGDKGDHGDRGDRGQKGHRGFTGLQGLPGPPGPNGEQGSAGIPGPFGPRGPPGPVG PSGKEGNPGPLGPIGPPGVRGSVGEAGPEGPPGEPGPPGPPGPPGHLTAALGDIMGHYDESMPDPLPEFTEDQAAPDDKNKTDPGVHATL KSLSSQIETMRSPDGSKKHPARTCDDLKLCHSAKQSGEYWIDPNQGSVEDAIKVYCNMETGETCISANPSSVPRKTWWASKSPDNKPVWY GLDMNRGSQFAYGDHQSPNTAITQMTFLRLLSKEASQNITYICKNSVGYMDDQAKNLKKAVVLKGANDLDIKAEGNIRFRYIVLQDTCSK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GTF3C3-COL5A2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GTF3C3-COL5A2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for GTF3C3-COL5A2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | GTF3C3 | C0036572 | Seizures | 1 | GENOMICS_ENGLAND |
| Hgene | GTF3C3 | C0557874 | Global developmental delay | 1 | GENOMICS_ENGLAND |
| Hgene | GTF3C3 | C3714756 | Intellectual Disability | 1 | GENOMICS_ENGLAND |
| Tgene | C4538407 | EHLERS-DANLOS SYNDROME, CLASSIC TYPE, 2 | 4 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C4552122 | EHLERS-DANLOS SYNDROME, CLASSIC TYPE, 1 | 3 | GENOMICS_ENGLAND | |
| Tgene | C0220679 | Ehlers-Danlos Syndrome, Autosomal Dominant, Type Unspecified | 2 | ORPHANET | |
| Tgene | C3151201 | MULTISYSTEMIC SMOOTH MUSCLE DYSFUNCTION SYNDROME | 2 | GENOMICS_ENGLAND | |
| Tgene | C0000786 | Spontaneous abortion | 1 | CTD_human | |
| Tgene | C0000822 | Abortion, Tubal | 1 | CTD_human | |
| Tgene | C0005779 | Blood Coagulation Disorders | 1 | GENOMICS_ENGLAND | |
| Tgene | C0023890 | Liver Cirrhosis | 1 | CTD_human | |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human | |
| Tgene | C0239946 | Fibrosis, Liver | 1 | CTD_human | |
| Tgene | C0268335 | Ehlers-Danlos syndrome type 1 | 1 | GENOMICS_ENGLAND | |
| Tgene | C0268336 | Ehlers-Danlos syndrome type 2 | 1 | GENOMICS_ENGLAND | |
| Tgene | C1458140 | Bleeding tendency | 1 | GENOMICS_ENGLAND | |
| Tgene | C1846545 | Autoimmune Lymphoproliferative Syndrome Type 2B | 1 | GENOMICS_ENGLAND | |
| Tgene | C3830362 | Early Pregnancy Loss | 1 | CTD_human | |
| Tgene | C4225429 | Ehlers-Danlos syndrome classic type | 1 | GENOMICS_ENGLAND | |
| Tgene | C4552766 | Miscarriage | 1 | CTD_human |
