
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CDKN2A-EXD2 (FusionGDB2 ID:HG1029TG55218) |
Fusion Gene Summary for CDKN2A-EXD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CDKN2A-EXD2 | Fusion gene ID: hg1029tg55218 | Hgene | Tgene | Gene symbol | CDKN2A | EXD2 | Gene ID | 1029 | 55218 |
| Gene name | cyclin dependent kinase inhibitor 2A | exonuclease 3'-5' domain containing 2 | |
| Synonyms | ARF|CDK4I|CDKN2|CMM2|INK4|INK4A|MLM|MTS-1|MTS1|P14|P14ARF|P16|P16-INK4A|P16INK4|P16INK4A|P19|P19ARF|TP16 | C14orf114|EXDL2 | |
| Cytomap | ('CDKN2A')('EXD2') 9p21.3 | 14q24.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cyclin-dependent kinase inhibitor 2ACDK4 inhibitor p16-INK4alternative reading framecell cycle negative regulator betacyclin-dependent kinase 4 inhibitor Acyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4)multiple tumor suppressor 1 | exonuclease 3'-5' domain-containing protein 23'-5' exoribonuclease EXD2exonuclease 3'-5' domain-like 2exonuclease 3'-5' domain-like-containing protein 2 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000304494, ENST00000446177, ENST00000498124, ENST00000579122, ENST00000361570, ENST00000470819, ENST00000479692, ENST00000494262, ENST00000497750, ENST00000498628, ENST00000530628, ENST00000578845, ENST00000579755, | ||
| Fusion gene scores | * DoF score | 14 X 9 X 11=1386 | 5 X 5 X 3=75 |
| # samples | 17 | 6 | |
| ** MAII score | log2(17/1386*10)=-3.02732060599951 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/75*10)=-0.321928094887362 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CDKN2A [Title/Abstract] AND EXD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CDKN2A(21974677)-EXD2(69695533), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CDKN2A-EXD2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDKN2A-EXD2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDKN2A-EXD2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CDKN2A-EXD2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CDKN2A | GO:0000082 | G1/S transition of mitotic cell cycle | 10208428 |
| Hgene | CDKN2A | GO:0007050 | cell cycle arrest | 15149599 |
| Hgene | CDKN2A | GO:0008285 | negative regulation of cell proliferation | 15149599 |
| Hgene | CDKN2A | GO:0030308 | negative regulation of cell growth | 10208428 |
| Hgene | CDKN2A | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 10353611 |
| Hgene | CDKN2A | GO:0042326 | negative regulation of phosphorylation | 8259215|10208428 |
| Hgene | CDKN2A | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 7739547|8259215 |
| Tgene | EXD2 | GO:0000724 | double-strand break repair via homologous recombination | 26807646 |
| Tgene | EXD2 | GO:0000729 | DNA double-strand break processing | 26807646 |
| Tgene | EXD2 | GO:0006302 | double-strand break repair | 26807646 |
| Tgene | EXD2 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 26807646 |
 Fusion gene breakpoints across CDKN2A (5'-gene) Fusion gene breakpoints across CDKN2A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
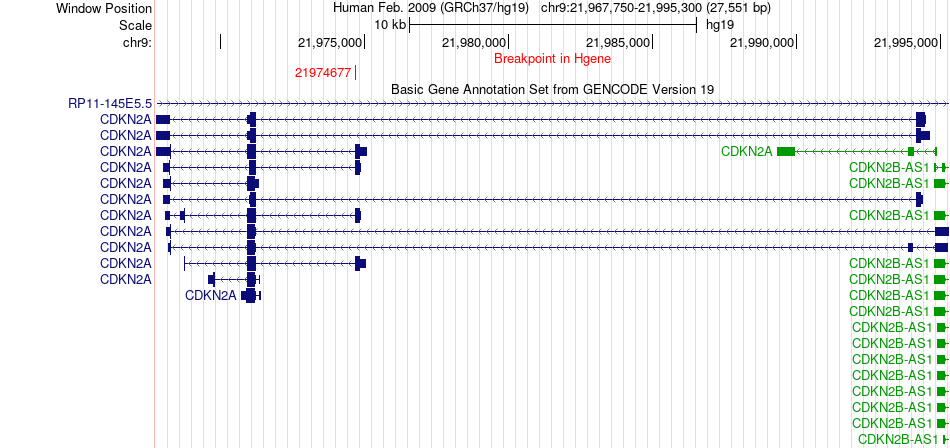 |
 Fusion gene breakpoints across EXD2 (3'-gene) Fusion gene breakpoints across EXD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
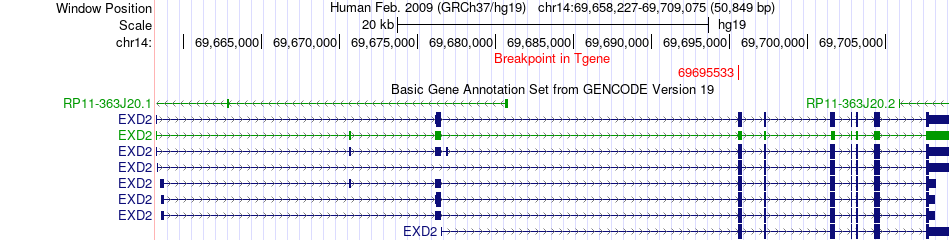 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-XF-AAMW-01A | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
Top |
Fusion Gene ORF analysis for CDKN2A-EXD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000304494 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-3UTR | ENST00000446177 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-3UTR | ENST00000498124 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-3UTR | ENST00000579122 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000304494 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000304494 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000304494 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000304494 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000304494 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000304494 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000446177 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000446177 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000446177 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000446177 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000446177 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000446177 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000498124 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000498124 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000498124 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000498124 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000498124 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000498124 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000579122 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000579122 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000579122 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000579122 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000579122 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| 5CDS-5UTR | ENST00000579122 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| In-frame | ENST00000304494 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| In-frame | ENST00000446177 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| In-frame | ENST00000498124 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| In-frame | ENST00000579122 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000361570 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000470819 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000479692 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000494262 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000497750 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000498628 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000530628 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000578845 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3CDS | ENST00000579755 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000361570 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000470819 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000479692 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000494262 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000497750 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000498628 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000530628 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000578845 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-3UTR | ENST00000579755 | ENST00000492815 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000361570 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000361570 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000361570 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000361570 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000361570 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000361570 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000470819 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000470819 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000470819 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000470819 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000470819 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000470819 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000479692 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000479692 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000479692 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000479692 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000479692 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000479692 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000494262 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000494262 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000494262 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000494262 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000494262 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000494262 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000497750 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000497750 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000497750 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000497750 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000497750 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000497750 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000498628 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000498628 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000498628 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000498628 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000498628 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000498628 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000530628 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000530628 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000530628 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000530628 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000530628 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000530628 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000578845 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000578845 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000578845 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000578845 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000578845 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000578845 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000579755 | ENST00000312994 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000579755 | ENST00000409014 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000579755 | ENST00000409242 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000579755 | ENST00000409675 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000579755 | ENST00000409949 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
| intron-5UTR | ENST00000579755 | ENST00000449989 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000304494 | CDKN2A | chr9 | 21974677 | - | ENST00000409018 | EXD2 | chr14 | 69695533 | + | 3212 | 421 | 124 | 1953 | 609 |
| ENST00000579122 | CDKN2A | chr9 | 21974677 | - | ENST00000409018 | EXD2 | chr14 | 69695533 | + | 2972 | 181 | 31 | 1713 | 560 |
| ENST00000498124 | CDKN2A | chr9 | 21974677 | - | ENST00000409018 | EXD2 | chr14 | 69695533 | + | 2980 | 189 | 39 | 1721 | 560 |
| ENST00000446177 | CDKN2A | chr9 | 21974677 | - | ENST00000409018 | EXD2 | chr14 | 69695533 | + | 3153 | 362 | 65 | 1894 | 609 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000304494 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + | 0.001888286 | 0.9981117 |
| ENST00000579122 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + | 0.001837231 | 0.9981628 |
| ENST00000498124 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + | 0.001796122 | 0.9982039 |
| ENST00000446177 | ENST00000409018 | CDKN2A | chr9 | 21974677 | - | EXD2 | chr14 | 69695533 | + | 0.001905661 | 0.9980944 |
Top |
Fusion Genomic Features for CDKN2A-EXD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CDKN2A | chr9 | 21974676 | - | EXD2 | chr14 | 69695532 | + | 0.000250769 | 0.99974924 |
| CDKN2A | chr9 | 21974676 | - | EXD2 | chr14 | 69695532 | + | 0.000250769 | 0.99974924 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
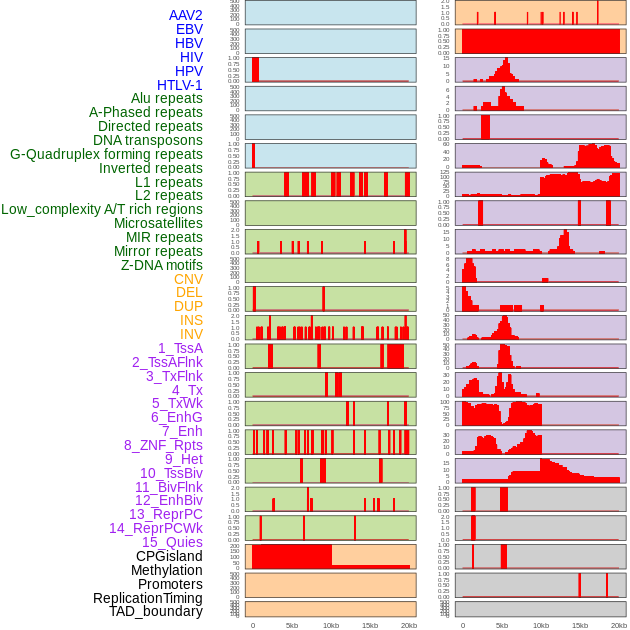 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
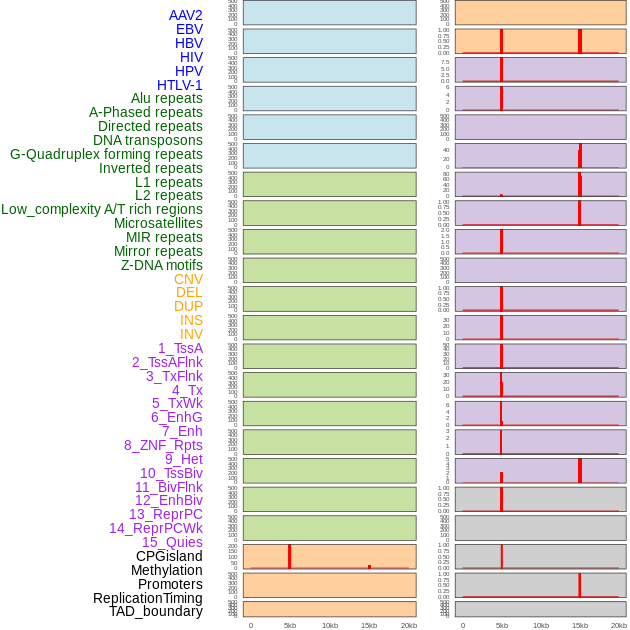 |
Top |
Fusion Protein Features for CDKN2A-EXD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:21974677/chr14:69695533) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000304494 | - | 1 | 3 | 11_40 | 50 | 157.0 | Repeat | Note=ANK 1 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000446177 | - | 1 | 3 | 11_40 | 50 | 168.0 | Repeat | Note=ANK 1 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498124 | - | 1 | 4 | 11_40 | 50 | 130.66666666666666 | Repeat | Note=ANK 1 |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000312994 | 1 | 9 | 155_247 | 111 | 622.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409014 | 3 | 11 | 155_247 | 0 | 497.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409018 | 1 | 9 | 155_247 | 111 | 622.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409242 | 1 | 9 | 155_247 | 0 | 497.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409675 | 0 | 8 | 155_247 | 0 | 497.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409949 | 2 | 10 | 155_247 | 0 | 497.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000449989 | 0 | 8 | 155_247 | 0 | 497.0 | Domain | 3'-5' exonuclease | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409014 | 3 | 11 | 1_4 | 0 | 497.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409014 | 3 | 11 | 26_621 | 0 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409242 | 1 | 9 | 1_4 | 0 | 497.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409242 | 1 | 9 | 26_621 | 0 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409675 | 0 | 8 | 1_4 | 0 | 497.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409675 | 0 | 8 | 26_621 | 0 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409949 | 2 | 10 | 1_4 | 0 | 497.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409949 | 2 | 10 | 26_621 | 0 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000449989 | 0 | 8 | 1_4 | 0 | 497.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000449989 | 0 | 8 | 26_621 | 0 | 497.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409014 | 3 | 11 | 5_25 | 0 | 497.0 | Transmembrane | Helical | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409242 | 1 | 9 | 5_25 | 0 | 497.0 | Transmembrane | Helical | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409675 | 0 | 8 | 5_25 | 0 | 497.0 | Transmembrane | Helical | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409949 | 2 | 10 | 5_25 | 0 | 497.0 | Transmembrane | Helical | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000449989 | 0 | 8 | 5_25 | 0 | 497.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000304494 | - | 1 | 3 | 110_139 | 50 | 157.0 | Repeat | Note=ANK 4 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000304494 | - | 1 | 3 | 44_72 | 50 | 157.0 | Repeat | Note=ANK 2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000304494 | - | 1 | 3 | 77_106 | 50 | 157.0 | Repeat | Note=ANK 3 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000446177 | - | 1 | 3 | 110_139 | 50 | 168.0 | Repeat | Note=ANK 4 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000446177 | - | 1 | 3 | 44_72 | 50 | 168.0 | Repeat | Note=ANK 2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000446177 | - | 1 | 3 | 77_106 | 50 | 168.0 | Repeat | Note=ANK 3 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000494262 | - | 1 | 4 | 110_139 | 0 | 106.0 | Repeat | Note=ANK 4 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000494262 | - | 1 | 4 | 11_40 | 0 | 106.0 | Repeat | Note=ANK 1 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000494262 | - | 1 | 4 | 44_72 | 0 | 106.0 | Repeat | Note=ANK 2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000494262 | - | 1 | 4 | 77_106 | 0 | 106.0 | Repeat | Note=ANK 3 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498124 | - | 1 | 4 | 110_139 | 50 | 130.66666666666666 | Repeat | Note=ANK 4 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498124 | - | 1 | 4 | 44_72 | 50 | 130.66666666666666 | Repeat | Note=ANK 2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498124 | - | 1 | 4 | 77_106 | 50 | 130.66666666666666 | Repeat | Note=ANK 3 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498628 | - | 1 | 3 | 110_139 | 0 | 106.0 | Repeat | Note=ANK 4 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498628 | - | 1 | 3 | 11_40 | 0 | 106.0 | Repeat | Note=ANK 1 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498628 | - | 1 | 3 | 44_72 | 0 | 106.0 | Repeat | Note=ANK 2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000498628 | - | 1 | 3 | 77_106 | 0 | 106.0 | Repeat | Note=ANK 3 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000578845 | - | 1 | 2 | 110_139 | 0 | 106.0 | Repeat | Note=ANK 4 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000578845 | - | 1 | 2 | 11_40 | 0 | 106.0 | Repeat | Note=ANK 1 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000578845 | - | 1 | 2 | 44_72 | 0 | 106.0 | Repeat | Note=ANK 2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000578845 | - | 1 | 2 | 77_106 | 0 | 106.0 | Repeat | Note=ANK 3 |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000312994 | 1 | 9 | 1_4 | 111 | 622.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000312994 | 1 | 9 | 26_621 | 111 | 622.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409018 | 1 | 9 | 1_4 | 111 | 622.0 | Topological domain | Mitochondrial intermembrane | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409018 | 1 | 9 | 26_621 | 111 | 622.0 | Topological domain | Cytoplasmic | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000312994 | 1 | 9 | 5_25 | 111 | 622.0 | Transmembrane | Helical | |
| Tgene | EXD2 | chr9:21974677 | chr14:69695533 | ENST00000409018 | 1 | 9 | 5_25 | 111 | 622.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CDKN2A-EXD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >15399_15399_1_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000304494_EXD2_chr14_69695533_ENST00000409018_length(transcript)=3212nt_BP=421nt CCCAACCTGGGGCGACTTCAGGGGTGCCACATTCGCTAAGTGCTCGGAGTTAATAGCACCTCCTCCGAGCACTCGCTCACGGCGTCCCCT TGCCTGGAAAGATACCGCGGTCCCTCCAGAGGATTTGAGGGACAGGGTCGGAGGGGGCTCTTCCGCCAGCACCGGAGGAAGAAAGAGGAG GGGCTGGCTGGTCACCAGAGGGTGGGGCGGACCGCGTGCGCTCGGCGGCTGCGGAGAGGGGGAGAGCAGGCAGCGGGCGGCGGGGAGCAG CATGGAGCCGGCGGCGGGGAGCAGCATGGAGCCTTCGGCTGACTGGCTGGCCACGGCCGCGGCCCGGGGTCGGGTAGAGGAGGTGCGGGC GCTGCTGGAGGCGGGGGCGCTGCCCAACGCACCGAATAGTTACGGTCGGAGGCCGATCCAGGTAAATTTGGAAGGCAAAGCCAGCCCTCT GTCACTTCTACAAATGGCCTCCCCAAGTGGCCTGTGTGTCTTGGTTCGCCTGCCCAAGCTAATCTGTGGAGGAAAAACACTACCAAGAAC GTTATTGGATATTTTGGCAGATGGCACCATTTTGAAAGTTGGAGTGGGATGCTCAGAAGATGCCAGCAAGCTTCTGCAGGATTATGGCCT CGTTGTTAGGGGGTGCCTGGACCTCCGATACCTAGCCATGCGGCAGAGAAACAATTTGCTCTGTAATGGGCTTAGCCTGAAGTCCCTCGC TGAGACTGTTTTGAACTTTCCCCTTGACAAGTCCCTTCTACTTCGTTGCAGCAACTGGGATGCTGAGACTCTCACAGAGGACCAGGTAAT TTATGCTGCCAGGGATGCCCAGATTTCAGTGGCTCTCTTTCTTCATCTTCTTGGATACCCTTTCTCTAGGAATTCACCTGGAGAAAAAAA CGATGACCACAGTAGCTGGAGAAAAGTCTTGGAAAAATGCCAGGGTGTGGTCGACATCCCATTTCGAAGCAAAGGAATGAGCAGATTGGG AGAAGAGGTTAATGGGGAAGCAACAGAATCTCAGCAGAAGCCAAGAAATAAGAAGTCTAAGATGGATGGGATGGTGCCAGGCAACCACCA AGGGAGAGACCCCAGAAAACATAAAAGAAAGCCTCTGGGGGTGGGCTATTCTGCCAGAAAATCACCTCTTTATGATAACTGCTTTCTCCA TGCTCCTGATGGACAGCCCCTCTGCACTTGTGATAGAAGAAAAGCTCAGTGGTACCTGGACAAAGGCATTGGTGAGCTGGTGAGTGAAGA GCCCTTTGTGGTGAAGCTACGGTTTGAACCTGCAGGAAGGCCCGAATCTCCTGGAGACTATTACTTGATGGTTAAAGAGAACCTGTGTGT AGTGTGTGGCAAGAGAGACTCCTACATTCGGAAGAACGTGATTCCACATGAGTACCGGAAGCACTTCCCCATCGAGATGAAGGACCACAA CTCCCACGATGTGCTGCTGCTCTGCACCTCCTGCCATGCCATTTCCAACTACTATGACAACCATCTGAAGCAGCAGCTGGCCAAGGAGTT CCAGGCCCCCATCGGCTCTGAGGAGGGCTTGCGCCTGCTGGAAGATCCTGAGCGCCGGCAGGTGCGTTCTGGGGCCAGGGCCCTGCTCAA CGCGGAGAGCCTGCCTACTCAGCGAAAGGAGGAGCTGCTGCAAGCACTCAGAGAGTTTTATAACACAGACGTGGTCACAGAGGAGATGCT TCAAGAGGCTGCCAGCCTGGAGACCAGAATCTCCAATGAAAACTATGTTCCTCACGGGCTGAAGGTGGTGCAGTGTCACAGCCAGGGTGG CCTGCGCTCCCTCATGCAGCTGGAGAGCCGCTGGCGTCAGCACTTCCTGGACTCCATGCAGCCCAAGCACCTGCCCCAGCAGTGGTCAGT GGACCACAACCATCAGAAGCTGCTCCGGAAATTCGGGGAAGATCTTCCCATCCAGCTGTCTTGATAGCTGCTTTCCTCCCAGTTAGGACA AGTGGGAAGCTGGAGCCAAGGTTGAAGAGTCACCTCTTCCCATTTTAGTACATCATTAATTGTCAAAGCCTGTGTGACACAACTCAGAAT ACTAACCTAGACTAATCCCAGGATGCTTCTGCTGGAGCAAAGATATTGTTTGAAGGAGAGTTTATGGTTTTGGATTTTAAACGGGCAGGG TCTTTTTTCCTCTCATTTTTGTGGACAAGAGAGGCCTTCGCCTTTATTTTTACTCTCCCTCTTCTGCTGTCCCTGTGCAGAGGAAAAATG AAGAATTCTCCCAGAAGTGACTTGTCAAGACTTAAAAAAAATGTTTTTAATGCATTTCTTCCTTGTCTAGTGCCTCGGTTTATCTCTAAC AGGGGCTGTCCAGTATATCGGTCCTGTTAGGAGGGGAGAAAAAGTTCTTCCAAAGGCTGGAGAAGTGAACAAGGAGTCAAATTTATTTTC CCAATTCAACTTCATAATTATCATTTCTTTGGCTTCATGCTCTCCCGTAACTCATGTGGTTGGGATCCATCCCATCTGGGTCACTTCAGT CTACTTCACGTACTTGAAAAGGCTTTCCTTTACACTTCCAGGACCAAACAGCAACTTCCTGCCACACACTTCCACCCTATCACTGGGAGA AATCCTTTTCTGGACATGAGCCTTTGACCTGGGTGGGGCAGAAAGAACCACAAACTCCATCTCCCAATAGAACTTTGAAATTCACTCAGC TTTTCCTTTCATGCTGTTTGTTGCCTGCTTGTTGCACTCCTCCTGCCCCAGAACTGCAAGATTTTTAGCTTCACCCCTTTCTGAGAGTAA TGTTATCTTTTATCAGAATCAGTATCAGTTCCCCTGTATTCTGTGCTTCATCGAATTTGCAAGACTGACCTCTTTTAAGCATTTAATTCA CTCCCAGAGTCATCTGGTCAGGTTGCAATATGAGGACTTCTCTGTCTCCTCTGAAGCCTGGGACACTGAGCTTACTTAATACATTAGATG TTCAAAAGAGGAGCGTTGTTTCATCTTTCAAAATGTTAGGCCATTACTTTGAGTATAAAATCGACTTATTAATGATTAGTAATTTTTCTA AAGTATTGGGAAAACTTTCTTATTTTATAAGATCTTAACAAGCTTAAAAAAGAATTTTATGACCAGAATCCAACAAGAGCTCTATTTTGG AATTGTGCCCAAGTTGGTGATGTTTACTCTAAAATTAATAATAAAACTACTTGTAAGCACAA >15399_15399_1_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000304494_EXD2_chr14_69695533_ENST00000409018_length(amino acids)=609AA_BP=98 MRDRVGGGSSASTGGRKRRGWLVTRGWGGPRALGGCGEGESRQRAAGSSMEPAAGSSMEPSADWLATAAARGRVEEVRALLEAGALPNAP NSYGRRPIQVNLEGKASPLSLLQMASPSGLCVLVRLPKLICGGKTLPRTLLDILADGTILKVGVGCSEDASKLLQDYGLVVRGCLDLRYL AMRQRNNLLCNGLSLKSLAETVLNFPLDKSLLLRCSNWDAETLTEDQVIYAARDAQISVALFLHLLGYPFSRNSPGEKNDDHSSWRKVLE KCQGVVDIPFRSKGMSRLGEEVNGEATESQQKPRNKKSKMDGMVPGNHQGRDPRKHKRKPLGVGYSARKSPLYDNCFLHAPDGQPLCTCD RRKAQWYLDKGIGELVSEEPFVVKLRFEPAGRPESPGDYYLMVKENLCVVCGKRDSYIRKNVIPHEYRKHFPIEMKDHNSHDVLLLCTSC HAISNYYDNHLKQQLAKEFQAPIGSEEGLRLLEDPERRQVRSGARALLNAESLPTQRKEELLQALREFYNTDVVTEEMLQEAASLETRIS NENYVPHGLKVVQCHSQGGLRSLMQLESRWRQHFLDSMQPKHLPQQWSVDHNHQKLLRKFGEDLPIQLS -------------------------------------------------------------- >15399_15399_2_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000446177_EXD2_chr14_69695533_ENST00000409018_length(transcript)=3153nt_BP=362nt CTCCTCCGAGCACTCGCTCACGGCGTCCCCTTGCCTGGAAAGATACCGCGGTCCCTCCAGAGGATTTGAGGGACAGGGTCGGAGGGGGCT CTTCCGCCAGCACCGGAGGAAGAAAGAGGAGGGGCTGGCTGGTCACCAGAGGGTGGGGCGGACCGCGTGCGCTCGGCGGCTGCGGAGAGG GGGAGAGCAGGCAGCGGGCGGCGGGGAGCAGCATGGAGCCGGCGGCGGGGAGCAGCATGGAGCCTTCGGCTGACTGGCTGGCCACGGCCG CGGCCCGGGGTCGGGTAGAGGAGGTGCGGGCGCTGCTGGAGGCGGGGGCGCTGCCCAACGCACCGAATAGTTACGGTCGGAGGCCGATCC AGGTAAATTTGGAAGGCAAAGCCAGCCCTCTGTCACTTCTACAAATGGCCTCCCCAAGTGGCCTGTGTGTCTTGGTTCGCCTGCCCAAGC TAATCTGTGGAGGAAAAACACTACCAAGAACGTTATTGGATATTTTGGCAGATGGCACCATTTTGAAAGTTGGAGTGGGATGCTCAGAAG ATGCCAGCAAGCTTCTGCAGGATTATGGCCTCGTTGTTAGGGGGTGCCTGGACCTCCGATACCTAGCCATGCGGCAGAGAAACAATTTGC TCTGTAATGGGCTTAGCCTGAAGTCCCTCGCTGAGACTGTTTTGAACTTTCCCCTTGACAAGTCCCTTCTACTTCGTTGCAGCAACTGGG ATGCTGAGACTCTCACAGAGGACCAGGTAATTTATGCTGCCAGGGATGCCCAGATTTCAGTGGCTCTCTTTCTTCATCTTCTTGGATACC CTTTCTCTAGGAATTCACCTGGAGAAAAAAACGATGACCACAGTAGCTGGAGAAAAGTCTTGGAAAAATGCCAGGGTGTGGTCGACATCC CATTTCGAAGCAAAGGAATGAGCAGATTGGGAGAAGAGGTTAATGGGGAAGCAACAGAATCTCAGCAGAAGCCAAGAAATAAGAAGTCTA AGATGGATGGGATGGTGCCAGGCAACCACCAAGGGAGAGACCCCAGAAAACATAAAAGAAAGCCTCTGGGGGTGGGCTATTCTGCCAGAA AATCACCTCTTTATGATAACTGCTTTCTCCATGCTCCTGATGGACAGCCCCTCTGCACTTGTGATAGAAGAAAAGCTCAGTGGTACCTGG ACAAAGGCATTGGTGAGCTGGTGAGTGAAGAGCCCTTTGTGGTGAAGCTACGGTTTGAACCTGCAGGAAGGCCCGAATCTCCTGGAGACT ATTACTTGATGGTTAAAGAGAACCTGTGTGTAGTGTGTGGCAAGAGAGACTCCTACATTCGGAAGAACGTGATTCCACATGAGTACCGGA AGCACTTCCCCATCGAGATGAAGGACCACAACTCCCACGATGTGCTGCTGCTCTGCACCTCCTGCCATGCCATTTCCAACTACTATGACA ACCATCTGAAGCAGCAGCTGGCCAAGGAGTTCCAGGCCCCCATCGGCTCTGAGGAGGGCTTGCGCCTGCTGGAAGATCCTGAGCGCCGGC AGGTGCGTTCTGGGGCCAGGGCCCTGCTCAACGCGGAGAGCCTGCCTACTCAGCGAAAGGAGGAGCTGCTGCAAGCACTCAGAGAGTTTT ATAACACAGACGTGGTCACAGAGGAGATGCTTCAAGAGGCTGCCAGCCTGGAGACCAGAATCTCCAATGAAAACTATGTTCCTCACGGGC TGAAGGTGGTGCAGTGTCACAGCCAGGGTGGCCTGCGCTCCCTCATGCAGCTGGAGAGCCGCTGGCGTCAGCACTTCCTGGACTCCATGC AGCCCAAGCACCTGCCCCAGCAGTGGTCAGTGGACCACAACCATCAGAAGCTGCTCCGGAAATTCGGGGAAGATCTTCCCATCCAGCTGT CTTGATAGCTGCTTTCCTCCCAGTTAGGACAAGTGGGAAGCTGGAGCCAAGGTTGAAGAGTCACCTCTTCCCATTTTAGTACATCATTAA TTGTCAAAGCCTGTGTGACACAACTCAGAATACTAACCTAGACTAATCCCAGGATGCTTCTGCTGGAGCAAAGATATTGTTTGAAGGAGA GTTTATGGTTTTGGATTTTAAACGGGCAGGGTCTTTTTTCCTCTCATTTTTGTGGACAAGAGAGGCCTTCGCCTTTATTTTTACTCTCCC TCTTCTGCTGTCCCTGTGCAGAGGAAAAATGAAGAATTCTCCCAGAAGTGACTTGTCAAGACTTAAAAAAAATGTTTTTAATGCATTTCT TCCTTGTCTAGTGCCTCGGTTTATCTCTAACAGGGGCTGTCCAGTATATCGGTCCTGTTAGGAGGGGAGAAAAAGTTCTTCCAAAGGCTG GAGAAGTGAACAAGGAGTCAAATTTATTTTCCCAATTCAACTTCATAATTATCATTTCTTTGGCTTCATGCTCTCCCGTAACTCATGTGG TTGGGATCCATCCCATCTGGGTCACTTCAGTCTACTTCACGTACTTGAAAAGGCTTTCCTTTACACTTCCAGGACCAAACAGCAACTTCC TGCCACACACTTCCACCCTATCACTGGGAGAAATCCTTTTCTGGACATGAGCCTTTGACCTGGGTGGGGCAGAAAGAACCACAAACTCCA TCTCCCAATAGAACTTTGAAATTCACTCAGCTTTTCCTTTCATGCTGTTTGTTGCCTGCTTGTTGCACTCCTCCTGCCCCAGAACTGCAA GATTTTTAGCTTCACCCCTTTCTGAGAGTAATGTTATCTTTTATCAGAATCAGTATCAGTTCCCCTGTATTCTGTGCTTCATCGAATTTG CAAGACTGACCTCTTTTAAGCATTTAATTCACTCCCAGAGTCATCTGGTCAGGTTGCAATATGAGGACTTCTCTGTCTCCTCTGAAGCCT GGGACACTGAGCTTACTTAATACATTAGATGTTCAAAAGAGGAGCGTTGTTTCATCTTTCAAAATGTTAGGCCATTACTTTGAGTATAAA ATCGACTTATTAATGATTAGTAATTTTTCTAAAGTATTGGGAAAACTTTCTTATTTTATAAGATCTTAACAAGCTTAAAAAAGAATTTTA TGACCAGAATCCAACAAGAGCTCTATTTTGGAATTGTGCCCAAGTTGGTGATGTTTACTCTAAAATTAATAATAAAACTACTTGTAAGCA CAA >15399_15399_2_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000446177_EXD2_chr14_69695533_ENST00000409018_length(amino acids)=609AA_BP=98 MRDRVGGGSSASTGGRKRRGWLVTRGWGGPRALGGCGEGESRQRAAGSSMEPAAGSSMEPSADWLATAAARGRVEEVRALLEAGALPNAP NSYGRRPIQVNLEGKASPLSLLQMASPSGLCVLVRLPKLICGGKTLPRTLLDILADGTILKVGVGCSEDASKLLQDYGLVVRGCLDLRYL AMRQRNNLLCNGLSLKSLAETVLNFPLDKSLLLRCSNWDAETLTEDQVIYAARDAQISVALFLHLLGYPFSRNSPGEKNDDHSSWRKVLE KCQGVVDIPFRSKGMSRLGEEVNGEATESQQKPRNKKSKMDGMVPGNHQGRDPRKHKRKPLGVGYSARKSPLYDNCFLHAPDGQPLCTCD RRKAQWYLDKGIGELVSEEPFVVKLRFEPAGRPESPGDYYLMVKENLCVVCGKRDSYIRKNVIPHEYRKHFPIEMKDHNSHDVLLLCTSC HAISNYYDNHLKQQLAKEFQAPIGSEEGLRLLEDPERRQVRSGARALLNAESLPTQRKEELLQALREFYNTDVVTEEMLQEAASLETRIS NENYVPHGLKVVQCHSQGGLRSLMQLESRWRQHFLDSMQPKHLPQQWSVDHNHQKLLRKFGEDLPIQLS -------------------------------------------------------------- >15399_15399_3_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000498124_EXD2_chr14_69695533_ENST00000409018_length(transcript)=2980nt_BP=189nt GGAGAGGGGGAGAGCAGGCAGCGGGCGGCGGGGAGCAGCATGGAGCCGGCGGCGGGGAGCAGCATGGAGCCTTCGGCTGACTGGCTGGCC ACGGCCGCGGCCCGGGGTCGGGTAGAGGAGGTGCGGGCGCTGCTGGAGGCGGGGGCGCTGCCCAACGCACCGAATAGTTACGGTCGGAGG CCGATCCAGGTAAATTTGGAAGGCAAAGCCAGCCCTCTGTCACTTCTACAAATGGCCTCCCCAAGTGGCCTGTGTGTCTTGGTTCGCCTG CCCAAGCTAATCTGTGGAGGAAAAACACTACCAAGAACGTTATTGGATATTTTGGCAGATGGCACCATTTTGAAAGTTGGAGTGGGATGC TCAGAAGATGCCAGCAAGCTTCTGCAGGATTATGGCCTCGTTGTTAGGGGGTGCCTGGACCTCCGATACCTAGCCATGCGGCAGAGAAAC AATTTGCTCTGTAATGGGCTTAGCCTGAAGTCCCTCGCTGAGACTGTTTTGAACTTTCCCCTTGACAAGTCCCTTCTACTTCGTTGCAGC AACTGGGATGCTGAGACTCTCACAGAGGACCAGGTAATTTATGCTGCCAGGGATGCCCAGATTTCAGTGGCTCTCTTTCTTCATCTTCTT GGATACCCTTTCTCTAGGAATTCACCTGGAGAAAAAAACGATGACCACAGTAGCTGGAGAAAAGTCTTGGAAAAATGCCAGGGTGTGGTC GACATCCCATTTCGAAGCAAAGGAATGAGCAGATTGGGAGAAGAGGTTAATGGGGAAGCAACAGAATCTCAGCAGAAGCCAAGAAATAAG AAGTCTAAGATGGATGGGATGGTGCCAGGCAACCACCAAGGGAGAGACCCCAGAAAACATAAAAGAAAGCCTCTGGGGGTGGGCTATTCT GCCAGAAAATCACCTCTTTATGATAACTGCTTTCTCCATGCTCCTGATGGACAGCCCCTCTGCACTTGTGATAGAAGAAAAGCTCAGTGG TACCTGGACAAAGGCATTGGTGAGCTGGTGAGTGAAGAGCCCTTTGTGGTGAAGCTACGGTTTGAACCTGCAGGAAGGCCCGAATCTCCT GGAGACTATTACTTGATGGTTAAAGAGAACCTGTGTGTAGTGTGTGGCAAGAGAGACTCCTACATTCGGAAGAACGTGATTCCACATGAG TACCGGAAGCACTTCCCCATCGAGATGAAGGACCACAACTCCCACGATGTGCTGCTGCTCTGCACCTCCTGCCATGCCATTTCCAACTAC TATGACAACCATCTGAAGCAGCAGCTGGCCAAGGAGTTCCAGGCCCCCATCGGCTCTGAGGAGGGCTTGCGCCTGCTGGAAGATCCTGAG CGCCGGCAGGTGCGTTCTGGGGCCAGGGCCCTGCTCAACGCGGAGAGCCTGCCTACTCAGCGAAAGGAGGAGCTGCTGCAAGCACTCAGA GAGTTTTATAACACAGACGTGGTCACAGAGGAGATGCTTCAAGAGGCTGCCAGCCTGGAGACCAGAATCTCCAATGAAAACTATGTTCCT CACGGGCTGAAGGTGGTGCAGTGTCACAGCCAGGGTGGCCTGCGCTCCCTCATGCAGCTGGAGAGCCGCTGGCGTCAGCACTTCCTGGAC TCCATGCAGCCCAAGCACCTGCCCCAGCAGTGGTCAGTGGACCACAACCATCAGAAGCTGCTCCGGAAATTCGGGGAAGATCTTCCCATC CAGCTGTCTTGATAGCTGCTTTCCTCCCAGTTAGGACAAGTGGGAAGCTGGAGCCAAGGTTGAAGAGTCACCTCTTCCCATTTTAGTACA TCATTAATTGTCAAAGCCTGTGTGACACAACTCAGAATACTAACCTAGACTAATCCCAGGATGCTTCTGCTGGAGCAAAGATATTGTTTG AAGGAGAGTTTATGGTTTTGGATTTTAAACGGGCAGGGTCTTTTTTCCTCTCATTTTTGTGGACAAGAGAGGCCTTCGCCTTTATTTTTA CTCTCCCTCTTCTGCTGTCCCTGTGCAGAGGAAAAATGAAGAATTCTCCCAGAAGTGACTTGTCAAGACTTAAAAAAAATGTTTTTAATG CATTTCTTCCTTGTCTAGTGCCTCGGTTTATCTCTAACAGGGGCTGTCCAGTATATCGGTCCTGTTAGGAGGGGAGAAAAAGTTCTTCCA AAGGCTGGAGAAGTGAACAAGGAGTCAAATTTATTTTCCCAATTCAACTTCATAATTATCATTTCTTTGGCTTCATGCTCTCCCGTAACT CATGTGGTTGGGATCCATCCCATCTGGGTCACTTCAGTCTACTTCACGTACTTGAAAAGGCTTTCCTTTACACTTCCAGGACCAAACAGC AACTTCCTGCCACACACTTCCACCCTATCACTGGGAGAAATCCTTTTCTGGACATGAGCCTTTGACCTGGGTGGGGCAGAAAGAACCACA AACTCCATCTCCCAATAGAACTTTGAAATTCACTCAGCTTTTCCTTTCATGCTGTTTGTTGCCTGCTTGTTGCACTCCTCCTGCCCCAGA ACTGCAAGATTTTTAGCTTCACCCCTTTCTGAGAGTAATGTTATCTTTTATCAGAATCAGTATCAGTTCCCCTGTATTCTGTGCTTCATC GAATTTGCAAGACTGACCTCTTTTAAGCATTTAATTCACTCCCAGAGTCATCTGGTCAGGTTGCAATATGAGGACTTCTCTGTCTCCTCT GAAGCCTGGGACACTGAGCTTACTTAATACATTAGATGTTCAAAAGAGGAGCGTTGTTTCATCTTTCAAAATGTTAGGCCATTACTTTGA GTATAAAATCGACTTATTAATGATTAGTAATTTTTCTAAAGTATTGGGAAAACTTTCTTATTTTATAAGATCTTAACAAGCTTAAAAAAG AATTTTATGACCAGAATCCAACAAGAGCTCTATTTTGGAATTGTGCCCAAGTTGGTGATGTTTACTCTAAAATTAATAATAAAACTACTT GTAAGCACAA >15399_15399_3_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000498124_EXD2_chr14_69695533_ENST00000409018_length(amino acids)=560AA_BP=49 MEPAAGSSMEPSADWLATAAARGRVEEVRALLEAGALPNAPNSYGRRPIQVNLEGKASPLSLLQMASPSGLCVLVRLPKLICGGKTLPRT LLDILADGTILKVGVGCSEDASKLLQDYGLVVRGCLDLRYLAMRQRNNLLCNGLSLKSLAETVLNFPLDKSLLLRCSNWDAETLTEDQVI YAARDAQISVALFLHLLGYPFSRNSPGEKNDDHSSWRKVLEKCQGVVDIPFRSKGMSRLGEEVNGEATESQQKPRNKKSKMDGMVPGNHQ GRDPRKHKRKPLGVGYSARKSPLYDNCFLHAPDGQPLCTCDRRKAQWYLDKGIGELVSEEPFVVKLRFEPAGRPESPGDYYLMVKENLCV VCGKRDSYIRKNVIPHEYRKHFPIEMKDHNSHDVLLLCTSCHAISNYYDNHLKQQLAKEFQAPIGSEEGLRLLEDPERRQVRSGARALLN AESLPTQRKEELLQALREFYNTDVVTEEMLQEAASLETRISNENYVPHGLKVVQCHSQGGLRSLMQLESRWRQHFLDSMQPKHLPQQWSV DHNHQKLLRKFGEDLPIQLS -------------------------------------------------------------- >15399_15399_4_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000579122_EXD2_chr14_69695533_ENST00000409018_length(transcript)=2972nt_BP=181nt GGAGAGCAGGCAGCGGGCGGCGGGGAGCAGCATGGAGCCGGCGGCGGGGAGCAGCATGGAGCCTTCGGCTGACTGGCTGGCCACGGCCGC GGCCCGGGGTCGGGTAGAGGAGGTGCGGGCGCTGCTGGAGGCGGGGGCGCTGCCCAACGCACCGAATAGTTACGGTCGGAGGCCGATCCA GGTAAATTTGGAAGGCAAAGCCAGCCCTCTGTCACTTCTACAAATGGCCTCCCCAAGTGGCCTGTGTGTCTTGGTTCGCCTGCCCAAGCT AATCTGTGGAGGAAAAACACTACCAAGAACGTTATTGGATATTTTGGCAGATGGCACCATTTTGAAAGTTGGAGTGGGATGCTCAGAAGA TGCCAGCAAGCTTCTGCAGGATTATGGCCTCGTTGTTAGGGGGTGCCTGGACCTCCGATACCTAGCCATGCGGCAGAGAAACAATTTGCT CTGTAATGGGCTTAGCCTGAAGTCCCTCGCTGAGACTGTTTTGAACTTTCCCCTTGACAAGTCCCTTCTACTTCGTTGCAGCAACTGGGA TGCTGAGACTCTCACAGAGGACCAGGTAATTTATGCTGCCAGGGATGCCCAGATTTCAGTGGCTCTCTTTCTTCATCTTCTTGGATACCC TTTCTCTAGGAATTCACCTGGAGAAAAAAACGATGACCACAGTAGCTGGAGAAAAGTCTTGGAAAAATGCCAGGGTGTGGTCGACATCCC ATTTCGAAGCAAAGGAATGAGCAGATTGGGAGAAGAGGTTAATGGGGAAGCAACAGAATCTCAGCAGAAGCCAAGAAATAAGAAGTCTAA GATGGATGGGATGGTGCCAGGCAACCACCAAGGGAGAGACCCCAGAAAACATAAAAGAAAGCCTCTGGGGGTGGGCTATTCTGCCAGAAA ATCACCTCTTTATGATAACTGCTTTCTCCATGCTCCTGATGGACAGCCCCTCTGCACTTGTGATAGAAGAAAAGCTCAGTGGTACCTGGA CAAAGGCATTGGTGAGCTGGTGAGTGAAGAGCCCTTTGTGGTGAAGCTACGGTTTGAACCTGCAGGAAGGCCCGAATCTCCTGGAGACTA TTACTTGATGGTTAAAGAGAACCTGTGTGTAGTGTGTGGCAAGAGAGACTCCTACATTCGGAAGAACGTGATTCCACATGAGTACCGGAA GCACTTCCCCATCGAGATGAAGGACCACAACTCCCACGATGTGCTGCTGCTCTGCACCTCCTGCCATGCCATTTCCAACTACTATGACAA CCATCTGAAGCAGCAGCTGGCCAAGGAGTTCCAGGCCCCCATCGGCTCTGAGGAGGGCTTGCGCCTGCTGGAAGATCCTGAGCGCCGGCA GGTGCGTTCTGGGGCCAGGGCCCTGCTCAACGCGGAGAGCCTGCCTACTCAGCGAAAGGAGGAGCTGCTGCAAGCACTCAGAGAGTTTTA TAACACAGACGTGGTCACAGAGGAGATGCTTCAAGAGGCTGCCAGCCTGGAGACCAGAATCTCCAATGAAAACTATGTTCCTCACGGGCT GAAGGTGGTGCAGTGTCACAGCCAGGGTGGCCTGCGCTCCCTCATGCAGCTGGAGAGCCGCTGGCGTCAGCACTTCCTGGACTCCATGCA GCCCAAGCACCTGCCCCAGCAGTGGTCAGTGGACCACAACCATCAGAAGCTGCTCCGGAAATTCGGGGAAGATCTTCCCATCCAGCTGTC TTGATAGCTGCTTTCCTCCCAGTTAGGACAAGTGGGAAGCTGGAGCCAAGGTTGAAGAGTCACCTCTTCCCATTTTAGTACATCATTAAT TGTCAAAGCCTGTGTGACACAACTCAGAATACTAACCTAGACTAATCCCAGGATGCTTCTGCTGGAGCAAAGATATTGTTTGAAGGAGAG TTTATGGTTTTGGATTTTAAACGGGCAGGGTCTTTTTTCCTCTCATTTTTGTGGACAAGAGAGGCCTTCGCCTTTATTTTTACTCTCCCT CTTCTGCTGTCCCTGTGCAGAGGAAAAATGAAGAATTCTCCCAGAAGTGACTTGTCAAGACTTAAAAAAAATGTTTTTAATGCATTTCTT CCTTGTCTAGTGCCTCGGTTTATCTCTAACAGGGGCTGTCCAGTATATCGGTCCTGTTAGGAGGGGAGAAAAAGTTCTTCCAAAGGCTGG AGAAGTGAACAAGGAGTCAAATTTATTTTCCCAATTCAACTTCATAATTATCATTTCTTTGGCTTCATGCTCTCCCGTAACTCATGTGGT TGGGATCCATCCCATCTGGGTCACTTCAGTCTACTTCACGTACTTGAAAAGGCTTTCCTTTACACTTCCAGGACCAAACAGCAACTTCCT GCCACACACTTCCACCCTATCACTGGGAGAAATCCTTTTCTGGACATGAGCCTTTGACCTGGGTGGGGCAGAAAGAACCACAAACTCCAT CTCCCAATAGAACTTTGAAATTCACTCAGCTTTTCCTTTCATGCTGTTTGTTGCCTGCTTGTTGCACTCCTCCTGCCCCAGAACTGCAAG ATTTTTAGCTTCACCCCTTTCTGAGAGTAATGTTATCTTTTATCAGAATCAGTATCAGTTCCCCTGTATTCTGTGCTTCATCGAATTTGC AAGACTGACCTCTTTTAAGCATTTAATTCACTCCCAGAGTCATCTGGTCAGGTTGCAATATGAGGACTTCTCTGTCTCCTCTGAAGCCTG GGACACTGAGCTTACTTAATACATTAGATGTTCAAAAGAGGAGCGTTGTTTCATCTTTCAAAATGTTAGGCCATTACTTTGAGTATAAAA TCGACTTATTAATGATTAGTAATTTTTCTAAAGTATTGGGAAAACTTTCTTATTTTATAAGATCTTAACAAGCTTAAAAAAGAATTTTAT GACCAGAATCCAACAAGAGCTCTATTTTGGAATTGTGCCCAAGTTGGTGATGTTTACTCTAAAATTAATAATAAAACTACTTGTAAGCAC AA >15399_15399_4_CDKN2A-EXD2_CDKN2A_chr9_21974677_ENST00000579122_EXD2_chr14_69695533_ENST00000409018_length(amino acids)=560AA_BP=49 MEPAAGSSMEPSADWLATAAARGRVEEVRALLEAGALPNAPNSYGRRPIQVNLEGKASPLSLLQMASPSGLCVLVRLPKLICGGKTLPRT LLDILADGTILKVGVGCSEDASKLLQDYGLVVRGCLDLRYLAMRQRNNLLCNGLSLKSLAETVLNFPLDKSLLLRCSNWDAETLTEDQVI YAARDAQISVALFLHLLGYPFSRNSPGEKNDDHSSWRKVLEKCQGVVDIPFRSKGMSRLGEEVNGEATESQQKPRNKKSKMDGMVPGNHQ GRDPRKHKRKPLGVGYSARKSPLYDNCFLHAPDGQPLCTCDRRKAQWYLDKGIGELVSEEPFVVKLRFEPAGRPESPGDYYLMVKENLCV VCGKRDSYIRKNVIPHEYRKHFPIEMKDHNSHDVLLLCTSCHAISNYYDNHLKQQLAKEFQAPIGSEEGLRLLEDPERRQVRSGARALLN AESLPTQRKEELLQALREFYNTDVVTEEMLQEAASLETRISNENYVPHGLKVVQCHSQGGLRSLMQLESRWRQHFLDSMQPKHLPQQWSV DHNHQKLLRKFGEDLPIQLS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CDKN2A-EXD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000361570 | - | 1 | 3 | 1_64 | 0 | 220.0 | CDK5RAP3 and MDM2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000530628 | - | 1 | 3 | 1_64 | 0 | 122.66666666666667 | CDK5RAP3 and MDM2 |
| Hgene | CDKN2A | chr9:21974677 | chr14:69695533 | ENST00000579755 | - | 1 | 3 | 1_64 | 0 | 134.33333333333334 | CDK5RAP3 and MDM2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CDKN2A-EXD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CDKN2A-EXD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CDKN2A | C1835044 | MELANOMA, CUTANEOUS MALIGNANT, SUSCEPTIBILITY TO, 2 | 13 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | CDKN2A | C1838547 | MELANOMA-PANCREATIC CANCER SYNDROME | 10 | CLINGEN;CTD_human |
| Hgene | CDKN2A | C0024121 | Lung Neoplasms | 4 | CTD_human;UNIPROT |
| Hgene | CDKN2A | C0242379 | Malignant neoplasm of lung | 4 | CTD_human |
| Hgene | CDKN2A | C0011570 | Mental Depression | 3 | PSYGENET |
| Hgene | CDKN2A | C0011581 | Depressive disorder | 3 | PSYGENET |
| Hgene | CDKN2A | C0023903 | Liver neoplasms | 3 | CTD_human |
| Hgene | CDKN2A | C0345904 | Malignant neoplasm of liver | 3 | CTD_human |
| Hgene | CDKN2A | C0345967 | Malignant mesothelioma | 3 | CTD_human |
| Hgene | CDKN2A | C1168401 | Squamous cell carcinoma of the head and neck | 3 | CTD_human |
| Hgene | CDKN2A | C0005684 | Malignant neoplasm of urinary bladder | 2 | CTD_human |
| Hgene | CDKN2A | C0005695 | Bladder Neoplasm | 2 | CTD_human;UNIPROT |
| Hgene | CDKN2A | C0006118 | Brain Neoplasms | 2 | CTD_human |
| Hgene | CDKN2A | C0006826 | Malignant Neoplasms | 2 | CGI;CTD_human |
| Hgene | CDKN2A | C0007131 | Non-Small Cell Lung Carcinoma | 2 | CTD_human;UNIPROT |
| Hgene | CDKN2A | C0017638 | Glioma | 2 | CGI;CTD_human |
| Hgene | CDKN2A | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human;GENOMICS_ENGLAND |
| Hgene | CDKN2A | C0025202 | melanoma | 2 | CGI;CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | CDKN2A | C0027651 | Neoplasms | 2 | CTD_human |
| Hgene | CDKN2A | C0030297 | Pancreatic Neoplasm | 2 | CGI;CTD_human;UNIPROT |
| Hgene | CDKN2A | C0040715 | Chromosomal translocation | 2 | CTD_human |
| Hgene | CDKN2A | C0041696 | Unipolar Depression | 2 | PSYGENET |
| Hgene | CDKN2A | C0085390 | Li-Fraumeni Syndrome | 2 | ORPHANET |
| Hgene | CDKN2A | C0086692 | Benign Neoplasm | 2 | CTD_human |
| Hgene | CDKN2A | C0153633 | Malignant neoplasm of brain | 2 | CTD_human |
| Hgene | CDKN2A | C0259783 | mixed gliomas | 2 | CTD_human |
| Hgene | CDKN2A | C0346647 | Malignant neoplasm of pancreas | 2 | CGI;CTD_human |
| Hgene | CDKN2A | C0496899 | Benign neoplasm of brain, unspecified | 2 | CTD_human |
| Hgene | CDKN2A | C0555198 | Malignant Glioma | 2 | CTD_human |
| Hgene | CDKN2A | C0750974 | Brain Tumor, Primary | 2 | CTD_human |
| Hgene | CDKN2A | C0750977 | Recurrent Brain Neoplasm | 2 | CTD_human |
| Hgene | CDKN2A | C0750979 | Primary malignant neoplasm of brain | 2 | CTD_human |
| Hgene | CDKN2A | C1269683 | Major Depressive Disorder | 2 | PSYGENET |
| Hgene | CDKN2A | C1527390 | Neoplasms, Intracranial | 2 | CTD_human |
| Hgene | CDKN2A | C2239176 | Liver carcinoma | 2 | CTD_human |
| Hgene | CDKN2A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Hgene | CDKN2A | C0001624 | Adrenal Gland Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0005940 | Bone Diseases | 1 | CTD_human |
| Hgene | CDKN2A | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Hgene | CDKN2A | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Hgene | CDKN2A | C0007137 | Squamous cell carcinoma | 1 | CTD_human;UNIPROT |
| Hgene | CDKN2A | C0008628 | Chromosome Deletion | 1 | CTD_human |
| Hgene | CDKN2A | C0014859 | Esophageal Neoplasms | 1 | CTD_human;UNIPROT |
| Hgene | CDKN2A | C0016325 | Fluoride Poisoning | 1 | CTD_human |
| Hgene | CDKN2A | C0017601 | Glaucoma | 1 | CTD_human |
| Hgene | CDKN2A | C0022783 | Vulvar Lichen Sclerosus | 1 | CTD_human |
| Hgene | CDKN2A | C0023449 | Acute lymphocytic leukemia | 1 | GENOMICS_ENGLAND |
| Hgene | CDKN2A | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | CDKN2A | C0024232 | Lymphatic Metastasis | 1 | CTD_human |
| Hgene | CDKN2A | C0024299 | Lymphoma | 1 | CTD_human |
| Hgene | CDKN2A | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Hgene | CDKN2A | C0025500 | Mesothelioma | 1 | CTD_human |
| Hgene | CDKN2A | C0026640 | Mouth Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0026764 | Multiple Myeloma | 1 | CTD_human |
| Hgene | CDKN2A | C0027626 | Neoplasm Invasiveness | 1 | CTD_human |
| Hgene | CDKN2A | C0027819 | Neuroblastoma | 1 | CTD_human |
| Hgene | CDKN2A | C0032927 | Precancerous Conditions | 1 | CTD_human |
| Hgene | CDKN2A | C0036920 | Sezary Syndrome | 1 | CTD_human |
| Hgene | CDKN2A | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0040100 | Thymoma | 1 | CTD_human |
| Hgene | CDKN2A | C0041107 | Trisomy | 1 | CTD_human |
| Hgene | CDKN2A | C0042065 | Genitourinary Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human |
| Hgene | CDKN2A | C0079773 | Lymphoma, T-Cell, Cutaneous | 1 | CTD_human |
| Hgene | CDKN2A | C0151779 | Cutaneous Melanoma | 1 | CGI;CTD_human |
| Hgene | CDKN2A | C0153381 | Malignant neoplasm of mouth | 1 | CTD_human |
| Hgene | CDKN2A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Hgene | CDKN2A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Hgene | CDKN2A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Hgene | CDKN2A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Hgene | CDKN2A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Hgene | CDKN2A | C0205969 | Thymic Carcinoma | 1 | CTD_human |
| Hgene | CDKN2A | C0206686 | Adrenocortical carcinoma | 1 | CTD_human |
| Hgene | CDKN2A | C0206727 | Nerve Sheath Tumors | 1 | CTD_human |
| Hgene | CDKN2A | C0279626 | Squamous cell carcinoma of esophagus | 1 | CTD_human |
| Hgene | CDKN2A | C0279628 | Adenocarcinoma Of Esophagus | 1 | CTD_human |
| Hgene | CDKN2A | C0282313 | Condition, Preneoplastic | 1 | CTD_human |
| Hgene | CDKN2A | C0376407 | Granulomatous Slack Skin | 1 | CTD_human |
| Hgene | CDKN2A | C0546837 | Malignant neoplasm of esophagus | 1 | CTD_human |
| Hgene | CDKN2A | C0596263 | Carcinogenesis | 1 | CTD_human |
| Hgene | CDKN2A | C0677866 | Brain Stem Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Hgene | CDKN2A | C0750887 | Adrenal Cancer | 1 | CTD_human |
| Hgene | CDKN2A | C0751569 | Genitourinary Cancer | 1 | CTD_human |
| Hgene | CDKN2A | C0751606 | Adult Acute Lymphocytic Leukemia | 1 | GENOMICS_ENGLAND |
| Hgene | CDKN2A | C0751689 | Peripheral Nerve Sheath Neoplasm | 1 | CTD_human |
| Hgene | CDKN2A | C0751691 | Perineurioma | 1 | CTD_human |
| Hgene | CDKN2A | C0751886 | Brain Stem Neoplasms, Primary | 1 | CTD_human |
| Hgene | CDKN2A | C0751887 | Medullary Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0751888 | Mesencephalic Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C0751889 | Pontine Tumors | 1 | CTD_human |
| Hgene | CDKN2A | C0887833 | Carcinoma, Pancreatic Ductal | 1 | CTD_human |
| Hgene | CDKN2A | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Hgene | CDKN2A | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Hgene | CDKN2A | C1297882 | Partial Trisomy | 1 | CTD_human |
| Hgene | CDKN2A | C1449861 | Micronuclei, Chromosome-Defective | 1 | CTD_human |
| Hgene | CDKN2A | C1449862 | Micronuclei, Genotoxicant-Induced | 1 | CTD_human |
| Hgene | CDKN2A | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Hgene | CDKN2A | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Hgene | CDKN2A | C1835042 | Melanoma astrocytoma syndrome | 1 | CTD_human;ORPHANET |
| Hgene | CDKN2A | C1961099 | Precursor T-Cell Lymphoblastic Leukemia-Lymphoma | 1 | ORPHANET |
| Hgene | CDKN2A | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Hgene | CDKN2A | C2314896 | Familial Atypical Mole Melanoma Syndrome | 1 | ORPHANET |
| Hgene | CDKN2A | C2930745 | Partial Monosomy | 1 | CTD_human |
| Hgene | CDKN2A | C2931038 | Pancreatic carcinoma, familial | 1 | ORPHANET |
| Hgene | CDKN2A | C2931822 | Nasopharyngeal carcinoma | 1 | CTD_human |
| Hgene | CDKN2A | C4283859 | Cutaneous Malignant Melanoma 2 | 1 | GENOMICS_ENGLAND |
| Hgene | CDKN2A | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
| Hgene | CDKN2A | C4721453 | Peripheral Nervous System Diseases | 1 | CTD_human |
