
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ADAR-CCT5 (FusionGDB2 ID:HG103TG22948) |
Fusion Gene Summary for ADAR-CCT5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ADAR-CCT5 | Fusion gene ID: hg103tg22948 | Hgene | Tgene | Gene symbol | ADAR | CCT5 | Gene ID | 103 | 22948 |
| Gene name | adenosine deaminase RNA specific | chaperonin containing TCP1 subunit 5 | |
| Synonyms | ADAR1|AGS6|DRADA|DSH|DSRAD|G1P1|IFI-4|IFI4|K88DSRBP|P136 | CCT-epsilon|CCTE|HEL-S-69|PNAS-102|TCP-1-epsilon | |
| Cytomap | ('ADAR')('CCT5') 1q21.3 | 5p15.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | double-stranded RNA-specific adenosine deaminase136 kDa double-stranded RNA-binding proteinadenosine deaminase acting on RNA 1-AdsRNA adenosine deaminasedsRNA adeonosine deaminaseinterferon-induced protein 4interferon-inducible protein 4 | T-complex protein 1 subunit epsilonchaperonin containing TCP1, subunit 5 (epsilon)epididymis secretory protein Li 69 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000292205, ENST00000368471, ENST00000368474, ENST00000471068, | ||
| Fusion gene scores | * DoF score | 9 X 10 X 7=630 | 58 X 14 X 17=13804 |
| # samples | 10 | 61 | |
| ** MAII score | log2(10/630*10)=-2.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(61/13804*10)=-4.50013332598527 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ADAR [Title/Abstract] AND CCT5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ADAR(154554538)-CCT5(10250033), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ADAR-CCT5 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ADAR-CCT5 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ADAR-CCT5 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ADAR-CCT5 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ADAR | GO:0006382 | adenosine to inosine editing | 15858013|19651874|21289159 |
| Hgene | ADAR | GO:0016553 | base conversion or substitution editing | 9020165 |
| Hgene | ADAR | GO:0031054 | pre-miRNA processing | 23622242 |
| Hgene | ADAR | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA | 23622242 |
| Hgene | ADAR | GO:0035455 | response to interferon-alpha | 16475990 |
| Hgene | ADAR | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation | 19651874 |
| Hgene | ADAR | GO:0045070 | positive regulation of viral genome replication | 19651874 |
 Fusion gene breakpoints across ADAR (5'-gene) Fusion gene breakpoints across ADAR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
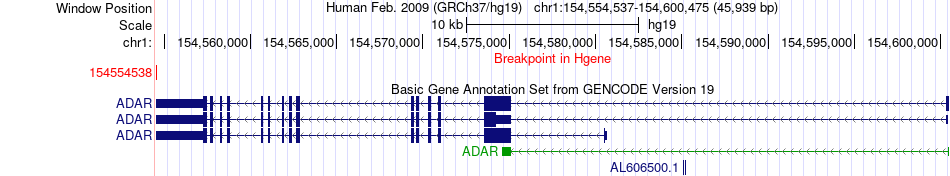 |
 Fusion gene breakpoints across CCT5 (3'-gene) Fusion gene breakpoints across CCT5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
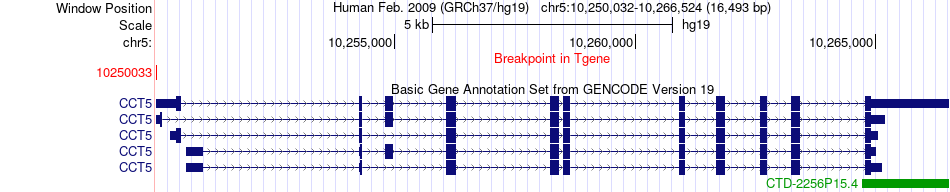 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KICH | TCGA-KM-8639-01A | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
Top |
Fusion Gene ORF analysis for ADAR-CCT5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000292205 | ENST00000503026 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000292205 | ENST00000506600 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000292205 | ENST00000515390 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000292205 | ENST00000515676 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368471 | ENST00000503026 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368471 | ENST00000506600 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368471 | ENST00000515390 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368471 | ENST00000515676 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368474 | ENST00000503026 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368474 | ENST00000506600 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368474 | ENST00000515390 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| 5CDS-intron | ENST00000368474 | ENST00000515676 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| Frame-shift | ENST00000292205 | ENST00000280326 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| Frame-shift | ENST00000368471 | ENST00000280326 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| In-frame | ENST00000368474 | ENST00000280326 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| intron-3CDS | ENST00000471068 | ENST00000280326 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000471068 | ENST00000503026 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000471068 | ENST00000506600 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000471068 | ENST00000515390 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
| intron-intron | ENST00000471068 | ENST00000515676 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000368474 | ADAR | chr1 | 154554538 | - | ENST00000280326 | CCT5 | chr5 | 10250033 | + | 10295 | 6620 | 128 | 3880 | 1250 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000368474 | ENST00000280326 | ADAR | chr1 | 154554538 | - | CCT5 | chr5 | 10250033 | + | 0.000435784 | 0.99956423 |
Top |
Fusion Genomic Features for ADAR-CCT5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ADAR | chr1 | 154554542 | - | CCT5 | chr5 | 10250032 | + | 0.012005751 | 0.9879943 |
| ADAR | chr1 | 154554542 | - | CCT5 | chr5 | 10250032 | + | 0.012005751 | 0.9879943 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
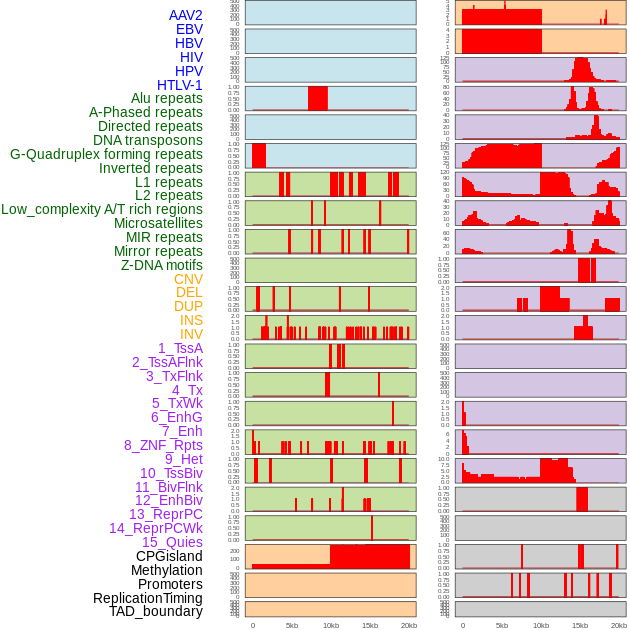 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
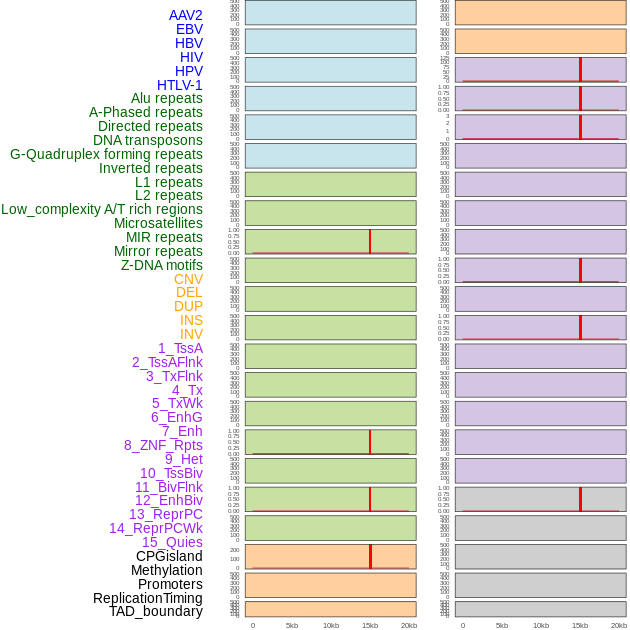 |
Top |
Fusion Protein Features for ADAR-CCT5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:154554538/chr5:10250033) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 133_199 | 1845 | 932.0 | Domain | Z-binding 1 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 293_357 | 1845 | 932.0 | Domain | Z-binding 2 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 503_571 | 1845 | 932.0 | Domain | DRBM 1 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 614_682 | 1845 | 932.0 | Domain | DRBM 2 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 726_794 | 1845 | 932.0 | Domain | DRBM 3 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 886_1221 | 1845 | 932.0 | Domain | A to I editase |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 133_199 | 2140 | 1227.0 | Domain | Z-binding 1 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 293_357 | 2140 | 1227.0 | Domain | Z-binding 2 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 503_571 | 2140 | 1227.0 | Domain | DRBM 1 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 614_682 | 2140 | 1227.0 | Domain | DRBM 2 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 726_794 | 2140 | 1227.0 | Domain | DRBM 3 |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 886_1221 | 2140 | 1227.0 | Domain | A to I editase |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 716_725 | 1845 | 932.0 | Region | N-terminal extension of DRBM 3 and constituent of a bi-partite nuclear localization signal |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 795_801 | 1845 | 932.0 | Region | C-terminal extension of DRBM 3 and constituent of a bi-partite nuclear localization signal |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 716_725 | 2140 | 1227.0 | Region | N-terminal extension of DRBM 3 and constituent of a bi-partite nuclear localization signal |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 795_801 | 2140 | 1227.0 | Region | C-terminal extension of DRBM 3 and constituent of a bi-partite nuclear localization signal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for ADAR-CCT5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2174_2174_1_ADAR-CCT5_ADAR_chr1_154554538_ENST00000368474_CCT5_chr5_10250033_ENST00000280326_length(transcript)=10295nt_BP=6620nt GAGGAAACGAAAGCGAAATTGAACCGGAGCCATCTTGGGCCCGGCGCGCAGACCCGCGGAGTTTCCCGTGCCGACGCCCCGGGGCCACTT CCAGTGCGGAGTAGCGGAGGCGTGGGGGCCTCGAGGGGCTGGCGCGGCCCAGCGGTCGGGCCAGGGTCGTGCCGCCGGCGGGTCGGGCCG GGCAATGCCTCGCGGGCGCAATGAATCCGCGGCAGGGGTATTCCCTCAGCGGATACTACACCCATCCATTTCAAGGCTATGAGCACAGAC AGCTCAGGTACCAGCAGCCTGGGCCAGGATCTTCCCCCAGTAGTTTCCTGCTTAAGCAAATAGAATTTCTCAAGGGGCAGCTCCCAGAAG CACCGGTGATTGGAAAGCAGACACCGTCACTGCCACCTTCCCTCCCAGGACTCCGGCCAAGGTTTCCAGTACTACTTGCCTCCAGTACCA GAGGCAGGCAAGTGGACATCAGGGGTGTCCCCAGGGGCGTGCATCTCAGAAGTCAGGGGCTCCAGAGAGGGTTCCAGCATCCTTCACCAC GTGGCAGGAGTCTGCCACAGAGAGGTGTTGATTGCCTTTCCTCACATTTCCAGGAACTGAGTATCTACCAAGATCAGGAACAAAGGATCT TAAAGTTCCTGGAAGAGCTTGGGGAAGGGAAGGCCACCACAGCACATGATCTGTCTGGGAAACTTGGGACTCCGAAGAAAGAAATCAATC GAGTTTTATACTCCCTGGCAAAGAAGGGCAAGCTACAGAAAGAGGCAGGAACACCCCCTTTGTGGAAAATCGCGGTCTCCACTCAGGCTT GGAACCAGCACAGCGGAGTGGTAAGACCAGACGGTCATAGCCAAGGAGCCCCAAACTCAGACCCGAGTTTGGAACCGGAAGACAGAAACT CCACATCTGTCTCAGAAGATCTTCTTGAGCCTTTTATTGCAGTCTCAGCTCAGGCTTGGAACCAGCACAGCGGAGTGGTAAGACCAGACA GTCATAGCCAAGGATCCCCAAACTCAGACCCAGGTTTGGAACCTGAAGACAGCAACTCCACATCTGCCTTGGAAGATCCTCTTGAGTTTT TAGACATGGCCGAGATCAAGGAGAAAATCTGCGACTATCTCTTCAATGTGTCTGACTCCTCTGCCCTGAATTTGGCTAAAAATATTGGCC TTACCAAGGCCCGAGATATAAATGCTGTGCTAATTGACATGGAAAGGCAGGGGGATGTCTATAGACAAGGGACAACCCCTCCCATATGGC ATTTGACAGACAAGAAGCGAGAGAGGATGCAAATCAAGAGAAATACGAACAGTGTTCCTGAAACCGCTCCAGCTGCAATCCCTGAGACCA AAAGAAACGCAGAGTTCCTCACCTGTAATATACCCACATCAAATGCCTCAAATAACATGGTAACCACAGAAAAAGTGGAGAATGGGCAGG AACCTGTCATAAAGTTAGAAAACAGGCAAGAGGCCAGACCAGAACCAGCAAGACTGAAACCACCTGTTCATTACAATGGCCCCTCAAAAG CAGGGTATGTTGACTTTGAAAATGGCCAGTGGGCCACAGATGACATCCCAGATGACTTGAATAGTATCCGCGCAGCACCAGGTGAGTTTC GAGCCATCATGGAGATGCCCTCCTTCTACAGTCATGGCTTGCCACGGTGTTCACCCTACAAGAAACTGACAGAGTGCCAGCTGAAGAACC CCATCAGCGGGCTGTTAGAATATGCCCAGTTCGCTAGTCAAACCTGTGAGTTCAACATGATAGAGCAGAGTGGACCACCCCATGAACCTC GATTTAAATTCCAGGTTGTCATCAATGGCCGAGAGTTTCCCCCAGCTGAAGCTGGAAGCAAGAAAGTGGCCAAGCAGGATGCAGCTATGA AAGCCATGACAATTCTGCTAGAGGAAGCCAAAGCCAAGGACAGTGGAAAATCAGAAGAATCATCCCACTATTCCACAGAGAAAGAATCAG AGAAGACTGCAGAGTCCCAGACCCCCACCCCTTCAGCCACATCCTTCTTTTCTGGGAAGAGCCCCGTCACCACACTGCTTGAGTGTATGC ACAAATTGGGGAACTCCTGCGAATTCCGTCTCCTGTCCAAAGAAGGCCCTGCCCATGAACCCAAGTTCCAATACTGTGTTGCAGTGGGAG CCCAAACTTTCCCCAGTGTGAGTGCTCCCAGCAAGAAAGTGGCAAAGCAGATGGCCGCAGAGGAAGCCATGAAGGCCCTGCATGGGGAGG CGACCAACTCCATGGCTTCTGATAACCAGCCTGAAGGTATGATCTCAGAGTCACTTGATAACTTGGAATCCATGATGCCCAACAAGGTCA GGAAGATTGGCGAGCTCGTGAGATACCTGAACACCAACCCTGTGGGTGGCCTTTTGGAGTACGCCCGCTCCCATGGCTTTGCTGCTGAAT TCAAGTTGGTCGACCAGTCCGGACCTCCTCACGAGCCCAAGTTCGTTTACCAAGCAAAAGTTGGGGGTCGCTGGTTCCCAGCCGTCTGCG CACACAGCAAGAAGCAAGGCAAGCAGGAAGCAGCAGATGCGGCTCTCCGTGTCTTGATTGGGGAGAACGAGAAGGCAGAACGCATGGGTT TCACAGAGGTAACCCCAGTGACAGGGGCCAGTCTCAGAAGAACTATGCTCCTCCTCTCAAGGTCCCCAGAAGCACAGCCAAAGACACTCC CTCTCACTGGCAGCACCTTCCATGACCAGATAGCCATGCTGAGCCACCGGTGCTTCAACACTCTGACTAACAGCTTCCAGCCCTCCTTGC TCGGCCGCAAGATTCTGGCCGCCATCATTATGAAAAAAGACTCTGAGGACATGGGTGTCGTCGTCAGCTTGGGAACAGGGAATCGCTGTG TGAAAGGAGATTCTCTCAGCCTAAAAGGAGAAACTGTCAATGACTGCCATGCAGAAATAATCTCCCGGAGAGGCTTCATCAGGTTTCTCT ACAGTGAGTTAATGAAATACAACTCCCAGACTGCGAAGGATAGTATATTTGAACCTGCTAAGGGAGGAGAAAAGCTCCAAATAAAAAAGA CTGTGTCATTCCATCTGTATATCAGCACTGCTCCGTGTGGAGATGGCGCCCTCTTTGACAAGTCCTGCAGCGACCGTGCTATGGAAAGCA CAGAATCCCGCCACTACCCTGTCTTCGAGAATCCCAAACAAGGAAAGCTCCGCACCAAGGTGGAGAACGGAGAAGGCACAATCCCTGTGG AATCCAGTGACATTGTGCCTACGTGGGATGGCATTCGGCTCGGGGAGAGACTCCGTACCATGTCCTGTAGTGACAAAATCCTACGCTGGA ACGTGCTGGGCCTGCAAGGGGCACTGTTGACCCACTTCCTGCAGCCCATTTATCTCAAATCTGTCACATTGGGTTACCTTTTCAGCCAAG GGCATCTGACCCGTGCTATTTGCTGTCGTGTGACAAGAGATGGGAGTGCATTTGAGGATGGACTACGACATCCCTTTATTGTCAACCACC CCAAGGTTGGCAGAGTCAGCATATATGATTCCAAAAGGCAATCCGGGAAGACTAAGGAGACAAGCGTCAACTGGTGTCTGGCTGATGGCT ATGACCTGGAGATCCTGGACGGTACCAGAGGCACTGTGGATGGGCCACGGAATGAATTGTCCCGGGTCTCCAAAAAGAACATTTTTCTTC TATTTAAGAAGCTCTGCTCCTTCCGTTACCGCAGGGATCTACTGAGACTCTCCTATGGTGAGGCCAAGAAAGCTGCCCGTGACTACGAGA CGGCCAAGAACTACTTCAAAAAAGGCCTGAAGGATATGGGCTATGGGAACTGGATTAGCAAACCCCAGGAGGAAAAGAACTTTTATCTCT GCCCAGTATAGTATGCTCCAGTGACAGATGGATTAGGGTGTGTCATACTAGGGTGTGAGAGAGGTAGGTCGTAGCATTCCTCATCACATG GTCAGGGGATTTTTTTTTCTCCTTTTTTTTTCTTTTTAAGCCATAATTGGTGATACTGAAAACTTTGGGTTCCCATTTATCCTGCTTTCT TTGGGATTGCTAGGCAAGGTCTGGCCAGGCCCCCCTTTTTTCCCCCAAGTGAAGAGGCAGAAACCTAAGAAGTTATCTTTTCTTTCTACC CAAAGCATACATAGTCACTGAGCACCTGCGGTCCATTTCCTCTTAAAAGTTTTGTTTTGATTTGTTTCCATTTCCTTTCCCTTTGTGTTT GCTACACTGACCTCTTGCGGTCTTGATTAGGTTTCAGTCAACTCTGGATCATGTCAGGGACTGATAATTTCATTTGTGGATTACGCAGAC CCCTCTACTTCCCCTCTTTCCCTTCTGAGATTCTTTCCTTGTGATCTGAATGTCTCCTTTTCCCCCTCAGAGGGCAAAGAGGTGAACATA AAGGATTTGGTGAAACATTTGTAAGGGTAGGAGTTGAAAACTGCAGTTCCCAGTGCCACGGAAGTGTGATTGGAGCCTGCAGATAATGCC CAGCCATCCTCCCATCCTGCACTTTAGCCAGCTGCAGGGCGGGCAAGGCAAGGAAAGCTGCTTCCCTGGAAGTGTATCACTTTCTCCGGC AGCTGGGAAGTCTAGAACCAGCCAGACTGGGTTAAGGGAGCTGCTCAAGCAATAGCAGAGGTTTCACCCGGCAGGATGACACAGACCACT TCCCAGGGAGCACGGGCATGCCTTGGAATATTGCCAAGCTTCCAGCTGCCTCTTCTCCTAAAGCATTCCTAGGAATATTTTCCCCGCCAA TGCTGGGCGTACACCCTAGCCAACGGGACAAATCCTAGAGGGTATAAAATCATCTCTGCTCAGATAATCATGACTTAGCAAGAATAAGGG CAAAAAATCCTGTTGGCTTAACGTCACTGTTCCACCCGGTGTAATATCTCTCATGACAGTGACACCAAGGGAAGTTGACTAAGTCACATG TAAATTAGGAGTGTTTTAAAGAATGCCATAGATGTTGATTCTTAACTGCTACAGATAACCTGTAATTGAGCAGATTTAAAATTCAGGCAT ACTTTTCCATTTATCCAAGTGCTTTCATTTTTCCAGATGGCTTCAGAAGTAGGCTCGTGGGCAGGGCGCAGACCTGATCTTTATAGGGTT GACATAGAAAGCAGTAGTTGTGGGTGAAAGGGCAGGTTGTCTTCAAACTCTGTGAGGTAGAATCCTTTGTCTATACCTCCATGAACATTG ACTCGTGTGTTCAGAGCCTTTGGCCTCTCTGTGGAGTCTGGCTCTCTGGCTCCTGTGCATTCTTTGAATAGTCACTCGTAAAAACTGTCA GTGCTTGAAACTGTTTCCTTTACTCATGTTGAAGGGACTTTGTTGGCTTTTAGAGTGTTGGTCATGACTCCAAGAGCAGAGCAGGGAAGA GCCCAAGCATAGACTTGGTGCCGTGGTGATGGCTGCAGTCCAGTTTTGTGATGCTGCTTTTACGTGTCCCTCGATAACAGTCAGCTAGAC ACACTCAGGAGGACTACTGAGGCTCTGCGACCTTCAGGAGCTGAGCCTGCCTCTCTCCTTTAGATGACAGACCTTCATCTGGGAACGTGC TGAGCCAGCACCCTCAGATGATTTCCCTCCAAACTGCTGACTAGGTCATCCTCTGTCTGGTAGAGACATTCACATCTTTGCTTTTATTCT ATGCTCTCTGTACTTTTGACCAAAAATTGACCAAAGTAAGAAAATGCAAGTTCTAAAAATAGACTAAGGATGCCTTTGCAGAACACCAAA GCATCCCAAGGAACTGGTAGGGAAGTGGCGCCTGTCTCCTGGAGTGGAAGAGGCCTGCTCCCTGGCTCTGGGTCTGCTGGGGGCACAGTA AATCAGTCTTGGCACCCACATCCAGGGCAGAGAGGTCTGTGGTTCTCAGCATCAGAAGGCAGCGCAGCCCCTCTCCTCTTCAGGCTACAG GGTTGTCACCTGCTGAGTCCTCAGGTTGTTTGGCCTCTCTGGTCCATCTTGGGCATTAGGTTCTCCAGCAGAGCTCTGGCCAGCTGCCTC TTCTTTAACTGGGAACACAGGCTCTCACAAGATCAGAACCCCCACTCACCCCCAAGATCTTATCTAGCAAGCCTGTAGTATTCAGTTTCT GTTGTAGGAAGAGAGCGAGGCATCCCTGAATTCCACGCATCTGCTGGAAACGAGCCGTGTCAGATCGCACATCCCTGCGCCCCCATGCCC CTCTGAGTCACACAGGACAGAGGAGGCAGAGCTTCTGCCCACTGTTATCTTCACTTTCTTTGTCCAGTCTTTTGTTTTTAATAAGCAGTG ACCCTCCCTACTCTTCTTTTTAATGATTTTTGTAGTTGATTTGTCTGAACTGTGGCTACTGTGCATTCCTTGAATAATCACTTGTAAAAA TTGTCAGTGCTTGAAGCTGTTTCCTTTACTCACATTGAAGGGACTTCGTTGGTTTTTTGGAGTCTTGGTTGTGACTCCAAGAGCAGAGTG AGGAAGACCCCCAAGCATAGACTCGGGTACTGTGATGATGGCTGCAGTCCAGTTTTATGATTCTGCTTTTATGTGTCCCTTGATAACAGT GACTTAACAATATACATTCCTCATAAATAAAAAAAAAACAAGAATCTGAAAAAAAAAAAAAAAACCGGAAATGGGTCCTACCATCTTCTC GGAGCCGGAGTGCGAAGAAATAAAGAAATAGTGCTTTAAGTCAATGAATTCCTCCTTGGGACCCACTATCGAGAAACTATCAGTGGTAAC GTTTTAAAAAATGACAAATTCAATCTGCTCTTGACTTGTGTGTCCTAAGATTTCCACTAAGTGTCTTCAAACCTCCCCCTCCCCGGCTTC CTGGATAATAGAAGTTCCCGAAGGCCGCCGATTCCAGAAGATACTGTCTGGCGTGAAATTAGTCTCAGTAGAAACATAAGTCCCGCGCGT CTTGTGCTGCGCGTGCGCAAGCTTTTGGGCCCTCCCGAGAAAGGGAAGTGCATTCTCGCTTCCGTAGCGGTCTCCGCCGGTTGGGGGGAA GTAATTCCGGTTGTTGCACCATGGCGTCCATGGGGACCCTCGCCTTCGATGAATATGGGCGCCCTTTCCTCATCATCAAGGATCAGGACC GCAAGTCCCGTCTTATGGGACTTGAGGCCCTCAAGTCTCATATAATGGCAGCAAAGGCTGTAGCAAATACAATGAGAACATCACTTGGAC CAAATGGGCTTGATAAGATGATGGTGGATAAGGATGGAGATGTGACTGTAACTAATGATGGGGCCACCATCTTAAGCATGATGGATGTTG ATCATCAGATTGCCAAGCTGATGGTGGAACTGTCCAAGTCTCAGGATGATGAAATTGGAGATGGAACCACAGGAGTGGTTGTCCTGGCTG GTGCCTTGTTAGAAGAAGCGGAGCAATTGCTAGACCGAGGCATTCACCCAATCAGAATAGCCGATGGCTATGAGCAGGCTGCTCGTGTTG CTATTGAACACCTGGACAAGATCAGCGATAGCGTCCTTGTTGACATAAAGGACACCGAACCCCTGATTCAGACAGCAAAAACCACGCTGG GCTCCAAAGTGGTCAACAGTTGTCACCGACAGATGGCTGAGATTGCTGTGAATGCCGTCCTCACTGTAGCAGATATGGAGCGGAGAGACG TTGACTTTGAGCTTATCAAAGTAGAAGGCAAAGTGGGCGGCAGGCTGGAGGACACTAAACTGATTAAGGGCGTGATTGTGGACAAGGATT TCAGTCACCCACAGATGCCAAAAAAAGTGGAAGATGCGAAGATTGCAATTCTCACATGTCCATTTGAACCACCCAAACCAAAAACAAAGC ATAAGCTGGATGTGACCTCTGTCGAAGATTATAAAGCCCTTCAGAAATACGAAAAGGAGAAATTTGAAGAGATGATTCAACAAATTAAAG AGACTGGTGCTAACCTAGCAATTTGTCAGTGGGGCTTTGATGATGAAGCAAATCACTTACTTCTTCAGAACAACTTGCCTGCGGTTCGCT GGGTAGGAGGACCTGAAATTGAGCTGATTGCCATCGCAACAGGAGGGCGGATCGTCCCCAGGTTCTCAGAGCTCACAGCCGAGAAGCTGG GCTTTGCTGGTCTTGTACAGGAGATCTCATTTGGGACAACTAAGGATAAAATGCTGGTCATCGAGCAGTGTAAGAACTCCAGAGCTGTAA CCATTTTTATTAGAGGAGGAAATAAGATGATCATTGAGGAGGCGAAACGATCCCTTCACGATGCTTTGTGTGTCATCCGGAACCTCATCC GCGATAATCGTGTGGTGTATGGAGGAGGGGCTGCTGAGATATCCTGTGCCCTGGCAGTTAGCCAAGAGGCGGATAAGTGCCCCACCTTAG AACAGTATGCCATGAGAGCGTTTGCCGACGCACTGGAGGTCATCCCCATGGCCCTCTCTGAAAACAGTGGCATGAATCCCATCCAGACTA TGACCGAAGTCCGAGCCAGACAGGTGAAGGAGATGAACCCTGCTCTTGGCATCGACTGTTTGCACAAGGGGACAAATGATATGAAGCAAC AGCATGTCATAGAAACCTTGATTGGCAAAAAGCAACAGATATCTCTTGCAACACAAATGGTTAGAATGATTTTGAAGATTGATGACATTC GTAAGCCTGGAGAATCTGAAGAATGAAGACATTGAGAAAACTATGTAGCAAGATCCACTTCTGTGATTAAGTAAATGGATGTCTCGTGAT GCATCTACAGTTATTTATTGTTACATCCTTTTCCAGACACTGTAGATGCTATAATAAAAATAGCTGTTTGGTAACCATAGTTTCACTTGT TCAAAGCTGTGTAATCGTGGGGGTACCATCTCAACTGCTTTTGTATTCATTGTATTAAAAGAATCTGTTTAAACAACCTTTATCTTCTCT TCGGGTTTAAGAAACGTTTATTGTAACAGTAATTAAATGCTGCCTTAATTGAAGGGGTTTGGGTGGATTTTTTTTTCTCAAAATAAGCTG TAGGGACTATTTTAACAGCTTAAACAGGAGCTCTCAAGATGCACTTTCATATTGAGAGGAATATGGGCTTGATCCTCTTCCTATCTAAAT GGGTGGGCCATTTGATTGTAGAGGGTCCACCACAGAATTATGGGATGCCTTAAGTGCTGTTACTAGGTTGCTCACAGCCTAACCTGGCGT GTTGTTTAGGGCTGATGGAGACCCATGTGAGCCTTTGCTTTCCTCTGGCCCCGGCCCCACCCTGAACACAGCTCATACACAGAATCAGGA CCAGCATGTGCAGAGCTGGCCACCAGCACAGGCTTAGGGCAGTTCAGAACCCACTTGTTTCCCTATCAGAGGGACACAGTGAAGTGGAGG TTAAAGTAAATTACAGGAAATAAGGGAGAAATCTTGCAGTTACCATGTTCAGATAGAGTGACTGAAATTAATTGTACTTACTAAAGTATT AACTAGCTAACAGTGATGGGCCAAGACGCTCCGAGAACTCTACCGGGATTGTCTGTTCTGACAACCCAGTGAGGCAGATACACTTTCTTA CTGCTCACATCTTACAGGTGAGTACTCATAATTGGCCAGCATCTCACCACCAGCAAGTAGTAGGGCCAGGTCAATCCCAGGCAGTCTGAC CCCAGAGTGGCCCAGCTCATCCCCTACTCTGTTATTTGCTTGTTAATGATTCTCTAGATTTTCTAAAATAATGTTTCTTAGCATTGTGAT GATAAAGCTCATGATGAACTTTATCACTAGTTATGCCACCTTAACTAGTCAGATTTCCTAGAATTAGGAAATGGTGACTCTTGTCTAAAT TTGGTTAAGTGATGAATTTGGGTTACCGTCTCATGTGAACCTGGAGATTCACCAGTCTTAACTTTTGGGTCATTGTGTTTTCTCTACATT CATGCATTGGATGTTTTGCTAAATAACTCCTGTGGATTTAGGAATGTGTGCTAATAGCAATCTTCCTAATTTTCATGTTTATATGGAACT ATGCAGTTGAGTATTGAAAGCTTTAAACTGAGTTTATTTACAAGGACTGAGTCTAGCCTACAGAGAACATACAGCAGCCTTCTTTGGACC ACAGTCTTATCCGAGGGGTCTGTGGTTGTATCAGAAGAGCCACTAAACCAATCCCCCTTTCCAAAATTGAACCTCACAGACGTTCCTGTT TTTTGTGATTGAGAAACTGGTCAATGAACAGGAAGACTTAGAGATGTTTCACAAGCCTTTGATTTTTGTTTCATTTATTTGTAATAAACC TCTTCCACGTGATGTCTTTTATTTTTCACGTTCCA >2174_2174_1_ADAR-CCT5_ADAR_chr1_154554538_ENST00000368474_CCT5_chr5_10250033_ENST00000280326_length(amino acids)=1250AA_BP= MARPSGRARVVPPAGRAGQCLAGAMNPRQGYSLSGYYTHPFQGYEHRQLRYQQPGPGSSPSSFLLKQIEFLKGQLPEAPVIGKQTPSLPP SLPGLRPRFPVLLASSTRGRQVDIRGVPRGVHLRSQGLQRGFQHPSPRGRSLPQRGVDCLSSHFQELSIYQDQEQRILKFLEELGEGKAT TAHDLSGKLGTPKKEINRVLYSLAKKGKLQKEAGTPPLWKIAVSTQAWNQHSGVVRPDGHSQGAPNSDPSLEPEDRNSTSVSEDLLEPFI AVSAQAWNQHSGVVRPDSHSQGSPNSDPGLEPEDSNSTSALEDPLEFLDMAEIKEKICDYLFNVSDSSALNLAKNIGLTKARDINAVLID MERQGDVYRQGTTPPIWHLTDKKRERMQIKRNTNSVPETAPAAIPETKRNAEFLTCNIPTSNASNNMVTTEKVENGQEPVIKLENRQEAR PEPARLKPPVHYNGPSKAGYVDFENGQWATDDIPDDLNSIRAAPGEFRAIMEMPSFYSHGLPRCSPYKKLTECQLKNPISGLLEYAQFAS QTCEFNMIEQSGPPHEPRFKFQVVINGREFPPAEAGSKKVAKQDAAMKAMTILLEEAKAKDSGKSEESSHYSTEKESEKTAESQTPTPSA TSFFSGKSPVTTLLECMHKLGNSCEFRLLSKEGPAHEPKFQYCVAVGAQTFPSVSAPSKKVAKQMAAEEAMKALHGEATNSMASDNQPEG MISESLDNLESMMPNKVRKIGELVRYLNTNPVGGLLEYARSHGFAAEFKLVDQSGPPHEPKFVYQAKVGGRWFPAVCAHSKKQGKQEAAD AALRVLIGENEKAERMGFTEVTPVTGASLRRTMLLLSRSPEAQPKTLPLTGSTFHDQIAMLSHRCFNTLTNSFQPSLLGRKILAAIIMKK DSEDMGVVVSLGTGNRCVKGDSLSLKGETVNDCHAEIISRRGFIRFLYSELMKYNSQTAKDSIFEPAKGGEKLQIKKTVSFHLYISTAPC GDGALFDKSCSDRAMESTESRHYPVFENPKQGKLRTKVENGEGTIPVESSDIVPTWDGIRLGERLRTMSCSDKILRWNVLGLQGALLTHF LQPIYLKSVTLGYLFSQGHLTRAICCRVTRDGSAFEDGLRHPFIVNHPKVGRVSIYDSKRQSGKTKETSVNWCLADGYDLEILDGTRGTV DGPRNELSRVSKKNIFLLFKKLCSFRYRRDLLRLSYGEAKKAARDYETAKNYFKKGLKDMGYGNWISKPQEEKNFYLCPV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ADAR-CCT5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368471 | - | 15 | 15 | 133_202 | 1845.0 | 932.0 | Z-DNA |
| Hgene | ADAR | chr1:154554538 | chr5:10250033 | ENST00000368474 | - | 15 | 15 | 133_202 | 2140.0 | 1227.0 | Z-DNA |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ADAR-CCT5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ADAR-CCT5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ADAR | C3539013 | AICARDI-GOUTIERES SYNDROME 6 | 6 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | ADAR | C0406775 | Symmetrical dyschromatosis of extremities | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ADAR | C0013421 | Dystonia | 3 | GENOMICS_ENGLAND |
| Hgene | ADAR | C0006142 | Malignant neoplasm of breast | 1 | CTD_human;UNIPROT |
| Hgene | ADAR | C0393591 | AICARDI-GOUTIERES SYNDROME | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | ADAR | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Hgene | ADAR | C0795996 | STRIATONIGRAL DEGENERATION, INFANTILE (disorder) | 1 | ORPHANET |
| Hgene | ADAR | C0796126 | AICARDI-GOUTIERES SYNDROME 1 | 1 | CTD_human |
| Hgene | ADAR | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Hgene | ADAR | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Hgene | ADAR | C3489724 | Aicardi-Goutieres Syndrome 2 | 1 | CTD_human |
| Hgene | ADAR | C3489725 | Pseudo-TORCH syndrome | 1 | CTD_human |
| Hgene | ADAR | C3860213 | Autoinflammatory disorder | 1 | GENOMICS_ENGLAND |
| Hgene | ADAR | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
| Tgene | C1850395 | Neuropathy, Hereditary Sensory, with Spastic Paraplegia, Autosomal Recessive | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0006142 | Malignant neoplasm of breast | 1 | CTD_human | |
| Tgene | C0019193 | Hepatitis, Toxic | 1 | CTD_human | |
| Tgene | C0019693 | HIV Infections | 1 | CTD_human | |
| Tgene | C0152013 | Adenocarcinoma of lung (disorder) | 1 | CTD_human | |
| Tgene | C0678222 | Breast Carcinoma | 1 | CTD_human | |
| Tgene | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human | |
| Tgene | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human | |
| Tgene | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human | |
| Tgene | C1458155 | Mammary Neoplasms | 1 | CTD_human | |
| Tgene | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human | |
| Tgene | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human | |
| Tgene | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human | |
| Tgene | C4505456 | HIV Coinfection | 1 | CTD_human | |
| Tgene | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
