
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:C12orf57-HABP4 (FusionGDB2 ID:HG113246TG22927) |
Fusion Gene Summary for C12orf57-HABP4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: C12orf57-HABP4 | Fusion gene ID: hg113246tg22927 | Hgene | Tgene | Gene symbol | C12orf57 | HABP4 | Gene ID | 113246 | 22927 |
| Gene name | chromosome 12 open reading frame 57 | hyaluronan binding protein 4 | |
| Synonyms | C10|GRCC10 | IHABP-4|IHABP4|Ki-1/57|SERBP1L | |
| Cytomap | ('C12orf57')('HABP4') 12p13.31 | 9q22.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein C10gene rich cluster C10likely ortholog of mouse gene rich cluster, C10 | intracellular hyaluronan-binding protein 4chromodomain helicase DNA binding protein 3 interacting proteinintracellular antigen detected by monoclonal antibody Ki-1ki-1/57 intracellular antigen | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | Q5JVS0 | |
| Ensembl transtripts involved in fusion gene | ENST00000229281, ENST00000537087, ENST00000544681, ENST00000540506, ENST00000542222, | ||
| Fusion gene scores | * DoF score | 4 X 2 X 2=16 | 8 X 8 X 5=320 |
| # samples | 4 | 9 | |
| ** MAII score | log2(4/16*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(9/320*10)=-1.83007499855769 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: C12orf57 [Title/Abstract] AND HABP4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | C12orf57(7053336)-HABP4(99220660), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | HABP4 | GO:0030578 | PML body organization | 28695742 |
| Tgene | HABP4 | GO:0043392 | negative regulation of DNA binding | 15862299 |
 Fusion gene breakpoints across C12orf57 (5'-gene) Fusion gene breakpoints across C12orf57 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
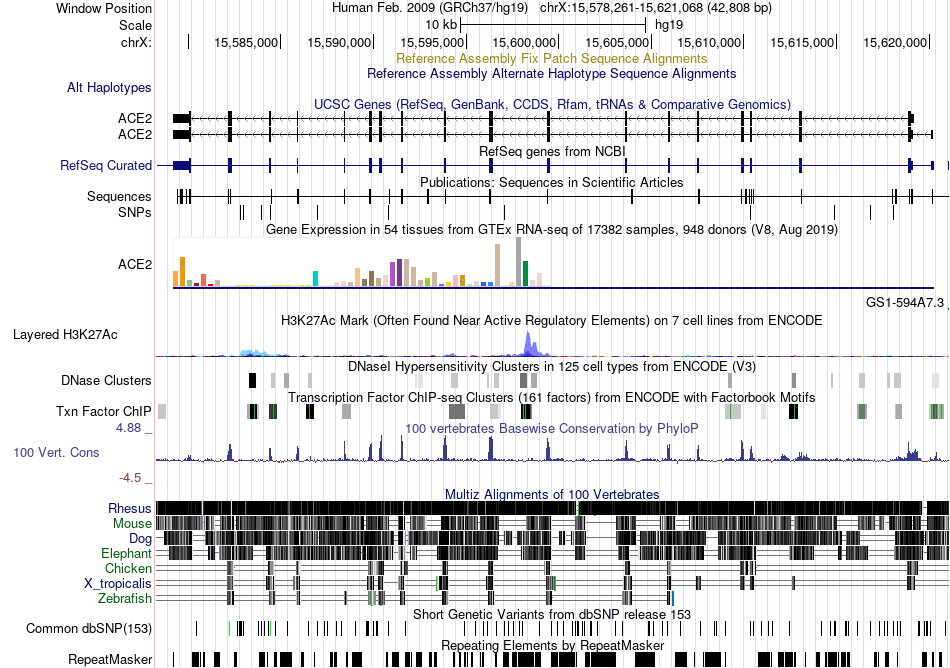 |
 Fusion gene breakpoints across HABP4 (3'-gene) Fusion gene breakpoints across HABP4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
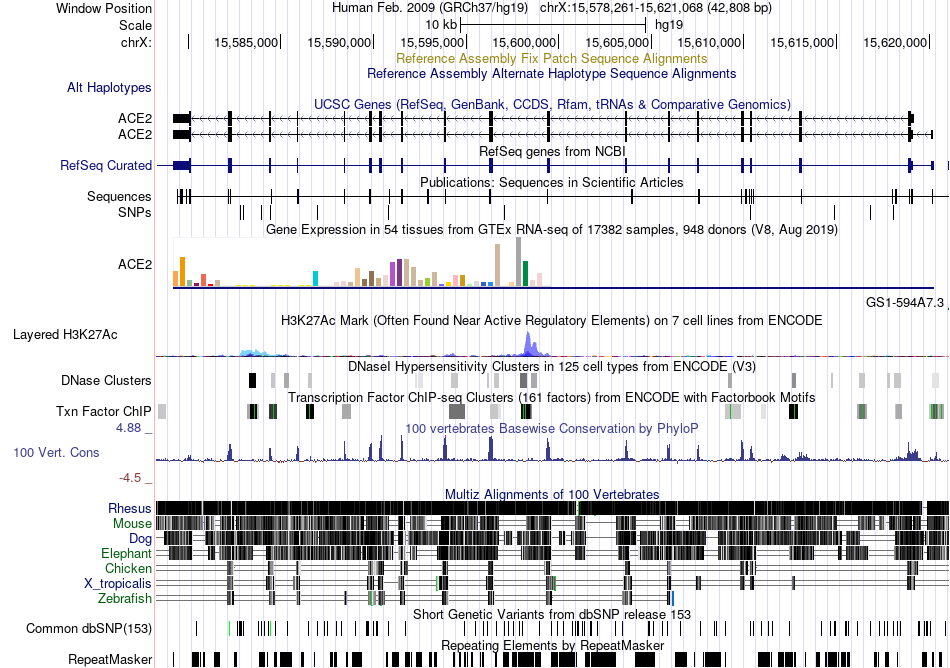 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 265N | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
Top |
Fusion Gene ORF analysis for C12orf57-HABP4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000229281 | ENST00000466976 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| 5CDS-intron | ENST00000537087 | ENST00000466976 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| 5CDS-intron | ENST00000544681 | ENST00000466976 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| 5UTR-3CDS | ENST00000540506 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| 5UTR-3CDS | ENST00000540506 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| 5UTR-intron | ENST00000540506 | ENST00000466976 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| In-frame | ENST00000229281 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| In-frame | ENST00000229281 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| In-frame | ENST00000537087 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| In-frame | ENST00000537087 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| In-frame | ENST00000544681 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| In-frame | ENST00000544681 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| intron-3CDS | ENST00000542222 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| intron-3CDS | ENST00000542222 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
| intron-intron | ENST00000542222 | ENST00000466976 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000544681 | C12orf57 | chr12 | 7053336 | + | ENST00000375251 | HABP4 | chr9 | 99220660 | + | 2233 | 357 | 305 | 934 | 209 |
| ENST00000544681 | C12orf57 | chr12 | 7053336 | + | ENST00000375249 | HABP4 | chr9 | 99220660 | + | 2548 | 357 | 305 | 1249 | 314 |
| ENST00000537087 | C12orf57 | chr12 | 7053336 | + | ENST00000375251 | HABP4 | chr9 | 99220660 | + | 2216 | 340 | 288 | 917 | 209 |
| ENST00000537087 | C12orf57 | chr12 | 7053336 | + | ENST00000375249 | HABP4 | chr9 | 99220660 | + | 2531 | 340 | 288 | 1232 | 314 |
| ENST00000229281 | C12orf57 | chr12 | 7053336 | + | ENST00000375251 | HABP4 | chr9 | 99220660 | + | 2027 | 151 | 99 | 728 | 209 |
| ENST00000229281 | C12orf57 | chr12 | 7053336 | + | ENST00000375249 | HABP4 | chr9 | 99220660 | + | 2342 | 151 | 99 | 1043 | 314 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000544681 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 0.002036907 | 0.9979631 |
| ENST00000544681 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 0.000666269 | 0.9993337 |
| ENST00000537087 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 0.001791002 | 0.998209 |
| ENST00000537087 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 0.000653198 | 0.9993468 |
| ENST00000229281 | ENST00000375251 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 0.001814168 | 0.9981858 |
| ENST00000229281 | ENST00000375249 | C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 0.000728143 | 0.9992718 |
Top |
Fusion Genomic Features for C12orf57-HABP4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 9.03E-06 | 0.99999094 |
| C12orf57 | chr12 | 7053336 | + | HABP4 | chr9 | 99220660 | + | 9.03E-06 | 0.99999094 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
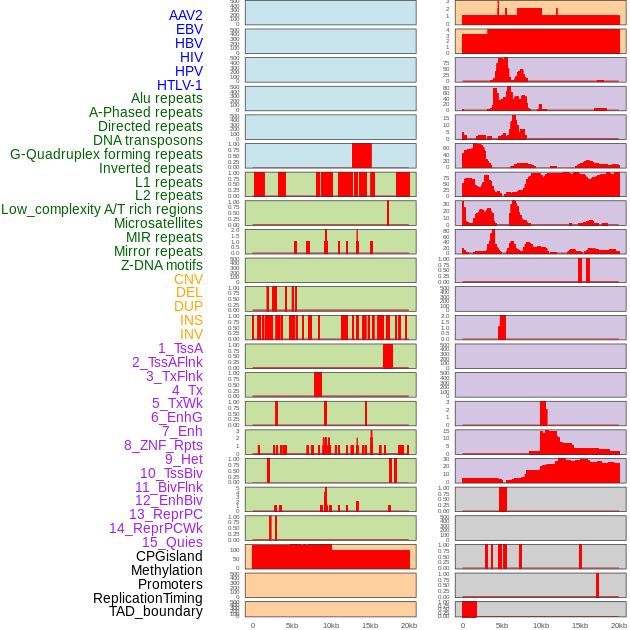 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
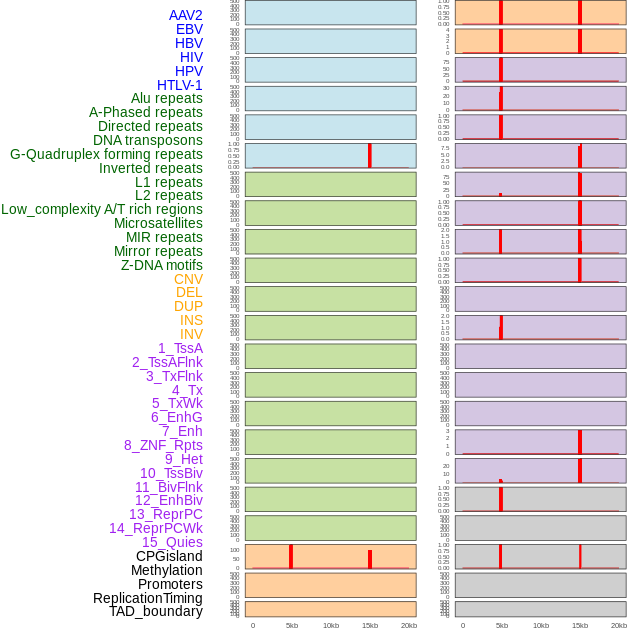 |
Top |
Fusion Protein Features for C12orf57-HABP4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:7053336/chr9:99220660) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | HABP4 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: RNA-binding protein that plays a role in the regulation of transcription, pre-mRNA splicing and mRNA translation (PubMed:14699138, PubMed:16455055, PubMed:19523114, PubMed:21771594). Negatively regulates DNA-binding activity of the transcription factor MEF2C in myocardial cells in response to mechanical stress (By similarity). Plays a role in pre-mRNA splicing regulation (PubMed:19523114). Binds (via C-terminus) to poly(U) RNA (PubMed:19523114). Involved in mRNA translation regulation, probably at the initiation step (PubMed:21771594). Seems to play a role in PML-nuclear bodies formation (PubMed:28695742). {ECO:0000250|UniProtKB:A1L1K8, ECO:0000269|PubMed:14699138, ECO:0000269|PubMed:16455055, ECO:0000269|PubMed:19523114, ECO:0000269|PubMed:21771594, ECO:0000269|PubMed:28695742}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | HABP4 | chr12:7053336 | chr9:99220660 | ENST00000375249 | 0 | 8 | 40_64 | 116 | 414.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | HABP4 | chr12:7053336 | chr9:99220660 | ENST00000375251 | 0 | 5 | 40_64 | 116 | 309.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for C12orf57-HABP4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >10865_10865_1_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000229281_HABP4_chr9_99220660_ENST00000375249_length(transcript)=2342nt_BP=151nt CTTTCCGCTCCCAGGGGCGTTGGGAACGGTTGTAGGACGTGGCTCTTTATTCGTGAGTTTTCCATTTACCTCCGCTGAACCTAGAGCTTC AGACGCCCTATGGCGTCCGCCTCGACCCAACCGGCGGCCTTGAGCGCTGAGCAAGCAAAGGGCCAGAAGCGGACTCCTAGAAGAGGGGAG CAGCAAGGATGGAATGACAGCCGTGGGCCGGAGGGGATGCTCGAAAGAGCTGAGCGGAGATCCTACAGGGAATACCGACCCTATGAGACA GAGAGGCAGGCAGACTTCACAGCTGAGAAGTTTCCAGATGAAAAACCAGGTGATAGGTTTGATCGAGACAGACCGTTGAGAGGACGTGGA GGCCCGAGAGGGGGTATGCGCGGCAGAGGCAGAGGTGGCCCTGGGAACAGAGTTTTTGACGCTTTTGACCAGAGAGGAAAGCGAGAATTT GAAAGATATGGTGGGAATGACAAAATAGCAGTCAGAACTGAAGACAACATGGGTGGATGTGGAGTTCGAACCTGGGGATCGGGTAAAGAT ACCAGTGATGTGGAGCCAACTGCACCGATGGAGGAACCCACAGTGGTGGAGGAGTCCCAGGGCACCCCGGAAGAGGAGTCTCCAGCCAAA GTTCCTGAGTTGGAGGTAGAAGAAGAAACCCAAGTTCAAGAGATGACTTTAGATGAGTGGAAAAATCTTCAAGAACAGACCAGACCAAAG CCTGAGTTTAACATCCGGAAACCAGAATCCACTGTTCCTTCCAAAGCCGTGGTGATTCACAAGTCAAAATACAGAGATGATATGGTAAAA GATGACTATGAGGACGATTCCCATGTTTTCCGGAAACCCGCCAATGACATCACATCCCAGCTGGAGATTAATTTTGGTAACCTCCCTCGT CCTGGGCGTGGAGCCAGAGGAGGCACCCGGGGAGGCCGGGGAAGGATCAGGAGGGCAGAGAACTATGGACCCAGAGCAGAAGTGGTGATG CAAGATGTTGCCCCCAACCCAGATGACCCGGAAGATTTCCCTGCGCTGTCTTGAAAGAGCCCTGTTTCCCAGCACCGCGGAGCTGCACTG CACACCTGTGGGGAGACTTTTCCAGCTGGGCCAAGGGAGTCAGACTCTAAGAACAATAGATGTTGCTTTTCCCGTGTCATGTAAATTTGT TGCACTTTTTTGGGCTGAGCTGTTAGAGGGGCTTCTCCAGAGGCTCGAGAGCAGGCCATTTCCCAAGAAGATGAAGAATGGTGACTGTGT TTTTATTGAAGGAATTTCAAATGAAGAATAATGTTTAAAATGTGTATATAGAGATAGTATAGACTCCTCCGCGGAAGCATGGAGGGAAAG GAGGTTGTAAAATAGACTCCATGGAGACTCTTAGGAAGCAGTAGATTCCCGGGGGCTGTGCCTTTAGCGTTAGAGGAAACACATAGAGCT GGAACTGTTAATGGAAAGCAGTCACAGCTGAGTTTTCGGAGACCAAGAAATTAAAATACAATTGCACTTACTGTTTACTTGTAAATGTTA TCTTCTCTCCTGGAATATTTTGAGTTCTGGTTTTCACTTAACCAATCATTTAAAAATCGCTATTGTGTAGCCACTGGCCACCACTCTGTG ACACAGAATCAGAGAAAGCGTTGATTTTTAATGGTGATTTAAACTACCTCGTGGTTTCTGTGTGTGTGCACACACACATCTAGTGTTTGG GTGGTTTTTAGGAAGAATATTAAGTATGTGACTTCAAAAACCATGTTATTTAACCTGCCAATCAAATGGAAAGGGATCATGGCAAAAGCA AATACCGCATCCTTTTCTTCCGCAGAACTAGAGTCAGAGTTCGGCAGCTGCGTACAAAGGCCTCTGCCCTCGGCTTGAGCAAAAGTTGTA AATAACTAGTATTTATTAAGCACTTATAAGCAGTTTTTCTCACTTAGTCCTGGCTCCAGTTCTAGAGTTCCTCTTTATTGCTTTTGGTGA AAGTTTGGGGTTGGGGGTACACCTCAGAGGTTCAGTAATTCACCTGGAGTCCCACCCTGAACAGAGCTGGACCCAGAGCCACACACGCCC GGGAACACACACTGTGCTGGGGAGGAAGTGGGCCTAGGAGGGCCTGCAGGTCCAGGCAGCTGTAGAGCTGCTAGAAGCTGGGGTGTTGCT CTCCCCGCTTTCTCATAGACACAGAGGTACTGTCTGCCTGTTGTCACACAGTTCATATGCTCGCTTGAGATGGAATCTGAACCTTGGTCC CAGGATCTGTGCTTTTTCCCACTTTGCCACACTGTCTAAGGTGGCTTTGAACTGGAACCCAAGTGCAAATAAAGGTTGGTATTCGCTCCT GA >10865_10865_1_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000229281_HABP4_chr9_99220660_ENST00000375249_length(amino acids)=314AA_BP=17 MASASTQPAALSAEQAKGQKRTPRRGEQQGWNDSRGPEGMLERAERRSYREYRPYETERQADFTAEKFPDEKPGDRFDRDRPLRGRGGPR GGMRGRGRGGPGNRVFDAFDQRGKREFERYGGNDKIAVRTEDNMGGCGVRTWGSGKDTSDVEPTAPMEEPTVVEESQGTPEEESPAKVPE LEVEEETQVQEMTLDEWKNLQEQTRPKPEFNIRKPESTVPSKAVVIHKSKYRDDMVKDDYEDDSHVFRKPANDITSQLEINFGNLPRPGR GARGGTRGGRGRIRRAENYGPRAEVVMQDVAPNPDDPEDFPALS -------------------------------------------------------------- >10865_10865_2_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000229281_HABP4_chr9_99220660_ENST00000375251_length(transcript)=2027nt_BP=151nt CTTTCCGCTCCCAGGGGCGTTGGGAACGGTTGTAGGACGTGGCTCTTTATTCGTGAGTTTTCCATTTACCTCCGCTGAACCTAGAGCTTC AGACGCCCTATGGCGTCCGCCTCGACCCAACCGGCGGCCTTGAGCGCTGAGCAAGCAAAGGGCCAGAAGCGGACTCCTAGAAGAGGGGAG CAGCAAGGATGGAATGACAGCCGTGGGCCGGAGGGGATGCTCGAAAGAGCTGAGCGGAGATCCTACAGGGAATACCGACCCTATGAGACA GAGAGGCAGGCAGACTTCACAGCTGAGAAGTTTCCAGATGAAAAAGTTCCTGAGTTGGAGGTAGAAGAAGAAACCCAAGTTCAAGAGATG ACTTTAGATGAGTGGAAAAATCTTCAAGAACAGACCAGACCAAAGCCTGAGTTTAACATCCGGAAACCAGAATCCACTGTTCCTTCCAAA GCCGTGGTGATTCACAAGTCAAAATACAGAGATGATATGGTAAAAGATGACTATGAGGACGATTCCCATGTTTTCCGGAAACCCGCCAAT GACATCACATCCCAGCTGGAGATTAATTTTGGTAACCTCCCTCGTCCTGGGCGTGGAGCCAGAGGAGGCACCCGGGGAGGCCGGGGAAGG ATCAGGAGGGCAGAGAACTATGGACCCAGAGCAGAAGTGGTGATGCAAGATGTTGCCCCCAACCCAGATGACCCGGAAGATTTCCCTGCG CTGTCTTGAAAGAGCCCTGTTTCCCAGCACCGCGGAGCTGCACTGCACACCTGTGGGGAGACTTTTCCAGCTGGGCCAAGGGAGTCAGAC TCTAAGAACAATAGATGTTGCTTTTCCCGTGTCATGTAAATTTGTTGCACTTTTTTGGGCTGAGCTGTTAGAGGGGCTTCTCCAGAGGCT CGAGAGCAGGCCATTTCCCAAGAAGATGAAGAATGGTGACTGTGTTTTTATTGAAGGAATTTCAAATGAAGAATAATGTTTAAAATGTGT ATATAGAGATAGTATAGACTCCTCCGCGGAAGCATGGAGGGAAAGGAGGTTGTAAAATAGACTCCATGGAGACTCTTAGGAAGCAGTAGA TTCCCGGGGGCTGTGCCTTTAGCGTTAGAGGAAACACATAGAGCTGGAACTGTTAATGGAAAGCAGTCACAGCTGAGTTTTCGGAGACCA AGAAATTAAAATACAATTGCACTTACTGTTTACTTGTAAATGTTATCTTCTCTCCTGGAATATTTTGAGTTCTGGTTTTCACTTAACCAA TCATTTAAAAATCGCTATTGTGTAGCCACTGGCCACCACTCTGTGACACAGAATCAGAGAAAGCGTTGATTTTTAATGGTGATTTAAACT ACCTCGTGGTTTCTGTGTGTGTGCACACACACATCTAGTGTTTGGGTGGTTTTTAGGAAGAATATTAAGTATGTGACTTCAAAAACCATG TTATTTAACCTGCCAATCAAATGGAAAGGGATCATGGCAAAAGCAAATACCGCATCCTTTTCTTCCGCAGAACTAGAGTCAGAGTTCGGC AGCTGCGTACAAAGGCCTCTGCCCTCGGCTTGAGCAAAAGTTGTAAATAACTAGTATTTATTAAGCACTTATAAGCAGTTTTTCTCACTT AGTCCTGGCTCCAGTTCTAGAGTTCCTCTTTATTGCTTTTGGTGAAAGTTTGGGGTTGGGGGTACACCTCAGAGGTTCAGTAATTCACCT GGAGTCCCACCCTGAACAGAGCTGGACCCAGAGCCACACACGCCCGGGAACACACACTGTGCTGGGGAGGAAGTGGGCCTAGGAGGGCCT GCAGGTCCAGGCAGCTGTAGAGCTGCTAGAAGCTGGGGTGTTGCTCTCCCCGCTTTCTCATAGACACAGAGGTACTGTCTGCCTGTTGTC ACACAGTTCATATGCTCGCTTGAGATGGAATCTGAACCTTGGTCCCAGGATCTGTGCTTTTTCCCACTTTGCCACACTGTCTAAGGTGGC TTTGAACTGGAACCCAAGTGCAAATAAAGGTTGGTATTCGCTCCTGA >10865_10865_2_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000229281_HABP4_chr9_99220660_ENST00000375251_length(amino acids)=209AA_BP=17 MASASTQPAALSAEQAKGQKRTPRRGEQQGWNDSRGPEGMLERAERRSYREYRPYETERQADFTAEKFPDEKVPELEVEEETQVQEMTLD EWKNLQEQTRPKPEFNIRKPESTVPSKAVVIHKSKYRDDMVKDDYEDDSHVFRKPANDITSQLEINFGNLPRPGRGARGGTRGGRGRIRR AENYGPRAEVVMQDVAPNPDDPEDFPALS -------------------------------------------------------------- >10865_10865_3_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000537087_HABP4_chr9_99220660_ENST00000375249_length(transcript)=2531nt_BP=340nt AGAATTTGTCTAGTAGGCTTTCTGGCTTTTTACCGGAAAGCCCCTCTTATGATGTTTGTTGCCAATGATAGATTGTTTTCACTGTGCAAA AATTATGGGTAGTTTTGGTGGTCTTGATGCAGTTGTAAGCTTGGGGTATGAAGGTTTGGGCCACGCCTGGGCGCTTCCGGCTGCGCCGGA TGCTGTTTCCTTTCCGCTCCCAGGGGCGTTGGGAACGGTTGTAGGACGTGGCTCTTTATTCGTGAGTTTTCCATTTACCTCCGCTGAACC TAGAGCTTCAGACGCCCTATGGCGTCCGCCTCGACCCAACCGGCGGCCTTGAGCGCTGAGCAAGCAAAGGGCCAGAAGCGGACTCCTAGA AGAGGGGAGCAGCAAGGATGGAATGACAGCCGTGGGCCGGAGGGGATGCTCGAAAGAGCTGAGCGGAGATCCTACAGGGAATACCGACCC TATGAGACAGAGAGGCAGGCAGACTTCACAGCTGAGAAGTTTCCAGATGAAAAACCAGGTGATAGGTTTGATCGAGACAGACCGTTGAGA GGACGTGGAGGCCCGAGAGGGGGTATGCGCGGCAGAGGCAGAGGTGGCCCTGGGAACAGAGTTTTTGACGCTTTTGACCAGAGAGGAAAG CGAGAATTTGAAAGATATGGTGGGAATGACAAAATAGCAGTCAGAACTGAAGACAACATGGGTGGATGTGGAGTTCGAACCTGGGGATCG GGTAAAGATACCAGTGATGTGGAGCCAACTGCACCGATGGAGGAACCCACAGTGGTGGAGGAGTCCCAGGGCACCCCGGAAGAGGAGTCT CCAGCCAAAGTTCCTGAGTTGGAGGTAGAAGAAGAAACCCAAGTTCAAGAGATGACTTTAGATGAGTGGAAAAATCTTCAAGAACAGACC AGACCAAAGCCTGAGTTTAACATCCGGAAACCAGAATCCACTGTTCCTTCCAAAGCCGTGGTGATTCACAAGTCAAAATACAGAGATGAT ATGGTAAAAGATGACTATGAGGACGATTCCCATGTTTTCCGGAAACCCGCCAATGACATCACATCCCAGCTGGAGATTAATTTTGGTAAC CTCCCTCGTCCTGGGCGTGGAGCCAGAGGAGGCACCCGGGGAGGCCGGGGAAGGATCAGGAGGGCAGAGAACTATGGACCCAGAGCAGAA GTGGTGATGCAAGATGTTGCCCCCAACCCAGATGACCCGGAAGATTTCCCTGCGCTGTCTTGAAAGAGCCCTGTTTCCCAGCACCGCGGA GCTGCACTGCACACCTGTGGGGAGACTTTTCCAGCTGGGCCAAGGGAGTCAGACTCTAAGAACAATAGATGTTGCTTTTCCCGTGTCATG TAAATTTGTTGCACTTTTTTGGGCTGAGCTGTTAGAGGGGCTTCTCCAGAGGCTCGAGAGCAGGCCATTTCCCAAGAAGATGAAGAATGG TGACTGTGTTTTTATTGAAGGAATTTCAAATGAAGAATAATGTTTAAAATGTGTATATAGAGATAGTATAGACTCCTCCGCGGAAGCATG GAGGGAAAGGAGGTTGTAAAATAGACTCCATGGAGACTCTTAGGAAGCAGTAGATTCCCGGGGGCTGTGCCTTTAGCGTTAGAGGAAACA CATAGAGCTGGAACTGTTAATGGAAAGCAGTCACAGCTGAGTTTTCGGAGACCAAGAAATTAAAATACAATTGCACTTACTGTTTACTTG TAAATGTTATCTTCTCTCCTGGAATATTTTGAGTTCTGGTTTTCACTTAACCAATCATTTAAAAATCGCTATTGTGTAGCCACTGGCCAC CACTCTGTGACACAGAATCAGAGAAAGCGTTGATTTTTAATGGTGATTTAAACTACCTCGTGGTTTCTGTGTGTGTGCACACACACATCT AGTGTTTGGGTGGTTTTTAGGAAGAATATTAAGTATGTGACTTCAAAAACCATGTTATTTAACCTGCCAATCAAATGGAAAGGGATCATG GCAAAAGCAAATACCGCATCCTTTTCTTCCGCAGAACTAGAGTCAGAGTTCGGCAGCTGCGTACAAAGGCCTCTGCCCTCGGCTTGAGCA AAAGTTGTAAATAACTAGTATTTATTAAGCACTTATAAGCAGTTTTTCTCACTTAGTCCTGGCTCCAGTTCTAGAGTTCCTCTTTATTGC TTTTGGTGAAAGTTTGGGGTTGGGGGTACACCTCAGAGGTTCAGTAATTCACCTGGAGTCCCACCCTGAACAGAGCTGGACCCAGAGCCA CACACGCCCGGGAACACACACTGTGCTGGGGAGGAAGTGGGCCTAGGAGGGCCTGCAGGTCCAGGCAGCTGTAGAGCTGCTAGAAGCTGG GGTGTTGCTCTCCCCGCTTTCTCATAGACACAGAGGTACTGTCTGCCTGTTGTCACACAGTTCATATGCTCGCTTGAGATGGAATCTGAA CCTTGGTCCCAGGATCTGTGCTTTTTCCCACTTTGCCACACTGTCTAAGGTGGCTTTGAACTGGAACCCAAGTGCAAATAAAGGTTGGTA TTCGCTCCTGA >10865_10865_3_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000537087_HABP4_chr9_99220660_ENST00000375249_length(amino acids)=314AA_BP=17 MASASTQPAALSAEQAKGQKRTPRRGEQQGWNDSRGPEGMLERAERRSYREYRPYETERQADFTAEKFPDEKPGDRFDRDRPLRGRGGPR GGMRGRGRGGPGNRVFDAFDQRGKREFERYGGNDKIAVRTEDNMGGCGVRTWGSGKDTSDVEPTAPMEEPTVVEESQGTPEEESPAKVPE LEVEEETQVQEMTLDEWKNLQEQTRPKPEFNIRKPESTVPSKAVVIHKSKYRDDMVKDDYEDDSHVFRKPANDITSQLEINFGNLPRPGR GARGGTRGGRGRIRRAENYGPRAEVVMQDVAPNPDDPEDFPALS -------------------------------------------------------------- >10865_10865_4_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000537087_HABP4_chr9_99220660_ENST00000375251_length(transcript)=2216nt_BP=340nt AGAATTTGTCTAGTAGGCTTTCTGGCTTTTTACCGGAAAGCCCCTCTTATGATGTTTGTTGCCAATGATAGATTGTTTTCACTGTGCAAA AATTATGGGTAGTTTTGGTGGTCTTGATGCAGTTGTAAGCTTGGGGTATGAAGGTTTGGGCCACGCCTGGGCGCTTCCGGCTGCGCCGGA TGCTGTTTCCTTTCCGCTCCCAGGGGCGTTGGGAACGGTTGTAGGACGTGGCTCTTTATTCGTGAGTTTTCCATTTACCTCCGCTGAACC TAGAGCTTCAGACGCCCTATGGCGTCCGCCTCGACCCAACCGGCGGCCTTGAGCGCTGAGCAAGCAAAGGGCCAGAAGCGGACTCCTAGA AGAGGGGAGCAGCAAGGATGGAATGACAGCCGTGGGCCGGAGGGGATGCTCGAAAGAGCTGAGCGGAGATCCTACAGGGAATACCGACCC TATGAGACAGAGAGGCAGGCAGACTTCACAGCTGAGAAGTTTCCAGATGAAAAAGTTCCTGAGTTGGAGGTAGAAGAAGAAACCCAAGTT CAAGAGATGACTTTAGATGAGTGGAAAAATCTTCAAGAACAGACCAGACCAAAGCCTGAGTTTAACATCCGGAAACCAGAATCCACTGTT CCTTCCAAAGCCGTGGTGATTCACAAGTCAAAATACAGAGATGATATGGTAAAAGATGACTATGAGGACGATTCCCATGTTTTCCGGAAA CCCGCCAATGACATCACATCCCAGCTGGAGATTAATTTTGGTAACCTCCCTCGTCCTGGGCGTGGAGCCAGAGGAGGCACCCGGGGAGGC CGGGGAAGGATCAGGAGGGCAGAGAACTATGGACCCAGAGCAGAAGTGGTGATGCAAGATGTTGCCCCCAACCCAGATGACCCGGAAGAT TTCCCTGCGCTGTCTTGAAAGAGCCCTGTTTCCCAGCACCGCGGAGCTGCACTGCACACCTGTGGGGAGACTTTTCCAGCTGGGCCAAGG GAGTCAGACTCTAAGAACAATAGATGTTGCTTTTCCCGTGTCATGTAAATTTGTTGCACTTTTTTGGGCTGAGCTGTTAGAGGGGCTTCT CCAGAGGCTCGAGAGCAGGCCATTTCCCAAGAAGATGAAGAATGGTGACTGTGTTTTTATTGAAGGAATTTCAAATGAAGAATAATGTTT AAAATGTGTATATAGAGATAGTATAGACTCCTCCGCGGAAGCATGGAGGGAAAGGAGGTTGTAAAATAGACTCCATGGAGACTCTTAGGA AGCAGTAGATTCCCGGGGGCTGTGCCTTTAGCGTTAGAGGAAACACATAGAGCTGGAACTGTTAATGGAAAGCAGTCACAGCTGAGTTTT CGGAGACCAAGAAATTAAAATACAATTGCACTTACTGTTTACTTGTAAATGTTATCTTCTCTCCTGGAATATTTTGAGTTCTGGTTTTCA CTTAACCAATCATTTAAAAATCGCTATTGTGTAGCCACTGGCCACCACTCTGTGACACAGAATCAGAGAAAGCGTTGATTTTTAATGGTG ATTTAAACTACCTCGTGGTTTCTGTGTGTGTGCACACACACATCTAGTGTTTGGGTGGTTTTTAGGAAGAATATTAAGTATGTGACTTCA AAAACCATGTTATTTAACCTGCCAATCAAATGGAAAGGGATCATGGCAAAAGCAAATACCGCATCCTTTTCTTCCGCAGAACTAGAGTCA GAGTTCGGCAGCTGCGTACAAAGGCCTCTGCCCTCGGCTTGAGCAAAAGTTGTAAATAACTAGTATTTATTAAGCACTTATAAGCAGTTT TTCTCACTTAGTCCTGGCTCCAGTTCTAGAGTTCCTCTTTATTGCTTTTGGTGAAAGTTTGGGGTTGGGGGTACACCTCAGAGGTTCAGT AATTCACCTGGAGTCCCACCCTGAACAGAGCTGGACCCAGAGCCACACACGCCCGGGAACACACACTGTGCTGGGGAGGAAGTGGGCCTA GGAGGGCCTGCAGGTCCAGGCAGCTGTAGAGCTGCTAGAAGCTGGGGTGTTGCTCTCCCCGCTTTCTCATAGACACAGAGGTACTGTCTG CCTGTTGTCACACAGTTCATATGCTCGCTTGAGATGGAATCTGAACCTTGGTCCCAGGATCTGTGCTTTTTCCCACTTTGCCACACTGTC TAAGGTGGCTTTGAACTGGAACCCAAGTGCAAATAAAGGTTGGTATTCGCTCCTGA >10865_10865_4_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000537087_HABP4_chr9_99220660_ENST00000375251_length(amino acids)=209AA_BP=17 MASASTQPAALSAEQAKGQKRTPRRGEQQGWNDSRGPEGMLERAERRSYREYRPYETERQADFTAEKFPDEKVPELEVEEETQVQEMTLD EWKNLQEQTRPKPEFNIRKPESTVPSKAVVIHKSKYRDDMVKDDYEDDSHVFRKPANDITSQLEINFGNLPRPGRGARGGTRGGRGRIRR AENYGPRAEVVMQDVAPNPDDPEDFPALS -------------------------------------------------------------- >10865_10865_5_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000544681_HABP4_chr9_99220660_ENST00000375249_length(transcript)=2548nt_BP=357nt AGTGTTACAGCTCTTTTAGAATTTGTCTAGTAGGCTTTCTGGCTTTTTACCGGAAAGCCCCTCTTATGATGTTTGTTGCCAATGATAGAT TGTTTTCACTGTGCAAAAATTATGGGTAGTTTTGGTGGTCTTGATGCAGTTGTAAGCTTGGGGTATGAAGGTTTGGGCCACGCCTGGGCG CTTCCGGCTGCGCCGGATGCTGTTTCCTTTCCGCTCCCAGGGGCGTTGGGAACGGTTGTAGGACGTGGCTCTTTATTCGTGAGTTTTCCA TTTACCTCCGCTGAACCTAGAGCTTCAGACGCCCTATGGCGTCCGCCTCGACCCAACCGGCGGCCTTGAGCGCTGAGCAAGCAAAGGGCC AGAAGCGGACTCCTAGAAGAGGGGAGCAGCAAGGATGGAATGACAGCCGTGGGCCGGAGGGGATGCTCGAAAGAGCTGAGCGGAGATCCT ACAGGGAATACCGACCCTATGAGACAGAGAGGCAGGCAGACTTCACAGCTGAGAAGTTTCCAGATGAAAAACCAGGTGATAGGTTTGATC GAGACAGACCGTTGAGAGGACGTGGAGGCCCGAGAGGGGGTATGCGCGGCAGAGGCAGAGGTGGCCCTGGGAACAGAGTTTTTGACGCTT TTGACCAGAGAGGAAAGCGAGAATTTGAAAGATATGGTGGGAATGACAAAATAGCAGTCAGAACTGAAGACAACATGGGTGGATGTGGAG TTCGAACCTGGGGATCGGGTAAAGATACCAGTGATGTGGAGCCAACTGCACCGATGGAGGAACCCACAGTGGTGGAGGAGTCCCAGGGCA CCCCGGAAGAGGAGTCTCCAGCCAAAGTTCCTGAGTTGGAGGTAGAAGAAGAAACCCAAGTTCAAGAGATGACTTTAGATGAGTGGAAAA ATCTTCAAGAACAGACCAGACCAAAGCCTGAGTTTAACATCCGGAAACCAGAATCCACTGTTCCTTCCAAAGCCGTGGTGATTCACAAGT CAAAATACAGAGATGATATGGTAAAAGATGACTATGAGGACGATTCCCATGTTTTCCGGAAACCCGCCAATGACATCACATCCCAGCTGG AGATTAATTTTGGTAACCTCCCTCGTCCTGGGCGTGGAGCCAGAGGAGGCACCCGGGGAGGCCGGGGAAGGATCAGGAGGGCAGAGAACT ATGGACCCAGAGCAGAAGTGGTGATGCAAGATGTTGCCCCCAACCCAGATGACCCGGAAGATTTCCCTGCGCTGTCTTGAAAGAGCCCTG TTTCCCAGCACCGCGGAGCTGCACTGCACACCTGTGGGGAGACTTTTCCAGCTGGGCCAAGGGAGTCAGACTCTAAGAACAATAGATGTT GCTTTTCCCGTGTCATGTAAATTTGTTGCACTTTTTTGGGCTGAGCTGTTAGAGGGGCTTCTCCAGAGGCTCGAGAGCAGGCCATTTCCC AAGAAGATGAAGAATGGTGACTGTGTTTTTATTGAAGGAATTTCAAATGAAGAATAATGTTTAAAATGTGTATATAGAGATAGTATAGAC TCCTCCGCGGAAGCATGGAGGGAAAGGAGGTTGTAAAATAGACTCCATGGAGACTCTTAGGAAGCAGTAGATTCCCGGGGGCTGTGCCTT TAGCGTTAGAGGAAACACATAGAGCTGGAACTGTTAATGGAAAGCAGTCACAGCTGAGTTTTCGGAGACCAAGAAATTAAAATACAATTG CACTTACTGTTTACTTGTAAATGTTATCTTCTCTCCTGGAATATTTTGAGTTCTGGTTTTCACTTAACCAATCATTTAAAAATCGCTATT GTGTAGCCACTGGCCACCACTCTGTGACACAGAATCAGAGAAAGCGTTGATTTTTAATGGTGATTTAAACTACCTCGTGGTTTCTGTGTG TGTGCACACACACATCTAGTGTTTGGGTGGTTTTTAGGAAGAATATTAAGTATGTGACTTCAAAAACCATGTTATTTAACCTGCCAATCA AATGGAAAGGGATCATGGCAAAAGCAAATACCGCATCCTTTTCTTCCGCAGAACTAGAGTCAGAGTTCGGCAGCTGCGTACAAAGGCCTC TGCCCTCGGCTTGAGCAAAAGTTGTAAATAACTAGTATTTATTAAGCACTTATAAGCAGTTTTTCTCACTTAGTCCTGGCTCCAGTTCTA GAGTTCCTCTTTATTGCTTTTGGTGAAAGTTTGGGGTTGGGGGTACACCTCAGAGGTTCAGTAATTCACCTGGAGTCCCACCCTGAACAG AGCTGGACCCAGAGCCACACACGCCCGGGAACACACACTGTGCTGGGGAGGAAGTGGGCCTAGGAGGGCCTGCAGGTCCAGGCAGCTGTA GAGCTGCTAGAAGCTGGGGTGTTGCTCTCCCCGCTTTCTCATAGACACAGAGGTACTGTCTGCCTGTTGTCACACAGTTCATATGCTCGC TTGAGATGGAATCTGAACCTTGGTCCCAGGATCTGTGCTTTTTCCCACTTTGCCACACTGTCTAAGGTGGCTTTGAACTGGAACCCAAGT GCAAATAAAGGTTGGTATTCGCTCCTGA >10865_10865_5_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000544681_HABP4_chr9_99220660_ENST00000375249_length(amino acids)=314AA_BP=17 MASASTQPAALSAEQAKGQKRTPRRGEQQGWNDSRGPEGMLERAERRSYREYRPYETERQADFTAEKFPDEKPGDRFDRDRPLRGRGGPR GGMRGRGRGGPGNRVFDAFDQRGKREFERYGGNDKIAVRTEDNMGGCGVRTWGSGKDTSDVEPTAPMEEPTVVEESQGTPEEESPAKVPE LEVEEETQVQEMTLDEWKNLQEQTRPKPEFNIRKPESTVPSKAVVIHKSKYRDDMVKDDYEDDSHVFRKPANDITSQLEINFGNLPRPGR GARGGTRGGRGRIRRAENYGPRAEVVMQDVAPNPDDPEDFPALS -------------------------------------------------------------- >10865_10865_6_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000544681_HABP4_chr9_99220660_ENST00000375251_length(transcript)=2233nt_BP=357nt AGTGTTACAGCTCTTTTAGAATTTGTCTAGTAGGCTTTCTGGCTTTTTACCGGAAAGCCCCTCTTATGATGTTTGTTGCCAATGATAGAT TGTTTTCACTGTGCAAAAATTATGGGTAGTTTTGGTGGTCTTGATGCAGTTGTAAGCTTGGGGTATGAAGGTTTGGGCCACGCCTGGGCG CTTCCGGCTGCGCCGGATGCTGTTTCCTTTCCGCTCCCAGGGGCGTTGGGAACGGTTGTAGGACGTGGCTCTTTATTCGTGAGTTTTCCA TTTACCTCCGCTGAACCTAGAGCTTCAGACGCCCTATGGCGTCCGCCTCGACCCAACCGGCGGCCTTGAGCGCTGAGCAAGCAAAGGGCC AGAAGCGGACTCCTAGAAGAGGGGAGCAGCAAGGATGGAATGACAGCCGTGGGCCGGAGGGGATGCTCGAAAGAGCTGAGCGGAGATCCT ACAGGGAATACCGACCCTATGAGACAGAGAGGCAGGCAGACTTCACAGCTGAGAAGTTTCCAGATGAAAAAGTTCCTGAGTTGGAGGTAG AAGAAGAAACCCAAGTTCAAGAGATGACTTTAGATGAGTGGAAAAATCTTCAAGAACAGACCAGACCAAAGCCTGAGTTTAACATCCGGA AACCAGAATCCACTGTTCCTTCCAAAGCCGTGGTGATTCACAAGTCAAAATACAGAGATGATATGGTAAAAGATGACTATGAGGACGATT CCCATGTTTTCCGGAAACCCGCCAATGACATCACATCCCAGCTGGAGATTAATTTTGGTAACCTCCCTCGTCCTGGGCGTGGAGCCAGAG GAGGCACCCGGGGAGGCCGGGGAAGGATCAGGAGGGCAGAGAACTATGGACCCAGAGCAGAAGTGGTGATGCAAGATGTTGCCCCCAACC CAGATGACCCGGAAGATTTCCCTGCGCTGTCTTGAAAGAGCCCTGTTTCCCAGCACCGCGGAGCTGCACTGCACACCTGTGGGGAGACTT TTCCAGCTGGGCCAAGGGAGTCAGACTCTAAGAACAATAGATGTTGCTTTTCCCGTGTCATGTAAATTTGTTGCACTTTTTTGGGCTGAG CTGTTAGAGGGGCTTCTCCAGAGGCTCGAGAGCAGGCCATTTCCCAAGAAGATGAAGAATGGTGACTGTGTTTTTATTGAAGGAATTTCA AATGAAGAATAATGTTTAAAATGTGTATATAGAGATAGTATAGACTCCTCCGCGGAAGCATGGAGGGAAAGGAGGTTGTAAAATAGACTC CATGGAGACTCTTAGGAAGCAGTAGATTCCCGGGGGCTGTGCCTTTAGCGTTAGAGGAAACACATAGAGCTGGAACTGTTAATGGAAAGC AGTCACAGCTGAGTTTTCGGAGACCAAGAAATTAAAATACAATTGCACTTACTGTTTACTTGTAAATGTTATCTTCTCTCCTGGAATATT TTGAGTTCTGGTTTTCACTTAACCAATCATTTAAAAATCGCTATTGTGTAGCCACTGGCCACCACTCTGTGACACAGAATCAGAGAAAGC GTTGATTTTTAATGGTGATTTAAACTACCTCGTGGTTTCTGTGTGTGTGCACACACACATCTAGTGTTTGGGTGGTTTTTAGGAAGAATA TTAAGTATGTGACTTCAAAAACCATGTTATTTAACCTGCCAATCAAATGGAAAGGGATCATGGCAAAAGCAAATACCGCATCCTTTTCTT CCGCAGAACTAGAGTCAGAGTTCGGCAGCTGCGTACAAAGGCCTCTGCCCTCGGCTTGAGCAAAAGTTGTAAATAACTAGTATTTATTAA GCACTTATAAGCAGTTTTTCTCACTTAGTCCTGGCTCCAGTTCTAGAGTTCCTCTTTATTGCTTTTGGTGAAAGTTTGGGGTTGGGGGTA CACCTCAGAGGTTCAGTAATTCACCTGGAGTCCCACCCTGAACAGAGCTGGACCCAGAGCCACACACGCCCGGGAACACACACTGTGCTG GGGAGGAAGTGGGCCTAGGAGGGCCTGCAGGTCCAGGCAGCTGTAGAGCTGCTAGAAGCTGGGGTGTTGCTCTCCCCGCTTTCTCATAGA CACAGAGGTACTGTCTGCCTGTTGTCACACAGTTCATATGCTCGCTTGAGATGGAATCTGAACCTTGGTCCCAGGATCTGTGCTTTTTCC CACTTTGCCACACTGTCTAAGGTGGCTTTGAACTGGAACCCAAGTGCAAATAAAGGTTGGTATTCGCTCCTGA >10865_10865_6_C12orf57-HABP4_C12orf57_chr12_7053336_ENST00000544681_HABP4_chr9_99220660_ENST00000375251_length(amino acids)=209AA_BP=17 MASASTQPAALSAEQAKGQKRTPRRGEQQGWNDSRGPEGMLERAERRSYREYRPYETERQADFTAEKFPDEKVPELEVEEETQVQEMTLD EWKNLQEQTRPKPEFNIRKPESTVPSKAVVIHKSKYRDDMVKDDYEDDSHVFRKPANDITSQLEINFGNLPRPGRGARGGTRGGRGRIRR AENYGPRAEVVMQDVAPNPDDPEDFPALS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for C12orf57-HABP4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for C12orf57-HABP4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Tgene | HABP4 | Q5JVS0 | DB08818 | Hyaluronic acid | Binder | Small molecule | Approved|Vet_approved |
Top |
Related Diseases for C12orf57-HABP4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | C12orf57 | C1857512 | Temtamy syndrome | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | C12orf57 | C0020796 | Profound Mental Retardation | 1 | CTD_human |
| Hgene | C12orf57 | C0025363 | Mental Retardation, Psychosocial | 1 | CTD_human |
| Hgene | C12orf57 | C0917816 | Mental deficiency | 1 | CTD_human |
| Hgene | C12orf57 | C3714756 | Intellectual Disability | 1 | CTD_human |
