
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ARAP1-FADD (FusionGDB2 ID:HG116985TG8772) |
Fusion Gene Summary for ARAP1-FADD |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ARAP1-FADD | Fusion gene ID: hg116985tg8772 | Hgene | Tgene | Gene symbol | ARAP1 | FADD | Gene ID | 116985 | 8772 |
| Gene name | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 | Fas associated via death domain | |
| Synonyms | CENTD2 | GIG3|MORT1 | |
| Cytomap | ('ARAP1')('FADD') 11q13.4 | 11q13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 1ARF-GAP, RHO-GAP, ankyrin repeat, and pleckstrin homology domains-containing protein 1centaurin-delta-2cnt-d2 | FAS-associated death domain proteinFas (TNFRSF6)-associated via death domainFas-associating death domain-containing proteinFas-associating protein with death domaingrowth-inhibiting gene 3 proteinmediator of receptor-induced toxicity | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000393609, ENST00000334211, ENST00000359373, ENST00000393605, ENST00000426523, ENST00000429686, ENST00000455638, ENST00000495878, | ||
| Fusion gene scores | * DoF score | 15 X 15 X 9=2025 | 5 X 2 X 4=40 |
| # samples | 17 | 6 | |
| ** MAII score | log2(17/2025*10)=-3.57431525652165 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/40*10)=0.584962500721156 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: ARAP1 [Title/Abstract] AND FADD [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ARAP1(72412694)-FADD(70052239), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | ARAP1-FADD seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARAP1-FADD seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARAP1-FADD seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ARAP1-FADD seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ARAP1-FADD seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ARAP1-FADD seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ARAP1 | GO:0043547 | positive regulation of GTPase activity | 11804590 |
| Tgene | FADD | GO:0032757 | positive regulation of interleukin-8 production | 16127453 |
| Tgene | FADD | GO:0032760 | positive regulation of tumor necrosis factor production | 16127453 |
| Tgene | FADD | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 21785459 |
| Tgene | FADD | GO:0043065 | positive regulation of apoptotic process | 11821383 |
| Tgene | FADD | GO:0045862 | positive regulation of proteolysis | 18387192 |
| Tgene | FADD | GO:0045944 | positive regulation of transcription by RNA polymerase II | 16127453 |
| Tgene | FADD | GO:0097190 | apoptotic signaling pathway | 11101870 |
| Tgene | FADD | GO:0097191 | extrinsic apoptotic signaling pathway | 21785459 |
| Tgene | FADD | GO:0097202 | activation of cysteine-type endopeptidase activity | 18387192 |
 Fusion gene breakpoints across ARAP1 (5'-gene) Fusion gene breakpoints across ARAP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
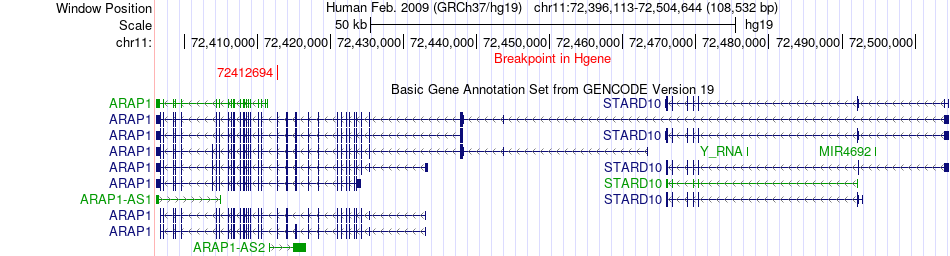 |
 Fusion gene breakpoints across FADD (3'-gene) Fusion gene breakpoints across FADD (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
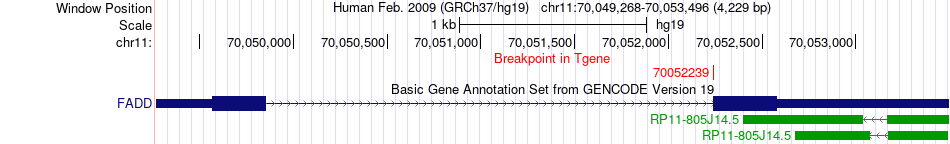 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-44-A4SU-01A | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
Top |
Fusion Gene ORF analysis for ARAP1-FADD |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000393609 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| In-frame | ENST00000334211 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| In-frame | ENST00000359373 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| In-frame | ENST00000393605 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| In-frame | ENST00000426523 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| In-frame | ENST00000429686 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| In-frame | ENST00000455638 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
| intron-3CDS | ENST00000495878 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000359373 | ARAP1 | chr11 | 72412694 | - | ENST00000301838 | FADD | chr11 | 70052239 | + | 4412 | 3154 | 810 | 3494 | 894 |
| ENST00000455638 | ARAP1 | chr11 | 72412694 | - | ENST00000301838 | FADD | chr11 | 70052239 | + | 3560 | 2302 | 0 | 2642 | 880 |
| ENST00000393605 | ARAP1 | chr11 | 72412694 | - | ENST00000301838 | FADD | chr11 | 70052239 | + | 3396 | 2138 | 544 | 2478 | 644 |
| ENST00000334211 | ARAP1 | chr11 | 72412694 | - | ENST00000301838 | FADD | chr11 | 70052239 | + | 3560 | 2302 | 510 | 2642 | 710 |
| ENST00000426523 | ARAP1 | chr11 | 72412694 | - | ENST00000301838 | FADD | chr11 | 70052239 | + | 3230 | 1972 | 180 | 2312 | 710 |
| ENST00000429686 | ARAP1 | chr11 | 72412694 | - | ENST00000301838 | FADD | chr11 | 70052239 | + | 3047 | 1789 | 180 | 2129 | 649 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000359373 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + | 0.004111399 | 0.99588865 |
| ENST00000455638 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + | 0.005432647 | 0.9945674 |
| ENST00000393605 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + | 0.004797612 | 0.9952024 |
| ENST00000334211 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + | 0.003253245 | 0.9967468 |
| ENST00000426523 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + | 0.005414498 | 0.9945856 |
| ENST00000429686 | ENST00000301838 | ARAP1 | chr11 | 72412694 | - | FADD | chr11 | 70052239 | + | 0.006160908 | 0.9938391 |
Top |
Fusion Genomic Features for ARAP1-FADD |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ARAP1 | chr11 | 72412693 | - | FADD | chr11 | 70052238 | + | 0.000196917 | 0.9998031 |
| ARAP1 | chr11 | 72412693 | - | FADD | chr11 | 70052238 | + | 0.000196917 | 0.9998031 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
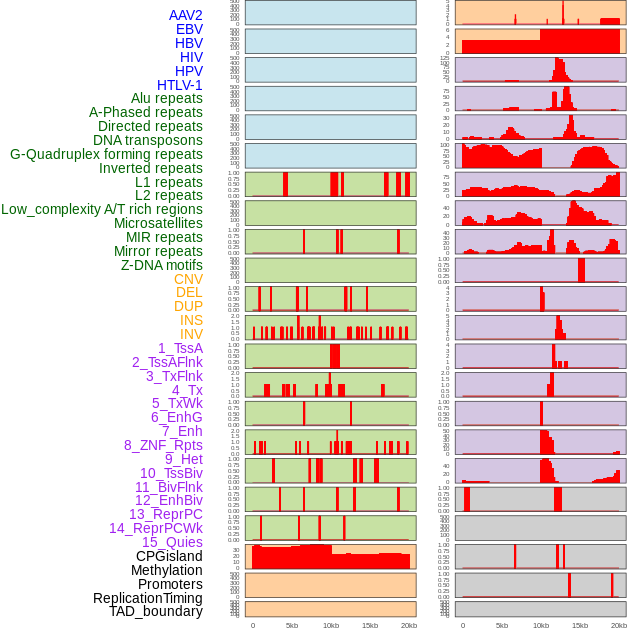 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
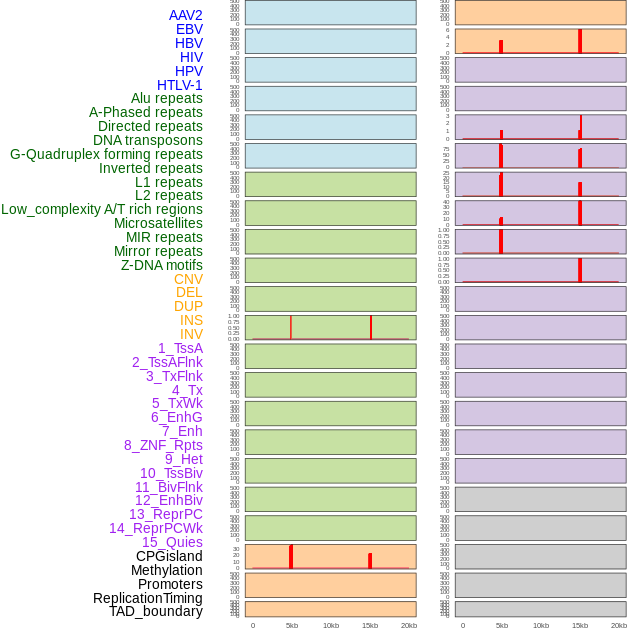 |
Top |
Fusion Protein Features for ARAP1-FADD |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:72412694/chr11:70052239) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 327_419 | 522 | 1206.0 | Domain | PH 1 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 6_70 | 522 | 1206.0 | Domain | SAM |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 327_419 | 767 | 1440.0 | Domain | PH 1 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 440_529 | 767 | 1440.0 | Domain | PH 2 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 535_660 | 767 | 1440.0 | Domain | Arf-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 6_70 | 767 | 1440.0 | Domain | SAM |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 327_419 | 527 | 1211.0 | Domain | PH 1 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 440_529 | 527 | 1211.0 | Domain | PH 2 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 6_70 | 527 | 1211.0 | Domain | SAM |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 327_419 | 767 | 1451.0 | Domain | PH 1 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 440_529 | 767 | 1451.0 | Domain | PH 2 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 535_660 | 767 | 1451.0 | Domain | Arf-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 6_70 | 767 | 1451.0 | Domain | SAM |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 327_419 | 461 | 1134.0 | Domain | PH 1 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 6_70 | 461 | 1134.0 | Domain | SAM |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 327_419 | 767 | 1440.0 | Domain | PH 1 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 440_529 | 767 | 1440.0 | Domain | PH 2 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 535_660 | 767 | 1440.0 | Domain | Arf-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 6_70 | 767 | 1440.0 | Domain | SAM |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 550_576 | 767 | 1440.0 | Zinc finger | C4-type |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 550_576 | 767 | 1451.0 | Zinc finger | C4-type |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 550_576 | 767 | 1440.0 | Zinc finger | C4-type |
| Tgene | FADD | chr11:72412694 | chr11:70052239 | ENST00000301838 | 0 | 2 | 97_181 | 95 | 209.0 | Domain | Death |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 1172_1261 | 522 | 1206.0 | Domain | Ras-associating |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 1274_1396 | 522 | 1206.0 | Domain | PH 4 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 440_529 | 522 | 1206.0 | Domain | PH 2 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 535_660 | 522 | 1206.0 | Domain | Arf-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 743_850 | 522 | 1206.0 | Domain | PH 3 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 954_1139 | 522 | 1206.0 | Domain | Rho-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 1172_1261 | 767 | 1440.0 | Domain | Ras-associating |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 1274_1396 | 767 | 1440.0 | Domain | PH 4 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 743_850 | 767 | 1440.0 | Domain | PH 3 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000359373 | - | 16 | 34 | 954_1139 | 767 | 1440.0 | Domain | Rho-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 1172_1261 | 527 | 1211.0 | Domain | Ras-associating |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 1274_1396 | 527 | 1211.0 | Domain | PH 4 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 535_660 | 527 | 1211.0 | Domain | Arf-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 743_850 | 527 | 1211.0 | Domain | PH 3 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 954_1139 | 527 | 1211.0 | Domain | Rho-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 1172_1261 | 767 | 1451.0 | Domain | Ras-associating |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 1274_1396 | 767 | 1451.0 | Domain | PH 4 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 743_850 | 767 | 1451.0 | Domain | PH 3 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393609 | - | 16 | 35 | 954_1139 | 767 | 1451.0 | Domain | Rho-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 1172_1261 | 461 | 1134.0 | Domain | Ras-associating |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 1274_1396 | 461 | 1134.0 | Domain | PH 4 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 440_529 | 461 | 1134.0 | Domain | PH 2 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 535_660 | 461 | 1134.0 | Domain | Arf-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 743_850 | 461 | 1134.0 | Domain | PH 3 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 954_1139 | 461 | 1134.0 | Domain | Rho-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 1172_1261 | 767 | 1440.0 | Domain | Ras-associating |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 1274_1396 | 767 | 1440.0 | Domain | PH 4 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 743_850 | 767 | 1440.0 | Domain | PH 3 |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000455638 | - | 14 | 32 | 954_1139 | 767 | 1440.0 | Domain | Rho-GAP |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000334211 | - | 14 | 33 | 550_576 | 522 | 1206.0 | Zinc finger | C4-type |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000393605 | - | 11 | 30 | 550_576 | 527 | 1211.0 | Zinc finger | C4-type |
| Hgene | ARAP1 | chr11:72412694 | chr11:70052239 | ENST00000429686 | - | 13 | 31 | 550_576 | 461 | 1134.0 | Zinc finger | C4-type |
| Tgene | FADD | chr11:72412694 | chr11:70052239 | ENST00000301838 | 0 | 2 | 3_81 | 95 | 209.0 | Domain | DED |
Top |
Fusion Gene Sequence for ARAP1-FADD |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >5757_5757_1_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000334211_FADD_chr11_70052239_ENST00000301838_length(transcript)=3560nt_BP=2302nt ACTTTTCGCTTTCTCAGTGTTTTCAGGAAGTGTGTGTGTTGGCCACGGCTGCAGGAGGTGGGGGAGACACAAAGCAGGAAGCCTCCGGGA GACCAGAGCTGGGTGCAGACATACACACACACATACACACAGACACACAGAGTCACACACACTCACACACACTCTCTCTCTCTCTCCCTC TGTCTTTCTCTCTCTCTCTCTCTCTCTGTCCTTTCCTCCTGGACAGATCCACAGTTATACACAGAAACAAACACACACGCACGTAGAGAA GTGATTCACAAACACTTAAAGACATAAATCACAGGTGCAAAGCCATACCTGGGCTCAAAAACCTCCAAGAGAACGCAGCCTCAGACCCAC CCAGGGGCCAGGGGCCAGGCTGTTTGCGAGACCAGCCCAGGCGGGACCCAGGCTTGCACGGGCAGGTACACGACATTCTTGGGCATACGC AGCCCGCCTGGCCGGAGCTGTGGGAGTCCTCAGCCCCAAGACCCAGCAGGCGTCTGAGGCCTGCCCACTAAGGAGGAGGAGTCATTGCTG CCATCATTATCATCCCCTCCCCAGCCACAGTCTGAGGAGCCCCTGTCCACCCTCCCCCAGGGGCCTCCCCAGCCTCCCTCTCCACCTCCC TGCCCCCCGGAGATACCTCCAAAGCCGGTACGCCTGTTCCCAGAGTTCGATGACTCTGACTACGATGAGGTCCCAGAGGAGGGGCCGGGG GCCCCAGCCAGAGTGATGACCAAGAAGGAGGAGCCCCCACCGAGCCGAGTCCCACGGGCCGTGCGCGTGGCCAGTCTGCTGAGCGAGGGA GAGGAACTGTCTGGGGACGACCAAGGGGATGAGGAAGAGGATGACCACGCCTATGAGGGCGTCCCCAATGGCGGATGGCATACCAGCAGC CTGAGCTTGTCCTTGCCCAGCACAATAGCTGCGCCACACCCCATGGACGGGCCGCCTGGGGGCTCCACCCCCGTCACACCAGTCATCAAG GCTGGCTGGCTGGACAAGAACCCACCGCAGGGATCTTACATCTATCAGAAACGATGGGTGAGACTGGATACTGATCACCTGCGATACTTT GACAGTAACAAGGACGCTTACTCTAAGCGCTTTATCTCTGTGGCCTGCATCTCCCACGTGGCTGCCATCGGGGACCAGAAGTTTGAAGTG ATCACAAACAACCGAACCTTTGCCTTCCGGGCAGAGAGTGATGTGGAGCGGAAGGAGTGGATGCAGGCCCTGCAGCAGGCCATGGCTGAG CAGCGTGCCCGGGCCCGGCTCTCTAGCGCTTATCTGCTGGGAGTTCCAGGCTCAGAGCAGCCTGACCGCGCTGGCAGCCTGGAGCTTCGT GGCTTCAAGAATAAGCTGTACGTGGCCGTGGTCGGGGACAAAGTGCAGCTCTACAAGAATCTAGAGGAGTACCACCTGGGCATTGGCATC ACCTTCATCGACATGAGCGTGGGCAACGTGAAGGAAGTGGACCGGCGCAGCTTCGACCTCACCACGCCCTACCGCATCTTCAGCTTCTCT GCTGACTCAGAGCTAGAGAAGGAGCAGTGGTTGGAGGCCATGCAGGGAGCCATCGCTGAGGCCCTGTCTACCTCGGAGGTGGCCGAGCGC ATCTGGGCTGCAGCCCCCAACAGGTTCTGTGCTGACTGCGGGGCTCCTCAGCCTGACTGGGCCTCCATCAACCTCTGTGTTGTTATCTGC AAGCGCTGTGCAGGGGAGCACCGTGGCCTGGGCGCTGGCGTCTCCAAGGTGCGGAGCCTGAAGATGGACAGGAAGGTGTGGACAGAAACA CTTATCGAGCTCTTCTTACAGCTGGGGAATGGCGCTGGGAACCGCTTCTGGGCAGCCAACGTGCCCCCCAGTGAGGCCCTGCAGCCCAGC AGCAGCCCCAGCACCCGGCGGTGCCACCTGGAGGCCAAGTACCGTGAGGGCAAGTACCGCCGCTACCACCCGCTCTTTGGCAACCAGGAG GAGCTGGACAAGGCCCTGTGTGCTGCAGTCACCACCACAGACCTGGCTGAGACCCAGGCGCTCCTGGGCTGTGGGGCTGGGATCAACTGC TTCTCGGGGGACCCTGAGGCCCCCACGCCCCTGGCTCTTGCAGAGCAGGCGGGGCAGACGCTGCAGATGGAATTCCTTCGGAACAACCGG ACCACAGAGGTGCCTCGGCTGGACTCGATGAAGCCCCTGGAAAAGCACTACTCAGTTGTCCTGCCGACCGTGAGCCACAGTGGCTTCCTC TACAAGACTGCCTCTGCCGGCAAGCTGCTACAGGACCGCCGGGCCCGGGAAGACCTGTGTGCAGCATTTAACGTCATATGTGATAATGTG GGGAAAGATTGGAGAAGGCTGGCTCGTCAGCTCAAAGTCTCAGACACCAAGATCGACAGCATCGAGGACAGATACCCCCGCAACCTGACA GAGCGTGTGCGGGAGTCACTGAGAATCTGGAAGAACACAGAGAAGGAGAACGCAACAGTGGCCCACCTGGTGGGGGCTCTCAGGTCCTGC CAGATGAACCTGGTGGCTGACCTGGTACAAGAGGTTCAGCAGGCCCGTGACCTCCAGAACAGGAGTGGGGCCATGTCCCCGATGTCATGG AACTCAGACGCATCTACCTCCGAAGCGTCCTGATGGGCCGCTGCTTTGCGCTGGTGGACCACAGGCATCTACACAGCCTGGACTTTGGTT CTCTCCAGGAAGGTAGCCCAGCACTGTGAAGACCCAGCAGGAAGCCAGGCTGAGTGAGCCACAGACCACCTGCTTCTGAACTCAAGCTGC GTTTATTAATGCCTCTCCCGCACCAGGCCGGGCTTGGGCCCTGCACAGATATTTCCATTTCTTCCTCACTATGACACTGAGCAAGATCTT GTCTCCACTAAATGAGCTCCTGCGGGAGTAGTTGGAAAGTTGGAACCGTGTCCAGCACAGAAGGAATCTGTGCAGATGAGCAGTCACACT GTTACTCCACAGCGGAGGAGACCAGCTCAGAGGCCCAGGAATCGGAGCGAAGCAGAGAGGTGGAGAACTGGGATTTGAACCCCCGCCATC CTTCACCAGAGCCCATGCTCAACCACTGTGGCGTTCTGCTGCCCCTGCAGTTGGCAGAAAGGATGTTTTGTCCCATTTCCTTGGAGGCCA CCGGGACAGACCTGGACACTAGGGTCAGGCGGGGTGCTGTGGTGGGGAGAGGCATGGCTGGGGTGGGGGTGGGGAGACCTGGTTGGCCGT GGTCCAGCTCTTGGCCCCTGTGTGAGTTGAGTCTCCTCTCTGAGACTGCTAAGTAGGGGCAGTGATGGTTGCCAGGACGAATTGAGATAA TATCTGTGAGGTGCTGATGAGTGATTGACACACAGCACTCTCTAAATCTTCCTTGTGAGGATTATGGGTCCTGCAATTCTACAGTTTCTT ACTGTTTTGTATCAAAATCACTATCTTTCTGATAACAGAATTGCCAAGGCAGCGGGATCTCGTATCTTTAAAAAGCAGTCCTCTTATTCC TAAGGTAATCCTATTAAAACACAGCTTTACAACTTCCATACTACAAAAAA >5757_5757_1_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000334211_FADD_chr11_70052239_ENST00000301838_length(amino acids)=710AA_BP=597 MPTKEEESLLPSLSSPPQPQSEEPLSTLPQGPPQPPSPPPCPPEIPPKPVRLFPEFDDSDYDEVPEEGPGAPARVMTKKEEPPPSRVPRA VRVASLLSEGEELSGDDQGDEEEDDHAYEGVPNGGWHTSSLSLSLPSTIAAPHPMDGPPGGSTPVTPVIKAGWLDKNPPQGSYIYQKRWV RLDTDHLRYFDSNKDAYSKRFISVACISHVAAIGDQKFEVITNNRTFAFRAESDVERKEWMQALQQAMAEQRARARLSSAYLLGVPGSEQ PDRAGSLELRGFKNKLYVAVVGDKVQLYKNLEEYHLGIGITFIDMSVGNVKEVDRRSFDLTTPYRIFSFSADSELEKEQWLEAMQGAIAE ALSTSEVAERIWAAAPNRFCADCGAPQPDWASINLCVVICKRCAGEHRGLGAGVSKVRSLKMDRKVWTETLIELFLQLGNGAGNRFWAAN VPPSEALQPSSSPSTRRCHLEAKYREGKYRRYHPLFGNQEELDKALCAAVTTTDLAETQALLGCGAGINCFSGDPEAPTPLALAEQAGQT LQMEFLRNNRTTEVPRLDSMKPLEKHYSVVLPTVSHSGFLYKTASAGKLLQDRRAREDLCAAFNVICDNVGKDWRRLARQLKVSDTKIDS IEDRYPRNLTERVRESLRIWKNTEKENATVAHLVGALRSCQMNLVADLVQEVQQARDLQNRSGAMSPMSWNSDASTSEAS -------------------------------------------------------------- >5757_5757_2_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000359373_FADD_chr11_70052239_ENST00000301838_length(transcript)=4412nt_BP=3154nt GATTCACAAGGAATGACCCTCTTCATCGCCTCTCCTAATTCAGTCCTCACAACAGTCCTTTTACAAATGGGACAACAGGTTAGAGGAAGT CAGGCAGATTTCCAGCATCATAGAGAGTAAAGGACCAGGGAAGGATCAGGATTCAAGGACTGCACCCAGGCTCTGCTTCCAGCTTGCTGT GTGACTTTGGGTAATTTTGTTCCCTTAGGGAACTGAGCTTTCTCATTTGTAAATGCAAACAGGCTGTTGGGAGGATCAAATGAGATCCAG GGGTGAAAACAGCTTAGTTTACTTTCAGGAATTTACCCACGCGGTATATAAAGGCAAAATATTATTATAGTCAGGTGATTGTAGATTGAG GAACCCATTTCCTCATTCTGCAAATTGCAAACCTGAGGGCCCAAAGAGGGACAGGGGCTTGCCCCAGGTCTCAGCAGGCTGTGAGCAAGA GCTAAAGCCTAATCCTCCTGCCTTTGGGCCTGGAGCCCTTCCTTGTACCCCAGGGGTCAGTGTCTTTGTTGGATACAGGCTTAGATTGAC TGACTGTACCCTGAGAACCTAGGGGAGTCCCTGTTCCCAATTCTTCTCCTACCCCCACCTTGGCCTGATGGAGGAAGACCCTGCTGTGTT GAGATGAGCACCAGAGCCAAGAAGCTGAGGAGGATCTGGAGAATTCTGGAGGAAGAGGAGAGTGTTGCTGGAGCTGTACAGACCCTGCTT CTCAGCTCCCGCCTCAGGCCGAGATGAGGAGCCCTTCAGAATAGCTGCTGTCTCTGGGAGGACCCGGGCGTCCTTGGCAGCCCAGCTGCT CTGGACAAAGCCCTGCCAGTCAGGCCTCCGCTGGCAGGAACCATGGCAGAGGCTGGGGATGCTGCGCTATCGGTGGCCGAGTGGCTGCGG GCATTGCACCTGGAGCAGTACACGGGGCTCTTTGAGCAGCATGGCCTGGTGTGGGCCACTGAGTGCCAAGGCCTCAGCGACACCCGCCTG ATGGACATGGGCATGCTACTCCCTGGTCACCGCCGCCGCATCCTGGCTGGCCTGCTCCGTGCCCATACCTCACCGGCCCCTGCACCCCGC CCCACCCCACGGCCTGTGCCCATGAAGCGCCACATCTTCCGCTCACCACCTGTGCCTGCCACTCCACCCGAGCCGCTGCCCACCACTACA GAGGATGAGGGGCTCCCCGCTGCCCCACCCATCCCGCCCCGGAGGAGCTGCCTTCCGCCCACCTGCTTCACCACCCCATCCACAGCTGCC CCAGACCCTGTGCTGCCCCCGCTGCCTGCTAAGCGGCATTTGGCAGAGCTGAGCGTTCCACCCGTGCCGCCCCGCACCGGACCCCCCCGC CTGCTGGTGAGCCTGCCCACTAAGGAGGAGGAGTCATTGCTGCCATCATTATCATCCCCTCCCCAGCCACAGTCTGAGGAGCCCCTGTCC ACCCTCCCCCAGGGGCCTCCCCAGCCTCCCTCTCCACCTCCCTGCCCCCCGGAGATACCTCCAAAGCCGGTACGCCTGTTCCCAGAGTTC GATGACTCTGACTACGATGAGGTCCCAGAGGAGGGGCCGGGGGCCCCAGCCAGAGTGATGACCAAGAAGGAGGAGCCCCCACCGAGCCGA GTCCCACGGGCCGTGCGCGTGGCCAGTCTGCTGAGCGAGGGAGAGGAACTGTCTGGGGACGACCAAGGGGATGAGGAAGAGGATGACCAC GCCTATGAGGGCGTCCCCAATGGCGGATGGCATACCAGCAGCCTGAGCTTGTCCTTGCCCAGCACAATAGCTGCGCCACACCCCATGGAC GGGCCGCCTGGGGGCTCCACCCCCGTCACACCAGTCATCAAGGCTGGCTGGCTGGACAAGAACCCACCGCAGGGATCTTACATCTATCAG AAACGATGGGTGAGACTGGATACTGATCACCTGCGATACTTTGACAGTAACAAGGACGCTTACTCTAAGCGCTTTATCTCTGTGGCCTGC ATCTCCCACGTGGCTGCCATCGGGGACCAGAAGTTTGAAGTGATCACAAACAACCGAACCTTTGCCTTCCGGGCAGAGAGTGATGTGGAG CGGAAGGAGTGGATGCAGGCCCTGCAGCAGGCCATGGCTGAGCAGCGTGCCCGGGCCCGGCTCTCTAGCGCTTATCTGCTGGGAGTTCCA GGCTCAGAGCAGCCTGACCGCGCTGGCAGCCTGGAGCTTCGTGGCTTCAAGAATAAGCTGTACGTGGCCGTGGTCGGGGACAAAGTGCAG CTCTACAAGAATCTAGAGGAGTACCACCTGGGCATTGGCATCACCTTCATCGACATGAGCGTGGGCAACGTGAAGGAAGTGGACCGGCGC AGCTTCGACCTCACCACGCCCTACCGCATCTTCAGCTTCTCTGCTGACTCAGAGCTAGAGAAGGAGCAGTGGTTGGAGGCCATGCAGGGA GCCATCGCTGAGGCCCTGTCTACCTCGGAGGTGGCCGAGCGCATCTGGGCTGCAGCCCCCAACAGGTTCTGTGCTGACTGCGGGGCTCCT CAGCCTGACTGGGCCTCCATCAACCTCTGTGTTGTTATCTGCAAGCGCTGTGCAGGGGAGCACCGTGGCCTGGGCGCTGGCGTCTCCAAG GTGCGGAGCCTGAAGATGGACAGGAAGGTGTGGACAGAAACACTTATCGAGCTCTTCTTACAGCTGGGGAATGGCGCTGGGAACCGCTTC TGGGCAGCCAACGTGCCCCCCAGTGAGGCCCTGCAGCCCAGCAGCAGCCCCAGCACCCGGCGGTGCCACCTGGAGGCCAAGTACCGTGAG GGCAAGTACCGCCGCTACCACCCGCTCTTTGGCAACCAGGAGGAGCTGGACAAGGCCCTGTGTGCTGCAGTCACCACCACAGACCTGGCT GAGACCCAGGCGCTCCTGGGCTGTGGGGCTGGGATCAACTGCTTCTCGGGGGACCCTGAGGCCCCCACGCCCCTGGCTCTTGCAGAGCAG GCGGGGCAGACGCTGCAGATGGAATTCCTTCGGAACAACCGGACCACAGAGGTGCCTCGGCTGGACTCGATGAAGCCCCTGGAAAAGCAC TACTCAGTTGTCCTGCCGACCGTGAGCCACAGTGGCTTCCTCTACAAGACTGCCTCTGCCGGCAAGCTGCTACAGGACCGCCGGGCCCGG GAAGACCTGTGTGCAGCATTTAACGTCATATGTGATAATGTGGGGAAAGATTGGAGAAGGCTGGCTCGTCAGCTCAAAGTCTCAGACACC AAGATCGACAGCATCGAGGACAGATACCCCCGCAACCTGACAGAGCGTGTGCGGGAGTCACTGAGAATCTGGAAGAACACAGAGAAGGAG AACGCAACAGTGGCCCACCTGGTGGGGGCTCTCAGGTCCTGCCAGATGAACCTGGTGGCTGACCTGGTACAAGAGGTTCAGCAGGCCCGT GACCTCCAGAACAGGAGTGGGGCCATGTCCCCGATGTCATGGAACTCAGACGCATCTACCTCCGAAGCGTCCTGATGGGCCGCTGCTTTG CGCTGGTGGACCACAGGCATCTACACAGCCTGGACTTTGGTTCTCTCCAGGAAGGTAGCCCAGCACTGTGAAGACCCAGCAGGAAGCCAG GCTGAGTGAGCCACAGACCACCTGCTTCTGAACTCAAGCTGCGTTTATTAATGCCTCTCCCGCACCAGGCCGGGCTTGGGCCCTGCACAG ATATTTCCATTTCTTCCTCACTATGACACTGAGCAAGATCTTGTCTCCACTAAATGAGCTCCTGCGGGAGTAGTTGGAAAGTTGGAACCG TGTCCAGCACAGAAGGAATCTGTGCAGATGAGCAGTCACACTGTTACTCCACAGCGGAGGAGACCAGCTCAGAGGCCCAGGAATCGGAGC GAAGCAGAGAGGTGGAGAACTGGGATTTGAACCCCCGCCATCCTTCACCAGAGCCCATGCTCAACCACTGTGGCGTTCTGCTGCCCCTGC AGTTGGCAGAAAGGATGTTTTGTCCCATTTCCTTGGAGGCCACCGGGACAGACCTGGACACTAGGGTCAGGCGGGGTGCTGTGGTGGGGA GAGGCATGGCTGGGGTGGGGGTGGGGAGACCTGGTTGGCCGTGGTCCAGCTCTTGGCCCCTGTGTGAGTTGAGTCTCCTCTCTGAGACTG CTAAGTAGGGGCAGTGATGGTTGCCAGGACGAATTGAGATAATATCTGTGAGGTGCTGATGAGTGATTGACACACAGCACTCTCTAAATC TTCCTTGTGAGGATTATGGGTCCTGCAATTCTACAGTTTCTTACTGTTTTGTATCAAAATCACTATCTTTCTGATAACAGAATTGCCAAG GCAGCGGGATCTCGTATCTTTAAAAAGCAGTCCTCTTATTCCTAAGGTAATCCTATTAAAACACAGCTTTACAACTTCCATACTACAAAA AA >5757_5757_2_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000359373_FADD_chr11_70052239_ENST00000301838_length(amino acids)=894AA_BP=781 MDKALPVRPPLAGTMAEAGDAALSVAEWLRALHLEQYTGLFEQHGLVWATECQGLSDTRLMDMGMLLPGHRRRILAGLLRAHTSPAPAPR PTPRPVPMKRHIFRSPPVPATPPEPLPTTTEDEGLPAAPPIPPRRSCLPPTCFTTPSTAAPDPVLPPLPAKRHLAELSVPPVPPRTGPPR LLVSLPTKEEESLLPSLSSPPQPQSEEPLSTLPQGPPQPPSPPPCPPEIPPKPVRLFPEFDDSDYDEVPEEGPGAPARVMTKKEEPPPSR VPRAVRVASLLSEGEELSGDDQGDEEEDDHAYEGVPNGGWHTSSLSLSLPSTIAAPHPMDGPPGGSTPVTPVIKAGWLDKNPPQGSYIYQ KRWVRLDTDHLRYFDSNKDAYSKRFISVACISHVAAIGDQKFEVITNNRTFAFRAESDVERKEWMQALQQAMAEQRARARLSSAYLLGVP GSEQPDRAGSLELRGFKNKLYVAVVGDKVQLYKNLEEYHLGIGITFIDMSVGNVKEVDRRSFDLTTPYRIFSFSADSELEKEQWLEAMQG AIAEALSTSEVAERIWAAAPNRFCADCGAPQPDWASINLCVVICKRCAGEHRGLGAGVSKVRSLKMDRKVWTETLIELFLQLGNGAGNRF WAANVPPSEALQPSSSPSTRRCHLEAKYREGKYRRYHPLFGNQEELDKALCAAVTTTDLAETQALLGCGAGINCFSGDPEAPTPLALAEQ AGQTLQMEFLRNNRTTEVPRLDSMKPLEKHYSVVLPTVSHSGFLYKTASAGKLLQDRRAREDLCAAFNVICDNVGKDWRRLARQLKVSDT KIDSIEDRYPRNLTERVRESLRIWKNTEKENATVAHLVGALRSCQMNLVADLVQEVQQARDLQNRSGAMSPMSWNSDASTSEAS -------------------------------------------------------------- >5757_5757_3_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000393605_FADD_chr11_70052239_ENST00000301838_length(transcript)=3396nt_BP=2138nt GCCTGTCAGGGTGTTGTGGGGGTGCCCAGCTGCCTGTCAGACCCTGAGGACTACCAGACAGAGAGTGGGCAGCCATTTCCCGGGAGGCTT CCCCTTCCCCCACCTCACTCTCTTGCCATACGGGGGTCAGGAGTCACGTGTAGCCACAGCGCTCTGGGCCAGGGGTCGGGGCTCTGGTGT GGCTCCCCCAGGCCCTGGCTGTATGTGGGTGGGCATGTGGCCTCCCCCCACCCTGTGTGTTAGGAACTCGTGTCCTGTCCCGGAGTGTCT GTCTCTGTTTCACTTCTTGTGTTCCCGTGTCTGTCTGTCTTGCATCCCTAGCCCGCAGTCTTTTTCTCTGTCCCTTTATCTTTCTGTTTT TGTCTGTGTCCTAGGTCGGTTTCTGCCACTGGCTCTCCTGTCCCTGTGATCTGGGTCTCTGTGTGGTCTCCGCTGGCAGGGGGAGGGGAG TGTGAGGAAACAGAACTAAAAGTGCTGGGCTGTGTGGGTCGGGGCTGTGTGGACAGAGGGGGCGTGGCAGCCCCGGGTGGGTGTGATCTG TGGCTTGAAGGGAAGGATGACGCTGAGTGGGTCTCGGGGGCAGGAGGAGCCCCCACCGAGCCGAGTCCCACGGGCCGTGCGCGTGGCCAG TCTGCTGAGCGAGGGAGAGGAACTGTCTGGGGACGACCAAGGGGATGAGGAAGAGGATGACCACGCCTATGAGGGCGTCCCCAATGGCGG ATGGCATACCAGCAGCCTGAGCTTGTCCTTGCCCAGCACAATAGCTGCGCCACACCCCATGGACGGGCCGCCTGGGGGCTCCACCCCCGT CACACCAGTCATCAAGGCTGGCTGGCTGGACAAGAACCCACCGCAGGGATCTTACATCTATCAGAAACGATGGGTGAGACTGGATACTGA TCACCTGCGATACTTTGACAGTAACAAGGACGCTTACTCTAAGCGCTTTATCTCTGTGGCCTGCATCTCCCACGTGGCTGCCATCGGGGA CCAGAAGTTTGAAGTGATCACAAACAACCGAACCTTTGCCTTCCGGGCAGAGAGTGATGTGGAGCGGAAGGAGTGGATGCAGGCCCTGCA GCAGGCCATGGCTGAGCAGCGTGCCCGGGCCCGGCTCTCTAGCGCTTATCTGCTGGGAGTTCCAGGCTCAGAGCAGCCTGACCGCGCTGG CAGCCTGGAGCTTCGTGGCTTCAAGAATAAGCTGTACGTGGCCGTGGTCGGGGACAAAGTGCAGCTCTACAAGAATCTAGAGGAGTACCA CCTGGGCATTGGCATCACCTTCATCGACATGAGCGTGGGCAACGTGAAGGAAGTGGACCGGCGCAGCTTCGACCTCACCACGCCCTACCG CATCTTCAGCTTCTCTGCTGACTCAGAGCTAGAGAAGGAGCAGTGGTTGGAGGCCATGCAGGGAGCCATCGCTGAGGCCCTGTCTACCTC GGAGGTGGCCGAGCGCATCTGGGCTGCAGCCCCCAACAGGTTCTGTGCTGACTGCGGGGCTCCTCAGCCTGACTGGGCCTCCATCAACCT CTGTGTTGTTATCTGCAAGCGCTGTGCAGGGGAGCACCGTGGCCTGGGCGCTGGCGTCTCCAAGGTGCGGAGCCTGAAGATGGACAGGAA GGTGTGGACAGAAACACTTATCGAGCTCTTCTTACAGCTGGGGAATGGCGCTGGGAACCGCTTCTGGGCAGCCAACGTGCCCCCCAGTGA GGCCCTGCAGCCCAGCAGCAGCCCCAGCACCCGGCGGTGCCACCTGGAGGCCAAGTACCGTGAGGGCAAGTACCGCCGCTACCACCCGCT CTTTGGCAACCAGGAGGAGCTGGACAAGGCCCTGTGTGCTGCAGTCACCACCACAGACCTGGCTGAGACCCAGGCGCTCCTGGGCTGTGG GGCTGGGATCAACTGCTTCTCGGGGGACCCTGAGGCCCCCACGCCCCTGGCTCTTGCAGAGCAGGCGGGGCAGACGCTGCAGATGGAATT CCTTCGGAACAACCGGACCACAGAGGTGCCTCGGCTGGACTCGATGAAGCCCCTGGAAAAGCACTACTCAGTTGTCCTGCCGACCGTGAG CCACAGTGGCTTCCTCTACAAGACTGCCTCTGCCGGCAAGCTGCTACAGGACCGCCGGGCCCGGGAAGACCTGTGTGCAGCATTTAACGT CATATGTGATAATGTGGGGAAAGATTGGAGAAGGCTGGCTCGTCAGCTCAAAGTCTCAGACACCAAGATCGACAGCATCGAGGACAGATA CCCCCGCAACCTGACAGAGCGTGTGCGGGAGTCACTGAGAATCTGGAAGAACACAGAGAAGGAGAACGCAACAGTGGCCCACCTGGTGGG GGCTCTCAGGTCCTGCCAGATGAACCTGGTGGCTGACCTGGTACAAGAGGTTCAGCAGGCCCGTGACCTCCAGAACAGGAGTGGGGCCAT GTCCCCGATGTCATGGAACTCAGACGCATCTACCTCCGAAGCGTCCTGATGGGCCGCTGCTTTGCGCTGGTGGACCACAGGCATCTACAC AGCCTGGACTTTGGTTCTCTCCAGGAAGGTAGCCCAGCACTGTGAAGACCCAGCAGGAAGCCAGGCTGAGTGAGCCACAGACCACCTGCT TCTGAACTCAAGCTGCGTTTATTAATGCCTCTCCCGCACCAGGCCGGGCTTGGGCCCTGCACAGATATTTCCATTTCTTCCTCACTATGA CACTGAGCAAGATCTTGTCTCCACTAAATGAGCTCCTGCGGGAGTAGTTGGAAAGTTGGAACCGTGTCCAGCACAGAAGGAATCTGTGCA GATGAGCAGTCACACTGTTACTCCACAGCGGAGGAGACCAGCTCAGAGGCCCAGGAATCGGAGCGAAGCAGAGAGGTGGAGAACTGGGAT TTGAACCCCCGCCATCCTTCACCAGAGCCCATGCTCAACCACTGTGGCGTTCTGCTGCCCCTGCAGTTGGCAGAAAGGATGTTTTGTCCC ATTTCCTTGGAGGCCACCGGGACAGACCTGGACACTAGGGTCAGGCGGGGTGCTGTGGTGGGGAGAGGCATGGCTGGGGTGGGGGTGGGG AGACCTGGTTGGCCGTGGTCCAGCTCTTGGCCCCTGTGTGAGTTGAGTCTCCTCTCTGAGACTGCTAAGTAGGGGCAGTGATGGTTGCCA GGACGAATTGAGATAATATCTGTGAGGTGCTGATGAGTGATTGACACACAGCACTCTCTAAATCTTCCTTGTGAGGATTATGGGTCCTGC AATTCTACAGTTTCTTACTGTTTTGTATCAAAATCACTATCTTTCTGATAACAGAATTGCCAAGGCAGCGGGATCTCGTATCTTTAAAAA GCAGTCCTCTTATTCCTAAGGTAATCCTATTAAAACACAGCTTTACAACTTCCATACTACAAAAAA >5757_5757_3_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000393605_FADD_chr11_70052239_ENST00000301838_length(amino acids)=644AA_BP=531 MKGRMTLSGSRGQEEPPPSRVPRAVRVASLLSEGEELSGDDQGDEEEDDHAYEGVPNGGWHTSSLSLSLPSTIAAPHPMDGPPGGSTPVT PVIKAGWLDKNPPQGSYIYQKRWVRLDTDHLRYFDSNKDAYSKRFISVACISHVAAIGDQKFEVITNNRTFAFRAESDVERKEWMQALQQ AMAEQRARARLSSAYLLGVPGSEQPDRAGSLELRGFKNKLYVAVVGDKVQLYKNLEEYHLGIGITFIDMSVGNVKEVDRRSFDLTTPYRI FSFSADSELEKEQWLEAMQGAIAEALSTSEVAERIWAAAPNRFCADCGAPQPDWASINLCVVICKRCAGEHRGLGAGVSKVRSLKMDRKV WTETLIELFLQLGNGAGNRFWAANVPPSEALQPSSSPSTRRCHLEAKYREGKYRRYHPLFGNQEELDKALCAAVTTTDLAETQALLGCGA GINCFSGDPEAPTPLALAEQAGQTLQMEFLRNNRTTEVPRLDSMKPLEKHYSVVLPTVSHSGFLYKTASAGKLLQDRRAREDLCAAFNVI CDNVGKDWRRLARQLKVSDTKIDSIEDRYPRNLTERVRESLRIWKNTEKENATVAHLVGALRSCQMNLVADLVQEVQQARDLQNRSGAMS PMSWNSDASTSEAS -------------------------------------------------------------- >5757_5757_4_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000426523_FADD_chr11_70052239_ENST00000301838_length(transcript)=3230nt_BP=1972nt ACCTCCAAGAGAACGCAGCCTCAGACCCACCCAGGGGCCAGGGGCCAGGCTGTTTGCGAGACCAGCCCAGGCGGGACCCAGGCTTGCACG GGCAGGTACACGACATTCTTGGGCATACGCAGCCCGCCTGGCCGGAGCTGTGGGAGTCCTCAGCCCCAAGACCCAGCAGGCGTCTGAGGC CTGCCCACTAAGGAGGAGGAGTCATTGCTGCCATCATTATCATCCCCTCCCCAGCCACAGTCTGAGGAGCCCCTGTCCACCCTCCCCCAG GGGCCTCCCCAGCCTCCCTCTCCACCTCCCTGCCCCCCGGAGATACCTCCAAAGCCGGTACGCCTGTTCCCAGAGTTCGATGACTCTGAC TACGATGAGGTCCCAGAGGAGGGGCCGGGGGCCCCAGCCAGAGTGATGACCAAGAAGGAGGAGCCCCCACCGAGCCGAGTCCCACGGGCC GTGCGCGTGGCCAGTCTGCTGAGCGAGGGAGAGGAACTGTCTGGGGACGACCAAGGGGATGAGGAAGAGGATGACCACGCCTATGAGGGC GTCCCCAATGGCGGATGGCATACCAGCAGCCTGAGCTTGTCCTTGCCCAGCACAATAGCTGCGCCACACCCCATGGACGGGCCGCCTGGG GGCTCCACCCCCGTCACACCAGTCATCAAGGCTGGCTGGCTGGACAAGAACCCACCGCAGGGATCTTACATCTATCAGAAACGATGGGTG AGACTGGATACTGATCACCTGCGATACTTTGACAGTAACAAGGACGCTTACTCTAAGCGCTTTATCTCTGTGGCCTGCATCTCCCACGTG GCTGCCATCGGGGACCAGAAGTTTGAAGTGATCACAAACAACCGAACCTTTGCCTTCCGGGCAGAGAGTGATGTGGAGCGGAAGGAGTGG ATGCAGGCCCTGCAGCAGGCCATGGCTGAGCAGCGTGCCCGGGCCCGGCTCTCTAGCGCTTATCTGCTGGGAGTTCCAGGCTCAGAGCAG CCTGACCGCGCTGGCAGCCTGGAGCTTCGTGGCTTCAAGAATAAGCTGTACGTGGCCGTGGTCGGGGACAAAGTGCAGCTCTACAAGAAT CTAGAGGAGTACCACCTGGGCATTGGCATCACCTTCATCGACATGAGCGTGGGCAACGTGAAGGAAGTGGACCGGCGCAGCTTCGACCTC ACCACGCCCTACCGCATCTTCAGCTTCTCTGCTGACTCAGAGCTAGAGAAGGAGCAGTGGTTGGAGGCCATGCAGGGAGCCATCGCTGAG GCCCTGTCTACCTCGGAGGTGGCCGAGCGCATCTGGGCTGCAGCCCCCAACAGGTTCTGTGCTGACTGCGGGGCTCCTCAGCCTGACTGG GCCTCCATCAACCTCTGTGTTGTTATCTGCAAGCGCTGTGCAGGGGAGCACCGTGGCCTGGGCGCTGGCGTCTCCAAGGTGCGGAGCCTG AAGATGGACAGGAAGGTGTGGACAGAAACACTTATCGAGCTCTTCTTACAGCTGGGGAATGGCGCTGGGAACCGCTTCTGGGCAGCCAAC GTGCCCCCCAGTGAGGCCCTGCAGCCCAGCAGCAGCCCCAGCACCCGGCGGTGCCACCTGGAGGCCAAGTACCGTGAGGGCAAGTACCGC CGCTACCACCCGCTCTTTGGCAACCAGGAGGAGCTGGACAAGGCCCTGTGTGCTGCAGTCACCACCACAGACCTGGCTGAGACCCAGGCG CTCCTGGGCTGTGGGGCTGGGATCAACTGCTTCTCGGGGGACCCTGAGGCCCCCACGCCCCTGGCTCTTGCAGAGCAGGCGGGGCAGACG CTGCAGATGGAATTCCTTCGGAACAACCGGACCACAGAGGTGCCTCGGCTGGACTCGATGAAGCCCCTGGAAAAGCACTACTCAGTTGTC CTGCCGACCGTGAGCCACAGTGGCTTCCTCTACAAGACTGCCTCTGCCGGCAAGCTGCTACAGGACCGCCGGGCCCGGGAAGACCTGTGT GCAGCATTTAACGTCATATGTGATAATGTGGGGAAAGATTGGAGAAGGCTGGCTCGTCAGCTCAAAGTCTCAGACACCAAGATCGACAGC ATCGAGGACAGATACCCCCGCAACCTGACAGAGCGTGTGCGGGAGTCACTGAGAATCTGGAAGAACACAGAGAAGGAGAACGCAACAGTG GCCCACCTGGTGGGGGCTCTCAGGTCCTGCCAGATGAACCTGGTGGCTGACCTGGTACAAGAGGTTCAGCAGGCCCGTGACCTCCAGAAC AGGAGTGGGGCCATGTCCCCGATGTCATGGAACTCAGACGCATCTACCTCCGAAGCGTCCTGATGGGCCGCTGCTTTGCGCTGGTGGACC ACAGGCATCTACACAGCCTGGACTTTGGTTCTCTCCAGGAAGGTAGCCCAGCACTGTGAAGACCCAGCAGGAAGCCAGGCTGAGTGAGCC ACAGACCACCTGCTTCTGAACTCAAGCTGCGTTTATTAATGCCTCTCCCGCACCAGGCCGGGCTTGGGCCCTGCACAGATATTTCCATTT CTTCCTCACTATGACACTGAGCAAGATCTTGTCTCCACTAAATGAGCTCCTGCGGGAGTAGTTGGAAAGTTGGAACCGTGTCCAGCACAG AAGGAATCTGTGCAGATGAGCAGTCACACTGTTACTCCACAGCGGAGGAGACCAGCTCAGAGGCCCAGGAATCGGAGCGAAGCAGAGAGG TGGAGAACTGGGATTTGAACCCCCGCCATCCTTCACCAGAGCCCATGCTCAACCACTGTGGCGTTCTGCTGCCCCTGCAGTTGGCAGAAA GGATGTTTTGTCCCATTTCCTTGGAGGCCACCGGGACAGACCTGGACACTAGGGTCAGGCGGGGTGCTGTGGTGGGGAGAGGCATGGCTG GGGTGGGGGTGGGGAGACCTGGTTGGCCGTGGTCCAGCTCTTGGCCCCTGTGTGAGTTGAGTCTCCTCTCTGAGACTGCTAAGTAGGGGC AGTGATGGTTGCCAGGACGAATTGAGATAATATCTGTGAGGTGCTGATGAGTGATTGACACACAGCACTCTCTAAATCTTCCTTGTGAGG ATTATGGGTCCTGCAATTCTACAGTTTCTTACTGTTTTGTATCAAAATCACTATCTTTCTGATAACAGAATTGCCAAGGCAGCGGGATCT CGTATCTTTAAAAAGCAGTCCTCTTATTCCTAAGGTAATCCTATTAAAACACAGCTTTACAACTTCCATACTACAAAAAA >5757_5757_4_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000426523_FADD_chr11_70052239_ENST00000301838_length(amino acids)=710AA_BP=597 MPTKEEESLLPSLSSPPQPQSEEPLSTLPQGPPQPPSPPPCPPEIPPKPVRLFPEFDDSDYDEVPEEGPGAPARVMTKKEEPPPSRVPRA VRVASLLSEGEELSGDDQGDEEEDDHAYEGVPNGGWHTSSLSLSLPSTIAAPHPMDGPPGGSTPVTPVIKAGWLDKNPPQGSYIYQKRWV RLDTDHLRYFDSNKDAYSKRFISVACISHVAAIGDQKFEVITNNRTFAFRAESDVERKEWMQALQQAMAEQRARARLSSAYLLGVPGSEQ PDRAGSLELRGFKNKLYVAVVGDKVQLYKNLEEYHLGIGITFIDMSVGNVKEVDRRSFDLTTPYRIFSFSADSELEKEQWLEAMQGAIAE ALSTSEVAERIWAAAPNRFCADCGAPQPDWASINLCVVICKRCAGEHRGLGAGVSKVRSLKMDRKVWTETLIELFLQLGNGAGNRFWAAN VPPSEALQPSSSPSTRRCHLEAKYREGKYRRYHPLFGNQEELDKALCAAVTTTDLAETQALLGCGAGINCFSGDPEAPTPLALAEQAGQT LQMEFLRNNRTTEVPRLDSMKPLEKHYSVVLPTVSHSGFLYKTASAGKLLQDRRAREDLCAAFNVICDNVGKDWRRLARQLKVSDTKIDS IEDRYPRNLTERVRESLRIWKNTEKENATVAHLVGALRSCQMNLVADLVQEVQQARDLQNRSGAMSPMSWNSDASTSEAS -------------------------------------------------------------- >5757_5757_5_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000429686_FADD_chr11_70052239_ENST00000301838_length(transcript)=3047nt_BP=1789nt ACCTCCAAGAGAACGCAGCCTCAGACCCACCCAGGGGCCAGGGGCCAGGCTGTTTGCGAGACCAGCCCAGGCGGGACCCAGGCTTGCACG GGCAGGTACACGACATTCTTGGGCATACGCAGCCCGCCTGGCCGGAGCTGTGGGAGTCCTCAGCCCCAAGACCCAGCAGGCGTCTGAGGC CTGCCCACTAAGGAGGAGGAGTCATTGCTGCCATCATTATCATCCCCTCCCCAGCCACAGTCTGAGGAGCCCCTGTCCACCCTCCCCCAG GGGCCTCCCCAGCCTCCCTCTCCACCTCCCTGCCCCCCGGAGATACCTCCAAAGCCGGTACGCCTGTTCCCAGAGTTCGATGACTCTGAC TACGATGAGGTCCCAGAGGAGGGGCCGGGGGCCCCAGCCAGAGTGATGACCAAGAAGGAGGAGCCCCCACCGAGCCGAGTCCCACGGGCC GTGCGCGTGGCCAGTCTGCTGAGCGAGGGAGAGGAACTGTCTGGGGACGACCAAGGGGATGAGGAAGAGGATGACCACGCCTATGAGGGC GTCCCCAATGGCGGATGGCATACCAGCAGCCTGAGCTTGTCCTTGCCCAGCACAATAGCTGCGCCACACCCCATGGACGGGCCGCCTGGG GGCTCCACCCCCGTCACACCAGTCATCAAGGCTGGCTGGCTGGACAAGAACCCACCGCAGGGATCTTACATCTATCAGAAACGATGGGTG AGACTGGATACTGATCACCTGCGATACTTTGACAGTAACAAGGACGCTTACTCTAAGCGCTTTATCTCTGTGGCCTGCATCTCCCACGTG GCTGCCATCGGGGACCAGAAGTTTGAAGTGATCACAAACAACCGAACCTTTGCCTTCCGGGCAGAGAGTGATGTGGAGCGGAAGGAGTGG ATGCAGGCCCTGCAGCAGGCCATGGCTGAGCAGCGTGCCCGGGCCCGGCTCTCTAGCGCTTATCTGCTGGGAGTTCCAGGCTCAGAGCAG CCTGACCGCGCTGGCAGCCTGGAGCTTCGTGGCTTCAAGAATAAGCTGTACGTGGCCGTGGTCGGGGACAAAGTGCAGCTCTACAAGAAT CTAGAGGAGTACCACCTGGGCATTGGCATCACCTTCATCGACATGAGCGTGGGCAACGTGAAGGAAGTGGACCGGCGCAGCTTCGACCTC ACCACGCCCTACCGCATCTTCAGCTTCTCTGCTGACTCAGAGCTAGAGAAGGAGCAGTGGTTGGAGGCCATGCAGGGAGCCATCGCTGAG GCCCTGTCTACCTCGGAGGTGGCCGAGCGCATCTGGGCTGCAGCCCCCAACAGGTTCTGTGCTGACTGCGGGGCTCCTCAGCCTGACTGG GCCTCCATCAACCTCTGTGTTGTTATCTGCAAGCGCTGTGCAGGGGAGCACCGTGGCCTGGGCGCTGGCGTCTCCAAGGTGCGGAGCCTG AAGATGGACAGGAAGGTGTGGACAGAAACACTTATCGAGGCCCTGTGTGCTGCAGTCACCACCACAGACCTGGCTGAGACCCAGGCGCTC CTGGGCTGTGGGGCTGGGATCAACTGCTTCTCGGGGGACCCTGAGGCCCCCACGCCCCTGGCTCTTGCAGAGCAGGCGGGGCAGACGCTG CAGATGGAATTCCTTCGGAACAACCGGACCACAGAGGTGCCTCGGCTGGACTCGATGAAGCCCCTGGAAAAGCACTACTCAGTTGTCCTG CCGACCGTGAGCCACAGTGGCTTCCTCTACAAGACTGCCTCTGCCGGCAAGCTGCTACAGGACCGCCGGGCCCGGGAAGACCTGTGTGCA GCATTTAACGTCATATGTGATAATGTGGGGAAAGATTGGAGAAGGCTGGCTCGTCAGCTCAAAGTCTCAGACACCAAGATCGACAGCATC GAGGACAGATACCCCCGCAACCTGACAGAGCGTGTGCGGGAGTCACTGAGAATCTGGAAGAACACAGAGAAGGAGAACGCAACAGTGGCC CACCTGGTGGGGGCTCTCAGGTCCTGCCAGATGAACCTGGTGGCTGACCTGGTACAAGAGGTTCAGCAGGCCCGTGACCTCCAGAACAGG AGTGGGGCCATGTCCCCGATGTCATGGAACTCAGACGCATCTACCTCCGAAGCGTCCTGATGGGCCGCTGCTTTGCGCTGGTGGACCACA GGCATCTACACAGCCTGGACTTTGGTTCTCTCCAGGAAGGTAGCCCAGCACTGTGAAGACCCAGCAGGAAGCCAGGCTGAGTGAGCCACA GACCACCTGCTTCTGAACTCAAGCTGCGTTTATTAATGCCTCTCCCGCACCAGGCCGGGCTTGGGCCCTGCACAGATATTTCCATTTCTT CCTCACTATGACACTGAGCAAGATCTTGTCTCCACTAAATGAGCTCCTGCGGGAGTAGTTGGAAAGTTGGAACCGTGTCCAGCACAGAAG GAATCTGTGCAGATGAGCAGTCACACTGTTACTCCACAGCGGAGGAGACCAGCTCAGAGGCCCAGGAATCGGAGCGAAGCAGAGAGGTGG AGAACTGGGATTTGAACCCCCGCCATCCTTCACCAGAGCCCATGCTCAACCACTGTGGCGTTCTGCTGCCCCTGCAGTTGGCAGAAAGGA TGTTTTGTCCCATTTCCTTGGAGGCCACCGGGACAGACCTGGACACTAGGGTCAGGCGGGGTGCTGTGGTGGGGAGAGGCATGGCTGGGG TGGGGGTGGGGAGACCTGGTTGGCCGTGGTCCAGCTCTTGGCCCCTGTGTGAGTTGAGTCTCCTCTCTGAGACTGCTAAGTAGGGGCAGT GATGGTTGCCAGGACGAATTGAGATAATATCTGTGAGGTGCTGATGAGTGATTGACACACAGCACTCTCTAAATCTTCCTTGTGAGGATT ATGGGTCCTGCAATTCTACAGTTTCTTACTGTTTTGTATCAAAATCACTATCTTTCTGATAACAGAATTGCCAAGGCAGCGGGATCTCGT ATCTTTAAAAAGCAGTCCTCTTATTCCTAAGGTAATCCTATTAAAACACAGCTTTACAACTTCCATACTACAAAAAA >5757_5757_5_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000429686_FADD_chr11_70052239_ENST00000301838_length(amino acids)=649AA_BP=536 MPTKEEESLLPSLSSPPQPQSEEPLSTLPQGPPQPPSPPPCPPEIPPKPVRLFPEFDDSDYDEVPEEGPGAPARVMTKKEEPPPSRVPRA VRVASLLSEGEELSGDDQGDEEEDDHAYEGVPNGGWHTSSLSLSLPSTIAAPHPMDGPPGGSTPVTPVIKAGWLDKNPPQGSYIYQKRWV RLDTDHLRYFDSNKDAYSKRFISVACISHVAAIGDQKFEVITNNRTFAFRAESDVERKEWMQALQQAMAEQRARARLSSAYLLGVPGSEQ PDRAGSLELRGFKNKLYVAVVGDKVQLYKNLEEYHLGIGITFIDMSVGNVKEVDRRSFDLTTPYRIFSFSADSELEKEQWLEAMQGAIAE ALSTSEVAERIWAAAPNRFCADCGAPQPDWASINLCVVICKRCAGEHRGLGAGVSKVRSLKMDRKVWTETLIEALCAAVTTTDLAETQAL LGCGAGINCFSGDPEAPTPLALAEQAGQTLQMEFLRNNRTTEVPRLDSMKPLEKHYSVVLPTVSHSGFLYKTASAGKLLQDRRAREDLCA AFNVICDNVGKDWRRLARQLKVSDTKIDSIEDRYPRNLTERVRESLRIWKNTEKENATVAHLVGALRSCQMNLVADLVQEVQQARDLQNR SGAMSPMSWNSDASTSEAS -------------------------------------------------------------- >5757_5757_6_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000455638_FADD_chr11_70052239_ENST00000301838_length(transcript)=3560nt_BP=2302nt ATGGCAGAGGCTGGGGATGCTGCGCTATCGGTGGCCGAGTGGCTGCGGGCATTGCACCTGGAGCAGTACACGGGGCTCTTTGAGCAGCAT GGCCTGGTGTGGGCCACTGAGTGCCAAGGCCTCAGCGACACCCGCCTGATGGACATGGGCATGCTACTCCCTGGTCACCGCCGCCGCATC CTGGCTGGCCTGCTCCGTGCCCATACCTCACCGGCCCCTGCACCCCGCCCCACCCCACGGCCTGTGCCCATGAAGCGCCACATCTTCCGC TCACCACCTGTGCCTGCCACTCCACCCGAGCCGCTGCCCACCACTACAGAGGATGAGGGGCTCCCCGCTGCCCCACCCATCCCGCCCCGG AGGAGCTGCCTTCCGCCCACCTGCTTCACCACCCCATCCACAGCTGCCCCAGACCCTGTGCTGCCCCCGCTGCCTGCTAAGCGGCATTTG GCAGAGCTGAGCGTTCCACCCGTGCCGCCCCGCACCGGACCCCCCCGCCTGCTGGTGAGCCTGCCCACTAAGGAGGAGGAGTCATTGCTG CCATCATTATCATCCCCTCCCCAGCCACAGTCTGAGGAGCCCCTGTCCACCCTCCCCCAGGGGCCTCCCCAGCCTCCCTCTCCACCTCCC TGCCCCCCGGAGATACCTCCAAAGCCGGTACGCCTGTTCCCAGAGTTCGATGACTCTGACTACGATGAGGTCCCAGAGGAGGGGCCGGGG GCCCCAGCCAGAGTGATGACCAAGAAGGAGGAGCCCCCACCGAGCCGAGTCCCACGGGCCGTGCGCGTGGCCAGTCTGCTGAGCGAGGGA GAGGAACTGTCTGGGGACGACCAAGGGGATGAGGAAGAGGATGACCACGCCTATGAGGGCGTCCCCAATGGCGGATGGCATACCAGCAGC CTGAGCTTGTCCTTGCCCAGCACAATAGCTGCGCCACACCCCATGGACGGGCCGCCTGGGGGCTCCACCCCCGTCACACCAGTCATCAAG GCTGGCTGGCTGGACAAGAACCCACCGCAGGGATCTTACATCTATCAGAAACGATGGGTGAGACTGGATACTGATCACCTGCGATACTTT GACAGTAACAAGGACGCTTACTCTAAGCGCTTTATCTCTGTGGCCTGCATCTCCCACGTGGCTGCCATCGGGGACCAGAAGTTTGAAGTG ATCACAAACAACCGAACCTTTGCCTTCCGGGCAGAGAGTGATGTGGAGCGGAAGGAGTGGATGCAGGCCCTGCAGCAGGCCATGGCTGAG CAGCGTGCCCGGGCCCGGCTCTCTAGCGCTTATCTGCTGGGAGTTCCAGGCTCAGAGCAGCCTGACCGCGCTGGCAGCCTGGAGCTTCGT GGCTTCAAGAATAAGCTGTACGTGGCCGTGGTCGGGGACAAAGTGCAGCTCTACAAGAATCTAGAGGAGTACCACCTGGGCATTGGCATC ACCTTCATCGACATGAGCGTGGGCAACGTGAAGGAAGTGGACCGGCGCAGCTTCGACCTCACCACGCCCTACCGCATCTTCAGCTTCTCT GCTGACTCAGAGCTAGAGAAGGAGCAGTGGTTGGAGGCCATGCAGGGAGCCATCGCTGAGGCCCTGTCTACCTCGGAGGTGGCCGAGCGC ATCTGGGCTGCAGCCCCCAACAGGTTCTGTGCTGACTGCGGGGCTCCTCAGCCTGACTGGGCCTCCATCAACCTCTGTGTTGTTATCTGC AAGCGCTGTGCAGGGGAGCACCGTGGCCTGGGCGCTGGCGTCTCCAAGGTGCGGAGCCTGAAGATGGACAGGAAGGTGTGGACAGAAACA CTTATCGAGCTCTTCTTACAGCTGGGGAATGGCGCTGGGAACCGCTTCTGGGCAGCCAACGTGCCCCCCAGTGAGGCCCTGCAGCCCAGC AGCAGCCCCAGCACCCGGCGGTGCCACCTGGAGGCCAAGTACCGTGAGGGCAAGTACCGCCGCTACCACCCGCTCTTTGGCAACCAGGAG GAGCTGGACAAGGCCCTGTGTGCTGCAGTCACCACCACAGACCTGGCTGAGACCCAGGCGCTCCTGGGCTGTGGGGCTGGGATCAACTGC TTCTCGGGGGACCCTGAGGCCCCCACGCCCCTGGCTCTTGCAGAGCAGGCGGGGCAGACGCTGCAGATGGAATTCCTTCGGAACAACCGG ACCACAGAGGTGCCTCGGCTGGACTCGATGAAGCCCCTGGAAAAGCACTACTCAGTTGTCCTGCCGACCGTGAGCCACAGTGGCTTCCTC TACAAGACTGCCTCTGCCGGCAAGCTGCTACAGGACCGCCGGGCCCGGGAAGACCTGTGTGCAGCATTTAACGTCATATGTGATAATGTG GGGAAAGATTGGAGAAGGCTGGCTCGTCAGCTCAAAGTCTCAGACACCAAGATCGACAGCATCGAGGACAGATACCCCCGCAACCTGACA GAGCGTGTGCGGGAGTCACTGAGAATCTGGAAGAACACAGAGAAGGAGAACGCAACAGTGGCCCACCTGGTGGGGGCTCTCAGGTCCTGC CAGATGAACCTGGTGGCTGACCTGGTACAAGAGGTTCAGCAGGCCCGTGACCTCCAGAACAGGAGTGGGGCCATGTCCCCGATGTCATGG AACTCAGACGCATCTACCTCCGAAGCGTCCTGATGGGCCGCTGCTTTGCGCTGGTGGACCACAGGCATCTACACAGCCTGGACTTTGGTT CTCTCCAGGAAGGTAGCCCAGCACTGTGAAGACCCAGCAGGAAGCCAGGCTGAGTGAGCCACAGACCACCTGCTTCTGAACTCAAGCTGC GTTTATTAATGCCTCTCCCGCACCAGGCCGGGCTTGGGCCCTGCACAGATATTTCCATTTCTTCCTCACTATGACACTGAGCAAGATCTT GTCTCCACTAAATGAGCTCCTGCGGGAGTAGTTGGAAAGTTGGAACCGTGTCCAGCACAGAAGGAATCTGTGCAGATGAGCAGTCACACT GTTACTCCACAGCGGAGGAGACCAGCTCAGAGGCCCAGGAATCGGAGCGAAGCAGAGAGGTGGAGAACTGGGATTTGAACCCCCGCCATC CTTCACCAGAGCCCATGCTCAACCACTGTGGCGTTCTGCTGCCCCTGCAGTTGGCAGAAAGGATGTTTTGTCCCATTTCCTTGGAGGCCA CCGGGACAGACCTGGACACTAGGGTCAGGCGGGGTGCTGTGGTGGGGAGAGGCATGGCTGGGGTGGGGGTGGGGAGACCTGGTTGGCCGT GGTCCAGCTCTTGGCCCCTGTGTGAGTTGAGTCTCCTCTCTGAGACTGCTAAGTAGGGGCAGTGATGGTTGCCAGGACGAATTGAGATAA TATCTGTGAGGTGCTGATGAGTGATTGACACACAGCACTCTCTAAATCTTCCTTGTGAGGATTATGGGTCCTGCAATTCTACAGTTTCTT ACTGTTTTGTATCAAAATCACTATCTTTCTGATAACAGAATTGCCAAGGCAGCGGGATCTCGTATCTTTAAAAAGCAGTCCTCTTATTCC TAAGGTAATCCTATTAAAACACAGCTTTACAACTTCCATACTACAAAAAA >5757_5757_6_ARAP1-FADD_ARAP1_chr11_72412694_ENST00000455638_FADD_chr11_70052239_ENST00000301838_length(amino acids)=880AA_BP=767 MAEAGDAALSVAEWLRALHLEQYTGLFEQHGLVWATECQGLSDTRLMDMGMLLPGHRRRILAGLLRAHTSPAPAPRPTPRPVPMKRHIFR SPPVPATPPEPLPTTTEDEGLPAAPPIPPRRSCLPPTCFTTPSTAAPDPVLPPLPAKRHLAELSVPPVPPRTGPPRLLVSLPTKEEESLL PSLSSPPQPQSEEPLSTLPQGPPQPPSPPPCPPEIPPKPVRLFPEFDDSDYDEVPEEGPGAPARVMTKKEEPPPSRVPRAVRVASLLSEG EELSGDDQGDEEEDDHAYEGVPNGGWHTSSLSLSLPSTIAAPHPMDGPPGGSTPVTPVIKAGWLDKNPPQGSYIYQKRWVRLDTDHLRYF DSNKDAYSKRFISVACISHVAAIGDQKFEVITNNRTFAFRAESDVERKEWMQALQQAMAEQRARARLSSAYLLGVPGSEQPDRAGSLELR GFKNKLYVAVVGDKVQLYKNLEEYHLGIGITFIDMSVGNVKEVDRRSFDLTTPYRIFSFSADSELEKEQWLEAMQGAIAEALSTSEVAER IWAAAPNRFCADCGAPQPDWASINLCVVICKRCAGEHRGLGAGVSKVRSLKMDRKVWTETLIELFLQLGNGAGNRFWAANVPPSEALQPS SSPSTRRCHLEAKYREGKYRRYHPLFGNQEELDKALCAAVTTTDLAETQALLGCGAGINCFSGDPEAPTPLALAEQAGQTLQMEFLRNNR TTEVPRLDSMKPLEKHYSVVLPTVSHSGFLYKTASAGKLLQDRRAREDLCAAFNVICDNVGKDWRRLARQLKVSDTKIDSIEDRYPRNLT ERVRESLRIWKNTEKENATVAHLVGALRSCQMNLVADLVQEVQQARDLQNRSGAMSPMSWNSDASTSEAS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ARAP1-FADD |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ARAP1-FADD |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ARAP1-FADD |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C3151062 | INFECTIONS, RECURRENT, WITH ENCEPHALOPATHY, HEPATIC DYSFUNCTION, AND CARDIOVASCULAR MALFORMATIONS | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0018784 | Sensorineural Hearing Loss (disorder) | 1 | CTD_human | |
| Tgene | C0027627 | Neoplasm Metastasis | 1 | CTD_human | |
| Tgene | C0038368 | Stomatognathic Diseases | 1 | CTD_human | |
| Tgene | C0376634 | Craniofacial Abnormalities | 1 | CTD_human | |
| Tgene | C1691779 | Sensory hearing loss | 1 | CTD_human | |
| Tgene | C1704330 | Dental Diseases | 1 | CTD_human | |
| Tgene | C2750325 | Oculootodental syndrome | 1 | ORPHANET | |
| Tgene | C3279150 | Functional hyposplenism | 1 | GENOMICS_ENGLAND | |
| Tgene | C4049796 | Abnormality of cardiovascular system morphology | 1 | GENOMICS_ENGLAND | |
| Tgene | C4317009 | Diverticular Diseases | 1 | CTD_human | |
| Tgene | C4505353 | Diverticular Bleeding | 1 | CTD_human |
