
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CRK-ABR (FusionGDB2 ID:HG1398TG29) |
Fusion Gene Summary for CRK-ABR |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CRK-ABR | Fusion gene ID: hg1398tg29 | Hgene | Tgene | Gene symbol | CRK | ABR | Gene ID | 1398 | 29 |
| Gene name | CRK proto-oncogene, adaptor protein | ABR activator of RhoGEF and GTPase | |
| Synonyms | CRKII|p38 | MDB | |
| Cytomap | ('CRK')('ABR') 17p13.3 | 17p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | adapter molecule crkproto-oncogene c-Crkv-crk avian sarcoma virus CT10 oncogene homologv-crk sarcoma virus CT10 oncogene-like protein | active breakpoint cluster region-related proteinABR, RhoGEF and GTPase activating proteinactive BCR-related | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P46108 | Q12979 | |
| Ensembl transtripts involved in fusion gene | ENST00000398970, ENST00000572145, ENST00000300574, ENST00000574295, | ENST00000572145, ENST00000300574, ENST00000398970, ENST00000574295, | |
| Fusion gene scores | * DoF score | 15 X 11 X 7=1155 | 17 X 14 X 9=2142 |
| # samples | 18 | 21 | |
| ** MAII score | log2(18/1155*10)=-2.68182403997375 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(21/2142*10)=-3.35049724708413 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CRK [Title/Abstract] AND ABR [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CRK(1340084)-ABR(1028702), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ABR-CRK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ABR-CRK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CRK-ABR seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CRK-ABR seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CRK-ABR seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CRK-ABR seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ABR-CRK seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CRK | GO:0009966 | regulation of signal transduction | 17515907 |
| Hgene | CRK | GO:0032956 | regulation of actin cytoskeleton organization | 11870224 |
| Hgene | CRK | GO:0043087 | regulation of GTPase activity | 11870224 |
| Hgene | CRK | GO:0048013 | ephrin receptor signaling pathway | 11870224 |
| Hgene | CRK | GO:0061045 | negative regulation of wound healing | 17515907 |
| Hgene | CRK | GO:2000146 | negative regulation of cell motility | 17515907 |
| Tgene | ABR | GO:0090630 | activation of GTPase activity | 7479768 |
 Fusion gene breakpoints across CRK (5'-gene) Fusion gene breakpoints across CRK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
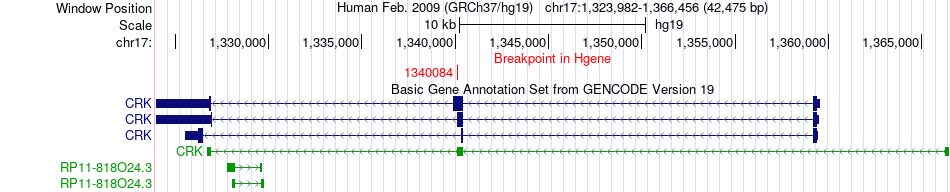 |
 Fusion gene breakpoints across ABR (3'-gene) Fusion gene breakpoints across ABR (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
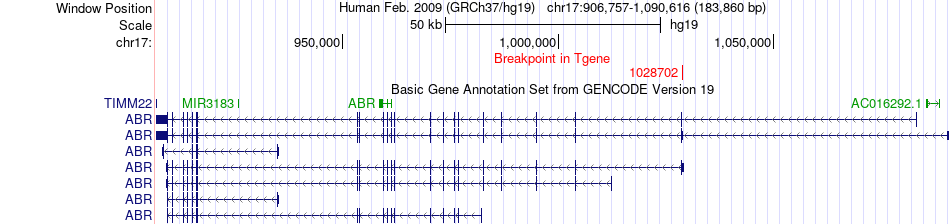 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A1KY-01A | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
Top |
Fusion Gene ORF analysis for CRK-ABR |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000398970 | ENST00000544583 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5CDS-5UTR | ENST00000398970 | ENST00000574437 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5CDS-intron | ENST00000398970 | ENST00000291107 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5CDS-intron | ENST00000398970 | ENST00000536794 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5CDS-intron | ENST00000398970 | ENST00000543210 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5CDS-intron | ENST00000398970 | ENST00000572441 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5CDS-intron | ENST00000398970 | ENST00000573895 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-3CDS | ENST00000572145 | ENST00000302538 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-5UTR | ENST00000572145 | ENST00000544583 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-5UTR | ENST00000572145 | ENST00000574437 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-intron | ENST00000572145 | ENST00000291107 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-intron | ENST00000572145 | ENST00000536794 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-intron | ENST00000572145 | ENST00000543210 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-intron | ENST00000572145 | ENST00000572441 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| 5UTR-intron | ENST00000572145 | ENST00000573895 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| In-frame | ENST00000398970 | ENST00000302538 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-3CDS | ENST00000300574 | ENST00000302538 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-3CDS | ENST00000574295 | ENST00000302538 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-5UTR | ENST00000300574 | ENST00000544583 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-5UTR | ENST00000300574 | ENST00000574437 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-5UTR | ENST00000574295 | ENST00000544583 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-5UTR | ENST00000574295 | ENST00000574437 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000300574 | ENST00000291107 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000300574 | ENST00000536794 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000300574 | ENST00000543210 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000300574 | ENST00000572441 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000300574 | ENST00000573895 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000574295 | ENST00000291107 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000574295 | ENST00000536794 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000574295 | ENST00000543210 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000574295 | ENST00000572441 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
| intron-intron | ENST00000574295 | ENST00000573895 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000398970 | CRK | chr17 | 1340084 | - | ENST00000302538 | ABR | chr17 | 1028702 | - | 5794 | 713 | 106 | 3231 | 1041 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000398970 | ENST00000302538 | CRK | chr17 | 1340084 | - | ABR | chr17 | 1028702 | - | 0.001456703 | 0.99854326 |
Top |
Fusion Genomic Features for CRK-ABR |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
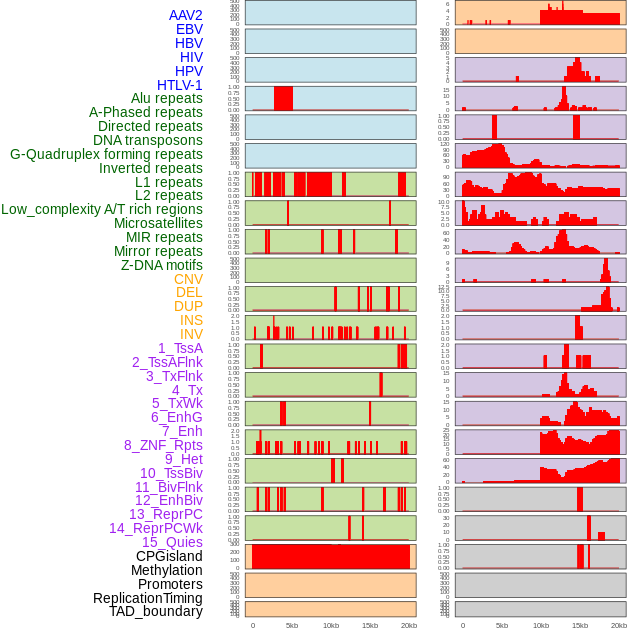 |
Top |
Fusion Protein Features for CRK-ABR |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:1340084/chr17:1028702) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CRK | ABR |
| FUNCTION: Involved in cell branching and adhesion mediated by BCAR1-CRK-RAPGEF1 signaling and activation of RAP1. {ECO:0000269|PubMed:12432078}.; FUNCTION: [Isoform Crk-II]: Regulates cell adhesion, spreading and migration (PubMed:31311869). Mediates attachment-induced MAPK8 activation, membrane ruffling and cell motility in a Rac-dependent manner. Involved in phagocytosis of apoptotic cells and cell motility via its interaction with DOCK1 and DOCK4 (PubMed:19004829). May regulate the EFNA5-EPHA3 signaling (By similarity). {ECO:0000250|UniProtKB:Q64010, ECO:0000269|PubMed:11870224, ECO:0000269|PubMed:1630456, ECO:0000269|PubMed:17515907, ECO:0000269|PubMed:19004829, ECO:0000269|PubMed:31311869}. | FUNCTION: Protein with a unique structure having two opposing regulatory activities toward small GTP-binding proteins. The C-terminus is a GTPase-activating protein domain which stimulates GTP hydrolysis by RAC1, RAC2 and CDC42. Accelerates the intrinsic rate of GTP hydrolysis of RAC1 or CDC42, leading to down-regulation of the active GTP-bound form (PubMed:7479768, PubMed:17116687). The central Dbl homology (DH) domain functions as guanine nucleotide exchange factor (GEF) that modulates the GTPases CDC42, RHOA and RAC1. Promotes the conversion of CDC42, RHOA and RAC1 from the GDP-bound to the GTP-bound form (PubMed:7479768). Functions as an important negative regulator of neuronal RAC1 activity (By similarity). Regulates macrophage functions such as CSF-1 directed motility and phagocytosis through the modulation of RAC1 activity (By similarity). {ECO:0000250|UniProtKB:Q5SSL4, ECO:0000269|PubMed:17116687, ECO:0000269|PubMed:7479768}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRK | chr17:1340084 | chr17:1028702 | ENST00000398970 | - | 2 | 3 | 132_192 | 202 | 205.0 | Domain | SH3 1 |
| Hgene | CRK | chr17:1340084 | chr17:1028702 | ENST00000398970 | - | 2 | 3 | 13_118 | 202 | 205.0 | Domain | SH2 |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000291107 | 0 | 22 | 417_420 | 0 | 823.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000302538 | 0 | 23 | 417_420 | 20 | 860.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000543210 | 0 | 8 | 417_420 | 0 | 311.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000544583 | 0 | 23 | 417_420 | 0 | 814.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000574437 | 0 | 22 | 417_420 | 0 | 814.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000291107 | 0 | 22 | 301_459 | 0 | 823.0 | Domain | PH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000291107 | 0 | 22 | 484_613 | 0 | 823.0 | Domain | C2 | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000291107 | 0 | 22 | 647_845 | 0 | 823.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000291107 | 0 | 22 | 91_284 | 0 | 823.0 | Domain | DH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000302538 | 0 | 23 | 301_459 | 20 | 860.0 | Domain | PH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000302538 | 0 | 23 | 484_613 | 20 | 860.0 | Domain | C2 | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000302538 | 0 | 23 | 647_845 | 20 | 860.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000302538 | 0 | 23 | 91_284 | 20 | 860.0 | Domain | DH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000543210 | 0 | 8 | 301_459 | 0 | 311.0 | Domain | PH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000543210 | 0 | 8 | 484_613 | 0 | 311.0 | Domain | C2 | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000543210 | 0 | 8 | 647_845 | 0 | 311.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000543210 | 0 | 8 | 91_284 | 0 | 311.0 | Domain | DH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000544583 | 0 | 23 | 301_459 | 0 | 814.0 | Domain | PH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000544583 | 0 | 23 | 484_613 | 0 | 814.0 | Domain | C2 | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000544583 | 0 | 23 | 647_845 | 0 | 814.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000544583 | 0 | 23 | 91_284 | 0 | 814.0 | Domain | DH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000574437 | 0 | 22 | 301_459 | 0 | 814.0 | Domain | PH | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000574437 | 0 | 22 | 484_613 | 0 | 814.0 | Domain | C2 | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000574437 | 0 | 22 | 647_845 | 0 | 814.0 | Domain | Rho-GAP | |
| Tgene | ABR | chr17:1340084 | chr17:1028702 | ENST00000574437 | 0 | 22 | 91_284 | 0 | 814.0 | Domain | DH |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CRK | chr17:1340084 | chr17:1028702 | ENST00000300574 | - | 1 | 3 | 132_192 | 0 | 305.0 | Domain | SH3 1 |
| Hgene | CRK | chr17:1340084 | chr17:1028702 | ENST00000300574 | - | 1 | 3 | 13_118 | 0 | 305.0 | Domain | SH2 |
| Hgene | CRK | chr17:1340084 | chr17:1028702 | ENST00000300574 | - | 1 | 3 | 235_296 | 0 | 305.0 | Domain | SH3 2 |
| Hgene | CRK | chr17:1340084 | chr17:1028702 | ENST00000398970 | - | 2 | 3 | 235_296 | 202 | 205.0 | Domain | SH3 2 |
Top |
Fusion Gene Sequence for CRK-ABR |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >19479_19479_1_CRK-ABR_CRK_chr17_1340084_ENST00000398970_ABR_chr17_1028702_ENST00000302538_length(transcript)=5794nt_BP=713nt GCTGCTGTGAAGCTGAAACCGGAGCCGGTCCGCTGGGCGGCGGGCGCCGGGGGCCGGAGGGGCGCGCGCGGCGGCGGCACCCCAGCGTTT AGGCGCGGAGGCAGCCATGGCGGGCAACTTCGACTCGGAGGAGCGGAGTAGCTGGTACTGGGGGAGGTTGAGTCGGCAGGAGGCGGTGGC GCTGCTGCAGGGCCAGCGGCACGGGGTGTTCCTGGTGCGGGACTCGAGCACCAGCCCCGGGGACTATGTGCTCAGCGTCTCAGAGAACTC GCGCGTCTCCCACTACATCATCAACAGCAGCGGCCCGCGCCCGCCGGTGCCACCGTCGCCCGCCCAGCCTCCGCCCGGGGTGAGCCCCTC CAGACTCCGAATAGGAGATCAAGAGTTTGATTCATTGCCTGCTTTACTGGAATTCTACAAAATACACTATTTGGACACTACAACGTTGAT AGAACCAGTTTCCAGATCCAGGCAGGGTAGTGGAGTGATTCTCAGGCAGGAGGAGGCGGAGTATGTGCGAGCCCTCTTTGACTTTAATGG GAATGATGAGGAAGATCTTCCCTTTAAGAAAGGAGACATCTTGAGAATCCGGGACAAGCCTGAAGAGCAGTGGTGGAATGCGGAGGACAG CGAAGGCAAGAGAGGGATGATTCCAGTCCCTTACGTCGAGAAGTATAGACCTGCCTCCGCCTCAGTATCGGCTCTGATTGGAGACTTCAG CTACGGGACGGACGAGTACGACGGAGAGGGGAATGAGGAGCAGAAGGGGCCCCCGGAGGGCTCAGAGACCATGCCGTACATCGATGAGTC GCCCACCATGTCCCCGCAGCTCAGCGCCCGCAGCCAGGGCGGGGGGGATGGCGTCTCCCCGACTCCACCTGAGGGACTGGCTCCTGGGGT GGAAGCAGGGAAAGGCCTGGAGATGAGGAAGCTGGTTCTCTCGGGGTTCTTGGCCAGCGAAGAGATCTACATTAACCAGCTGGAAGCCCT GTTGCTGCCCATGAAACCCCTGAAGGCCACCGCCACCACCTCCCAGCCCGTGCTCACCATCCAGCAGATCGAGACCATCTTCTACAAGAT CCAGGACATCTATGAGATCCACAAGGAGTTCTATGACAACCTGTGCCCCAAGGTGCAACAGTGGGACAGCCAGGTCACCATGGGCCACCT CTTCCAGAAGCTGGCCAGCCAGCTCGGTGTGTACAAAGCGTTTGTCGATAACTATAAAGTCGCTCTGGAGACAGCTGAGAAGTGCAGCCA GTCCAACAACCAGTTCCAGAAGATCTCAGAGGAACTCAAAGTGAAAGGTCCCAAGGACTCCAAGGACAGCCACACGTCTGTCACCATGGA AGCTCTGCTCTACAAGCCCATTGACCGGGTCACTCGGAGCACCCTAGTCCTACACGACCTGCTGAAGCACACACCTGTGGACCACCCCGA CTACCCGCTGCTGCAGGATGCCCTCCGCATCTCCCAGAACTTCCTGTCCAGCATCAACGAGGACATCGACCCCCGCCGGACTGCAGTGAC AACGCCCAAGGGGGAGACGCGACAGCTGGTGAAGGACGGCTTCCTGGTGGAAGTGTCAGAGAGCTCCCGGAAGCTGCGGCACGTCTTCCT CTTTACAGATGTCCTACTGTGTGCCAAGCTGAAGAAGACCTCTGCAGGGAAGCACCAGCAGTATGACTGTAAGTGGTACATCCCCCTGGC CGACCTGGTGTTTCCATCCCCCGAGGAGTCTGAGGCCAGCCCCCAGGTGCACCCCTTCCCAGACCATGAGCTGGAGGACATGAAGATGAA GATCTCTGCCCTCAAGAGTGAAATCCAGAAGGAGAAAGCCAACAAAGGCCAGAGCCGGGCCATCGAGCGCCTGAAGAAGAAGATGTTTGA GAATGAGTTCCTGCTGCTGCTCAACTCCCCCACAATCCCGTTCAGGATCCACAATCGGAATGGAAAGAGTTACCTGTTCCTACTGTCCTC GGACTACGAGAGGTCAGAGTGGAGAGAAGCAATTCAGAAACTACAGAAGAAGGATCTCCAGGCCTTTGTCCTGAGCTCAGTGGAGCTCCA GGTGCTCACAGGATCCTGTTTCAAGCTTAGGACTGTACACAACATTCCTGTCACCAGCAATAAAGACGACGATGAGTCTCCAGGACTCTA TGGCTTCCTTCATGTCATCGTCCACTCTGCCAAGGGATTTAAGCAATCAGCCAACCTGTACTGTACCCTGGAGGTGGATTCCTTCGGCTA TTTTGTCAGCAAAGCCAAAACCAGGGTGTTCCGGGACACAGCGGAGCCCAAGTGGGATGAGGAGTTTGAGATCGAGCTGGAGGGCTCCCA GTCCCTGAGGATCCTGTGCTATGAGAAGTGCTATGACAAGACCAAGGTCAACAAGGACAACAATGAGATCGTGGACAAGATCATGGGCAA AGGACAGATCCAGCTGGACCCACAAACCGTGGAGACCAAGAACTGGCACACGGACGTGATTGAGATGAACGGGATCAAAGTGGAATTTTC CATGAAATTCACCAGCCGAGATATGAGCCTGAAGAGGACCCCGTCCAAAAAGCAGACCGGCGTCTTCGGTGTGAAGATCAGCGTGGTGAC GAAGCGGGAGCGCTCCAAGGTGCCCTACATCGTCCGGCAGTGTGTGGAGGAGGTGGAGAAGAGGGGTATCGAGGAGGTTGGCATCTACAG GATATCGGGCGTGGCCACGGACATCCAGGCGCTCAAGGCCGTCTTCGATGCCAATAACAAGGACATCCTGCTGATGCTGAGTGACATGGA CATCAACGCCATCGCCGGGACGCTCAAGCTGTACTTCCGGGAACTGCCCGAGCCGCTCCTCACGGACCGACTCTACCCAGCCTTCATGGA GGGCATCGCCCTGTCAGACCCTGCTGCCAAGGAAAACTGCATGATGCACCTGCTCCGCTCCCTGCCCGACCCCAACCTCATCACCTTCCT CTTCCTGCTGGAACACTTGAAAAGGGTTGCCGAGAAGGAGCCCATCAACAAAATGTCACTTCACAACCTGGCTACCGTGTTTGGACCCAC GTTACTGAGACCCTCAGAAGTGGAGAGCAAAGCACACCTCACCTCGGCTGCGGACATCTGGTCCCATGACGTCATGGCGCAGGTCCAGGT CCTCCTCTACTACCTGCAGCACCCCCCCATTTCCTTCGCAGAACTCAAGCGGAACACACTGTACTTCTCCACCGACGTGTAGCCCGAGGC AGGGTGGCTGCGGGCGGGTGGTGGAACCAGCCCCTCCAGCCTGGGGTCCAACTCAGACTTGAAAGACTGCAATAGAAAACTCCCAAACCC AGCACTCCAGACTCGAGGGAAGCCAGCTTCCAAGAACTGGAATGCGTACGTCTTTTGTGCCACCTTGTACAAAGCCGGCTGCCCAGCCCC AGCCTCACCACCGCATCCCACCTCCTGCCCTCCATACCTCTAGTTGTGTCTGATGCTCCGTGCTGTTCGGGAATTGTTTTATGTACACTT GTCAGGCAGAAAAGGTAGTGACCGGCCCGGCGTGGGCACACAGACAGCCCGCTTTGTTCTTTCATTTCCTCCAGCACTTTCTTTCCGCCT GAGTCCAGCCCAAGGCCTTTTATTTTGCGCTGTGTAACTGCTGCCAGCTTCTCTCTTGGCCCTGCTCCCAGATGGCGGTCTCCTGGCAGC CTCCCCTCAGTCTTCCTCCACCCGCTCTTCCTTCCCAGCCTGCCTGCATGCATGTGCACCCTTGGTCTTCGCTCCATCGCCTTGAAAGCT CTGAAGAGGCCCTGGGTTGCCGCGGCAGCAGTGGTCTGTTTGATGCTGCCGTTTGCCGCTGCCGGCCCCTCCTCAGACTCCGCCTTTGGG AGCACACCTGCTTTGCCTTGCTGCCTGTGCAAATGTTGGACAAGCAGACACACTCACACTCGTCCCCAGCTTAGCACAGAGCTGGAGCGC CCATTTCTGGAATTTTCCGTTTGGGAATCTCCACTTCTGGGGTTTACCTGTTCGGCCTCCTGTCTATCAGTGAGGCATCTCTGACTGTTT CTTCTACTGCTTTTCAGTTCCCTTCCCTGCTGTTCTATTTCCTTTGAGTGTAAAGACTCACAGGTGACCTGCTATCGAGATAGCCAGAGG GTCAGGAGAGAATGGGGGAGGAGGCGGTCAGGCTGCTGAGGAAACACCACAGGCTGAACGGGGGAGGAATGCACATGCCACGCTGGGTGT CCCGGGTCGCGGGGAGGCAGCTCAGCTCTTAGGAGCAAGTTGTGGGGGCTTTTCAAGAGGGGCCAGGCTTCCTGGAGGGTGACTGATGTG GCCGAAGCAGGTGTCCAGGCAGGTAGGCTGCAGCCAGGAGCTCCCTGGCACCGCAGGACCTCGTGGTACTCTTGCCTTAGATTTTACACA CACTCCACAGCCAAGCACTGCCACGGTCCTCCAGGACCTGGGAAGCAAAGGCACAGGCCCACGGTGGCCAGCCATTGTGGTGCCGCCCCA GCTTCTGGATACAGCCTTTTGGGTAAACACTGGGAACTCCAGAAGTTGTGGGGAGAGTGGGGAATCAGACAGCCGCCTCTAGGGGCTGGG TTCTGCTGGGGCCTCCTTGTTGGTGCTGTAGGCACCCGCCAGGGAGCAGGGACCCGACTTGCAGACGCATTGCCCGGTACTAGGAAGGAG TGAGGTGTGTTCCCACCGTACACTTCCCACACGAGCTGCGGCTGCCAGCCTCGGGCCATCAGCCTAGGAGAGCAGATGCAGCTCCAGGGG CTCGACTTATAGCCAGTTACAGCTCCCCGGCTCTTCTGTGTGGCAGAGCGTCGTTTCCGGGCCCTCAGGGCTGGGGAGCTCAGTTCCCAT TGCTTGTGCTCAGGGCTGAGTCTTAAAGAAGGGTTTGCCGGCCCTAACGCTGCAGCGCGTGCGCGGTGAGAGGCCCTTTTTGAGCCTGTT TACTCCTGTGGCCTTGGGCAGAACAGTAAATACTCTGTGCACGGAGGAAAGACATGCCCAAGAGGAAGGAAGTACTGACCATCGGCTGCC TGTGAGCAGCTTAGCAAGGAGCCCTTGCTCCCTGGGAAAGGCGGTGAACTTGAGTCTAAAGATGCAGTGCCTGGCCCTTCCTAAGGTCCC TGCCTGGCATCCGAGTGTCGGTGTGTGGCACAGAAGGCTCCTGCTTGCTTCCAAAGTGATGGACAGGAAGGGGCAGAGTGAGTCACGGCC CAGACTGGGCACCTTCGCGTCTCAGCCTCAGGGAGCCCCACAGCCCCAAGCTCGCTGAGGCAACGTGAGAACAGGCTATGGGAAGGCTGC AAAGGCTGAGAAATGCAAAGGCTCATATTTATAAATCCCACCCCCAGAGTGGGGAGGGTCAGGTGCCAGACCTGGACTAAACTGCACCAA GGAAACACCCAGCAGGGTCTCCTGTGAGCCGGGGACCATGCAGCCCGAAACCTCCAGTCACTGCGCCCGGCAGGAGTCAGGAGCCAGGGA CTGTGCAGCCTGGAACCTCCAGTCACTGTGCCCAGCAGGGTGGGCTGTGCCCAGCAGGAGTCAGGCTAAGAAACGCCAGGTCTGCCTGTT CTTGCTGGGCAATGGCTGATGGCTGCCAGTTTCTGCTGATACACAGGTAGGATGGGACCCTTCATGAATATCTGACTTTAATAAGTTGGT AAGGATATATTTTTTTGTCTATGTTCTGTTTCAACTTATGTAGATTATTATAAATTGATGTAAACCACGTGAGAGGAAAATGTTAATAAA AAATGCAAAGCCCCATCATTTGCACAAAACTCAG >19479_19479_1_CRK-ABR_CRK_chr17_1340084_ENST00000398970_ABR_chr17_1028702_ENST00000302538_length(amino acids)=1041AA_BP=0 MAGNFDSEERSSWYWGRLSRQEAVALLQGQRHGVFLVRDSSTSPGDYVLSVSENSRVSHYIINSSGPRPPVPPSPAQPPPGVSPSRLRIG DQEFDSLPALLEFYKIHYLDTTTLIEPVSRSRQGSGVILRQEEAEYVRALFDFNGNDEEDLPFKKGDILRIRDKPEEQWWNAEDSEGKRG MIPVPYVEKYRPASASVSALIGDFSYGTDEYDGEGNEEQKGPPEGSETMPYIDESPTMSPQLSARSQGGGDGVSPTPPEGLAPGVEAGKG LEMRKLVLSGFLASEEIYINQLEALLLPMKPLKATATTSQPVLTIQQIETIFYKIQDIYEIHKEFYDNLCPKVQQWDSQVTMGHLFQKLA SQLGVYKAFVDNYKVALETAEKCSQSNNQFQKISEELKVKGPKDSKDSHTSVTMEALLYKPIDRVTRSTLVLHDLLKHTPVDHPDYPLLQ DALRISQNFLSSINEDIDPRRTAVTTPKGETRQLVKDGFLVEVSESSRKLRHVFLFTDVLLCAKLKKTSAGKHQQYDCKWYIPLADLVFP SPEESEASPQVHPFPDHELEDMKMKISALKSEIQKEKANKGQSRAIERLKKKMFENEFLLLLNSPTIPFRIHNRNGKSYLFLLSSDYERS EWREAIQKLQKKDLQAFVLSSVELQVLTGSCFKLRTVHNIPVTSNKDDDESPGLYGFLHVIVHSAKGFKQSANLYCTLEVDSFGYFVSKA KTRVFRDTAEPKWDEEFEIELEGSQSLRILCYEKCYDKTKVNKDNNEIVDKIMGKGQIQLDPQTVETKNWHTDVIEMNGIKVEFSMKFTS RDMSLKRTPSKKQTGVFGVKISVVTKRERSKVPYIVRQCVEEVEKRGIEEVGIYRISGVATDIQALKAVFDANNKDILLMLSDMDINAIA GTLKLYFRELPEPLLTDRLYPAFMEGIALSDPAAKENCMMHLLRSLPDPNLITFLFLLEHLKRVAEKEPINKMSLHNLATVFGPTLLRPS EVESKAHLTSAADIWSHDVMAQVQVLLYYLQHPPISFAELKRNTLYFSTDV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CRK-ABR |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CRK-ABR |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CRK-ABR |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CRK | C0151744 | Myocardial Ischemia | 1 | CTD_human |
| Tgene | C0033578 | Prostatic Neoplasms | 1 | CTD_human | |
| Tgene | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
