
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CTNNA1-MGME1 (FusionGDB2 ID:HG1495TG92667) |
Fusion Gene Summary for CTNNA1-MGME1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CTNNA1-MGME1 | Fusion gene ID: hg1495tg92667 | Hgene | Tgene | Gene symbol | CTNNA1 | MGME1 | Gene ID | 1495 | 92667 |
| Gene name | catenin alpha 1 | mitochondrial genome maintenance exonuclease 1 | |
| Synonyms | CAP102|MDPT2 | C20orf72|DDK1|MTDPS11|bA504H3.4 | |
| Cytomap | ('CTNNA1','CTNNA1')('C20orf72','MGME1') 5q31.2 | 20p11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | catenin alpha-1alpha-E-catenincatenin (cadherin-associated protein), alpha 1, 102kDaepididymis secretory sperm binding proteinrenal carcinoma antigen NY-REN-13 | mitochondrial genome maintenance exonuclease 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P35221 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000302763, ENST00000355078, ENST00000518825, ENST00000520400, ENST00000540387, | ||
| Fusion gene scores | * DoF score | 25 X 19 X 12=5700 | 6 X 5 X 4=120 |
| # samples | 34 | 7 | |
| ** MAII score | log2(34/5700*10)=-4.06735526780176 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/120*10)=-0.777607578663552 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CTNNA1 [Title/Abstract] AND MGME1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CTNNA1(138163407)-C20orf72(17968808), # samples:1 CTNNA1(138163407)-MGME1(17970582), # samples:1 CTNNA1(138163407)-MGME1(17968809), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CTNNA1-MGME1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CTNNA1-MGME1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CTNNA1-MGME1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CTNNA1-MGME1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CTNNA1 | GO:0071681 | cellular response to indole-3-methanol | 10868478 |
 Fusion gene breakpoints across CTNNA1 (5'-gene) Fusion gene breakpoints across CTNNA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
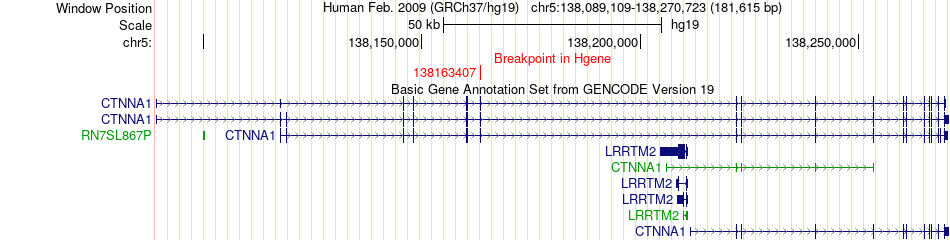 |
 Fusion gene breakpoints across MGME1 (3'-gene) Fusion gene breakpoints across MGME1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
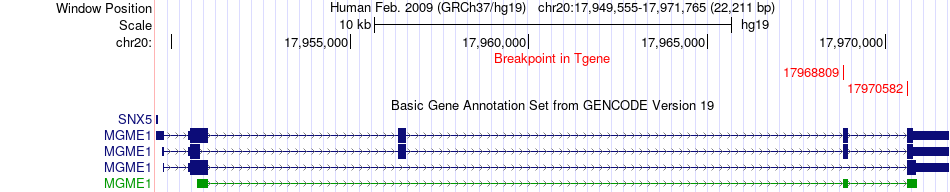 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-AP-A0LI | CTNNA1 | chr5 | 138163407 | + | C20orf72 | chr20 | 17968808 | + |
| ChimerDB4 | UCEC | TCGA-AP-A0LI-01A | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| ChimerDB4 | UCEC | TCGA-AP-A0LI-01A | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
Top |
Fusion Gene ORF analysis for CTNNA1-MGME1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000302763 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| 5CDS-3UTR | ENST00000302763 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| 5CDS-3UTR | ENST00000355078 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| 5CDS-3UTR | ENST00000355078 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| 5CDS-3UTR | ENST00000518825 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| 5CDS-3UTR | ENST00000518825 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| 5CDS-intron | ENST00000302763 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| 5CDS-intron | ENST00000355078 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| 5CDS-intron | ENST00000518825 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000302763 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000302763 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| Frame-shift | ENST00000302763 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000302763 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| Frame-shift | ENST00000355078 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000355078 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| Frame-shift | ENST00000355078 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000355078 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| Frame-shift | ENST00000518825 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000518825 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| Frame-shift | ENST00000518825 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| Frame-shift | ENST00000518825 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| In-frame | ENST00000302763 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| In-frame | ENST00000355078 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| In-frame | ENST00000518825 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3CDS | ENST00000520400 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3CDS | ENST00000520400 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-3CDS | ENST00000520400 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3CDS | ENST00000520400 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-3CDS | ENST00000520400 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3CDS | ENST00000540387 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3CDS | ENST00000540387 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-3CDS | ENST00000540387 | ENST00000377709 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3CDS | ENST00000540387 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-3CDS | ENST00000540387 | ENST00000377710 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3UTR | ENST00000520400 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-3UTR | ENST00000520400 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-3UTR | ENST00000540387 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-3UTR | ENST00000540387 | ENST00000467391 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + |
| intron-intron | ENST00000520400 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
| intron-intron | ENST00000540387 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968809 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000355078 | CTNNA1 | chr5 | 138163407 | + | ENST00000377704 | MGME1 | chr20 | 17970582 | + | 2142 | 958 | 202 | 1128 | 308 |
| ENST00000302763 | CTNNA1 | chr5 | 138163407 | + | ENST00000377704 | MGME1 | chr20 | 17970582 | + | 2336 | 1152 | 90 | 1322 | 410 |
| ENST00000518825 | CTNNA1 | chr5 | 138163407 | + | ENST00000377704 | MGME1 | chr20 | 17970582 | + | 2248 | 1064 | 2 | 1234 | 410 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000355078 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + | 0.00094101 | 0.999059 |
| ENST00000302763 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + | 0.000895224 | 0.9991048 |
| ENST00000518825 | ENST00000377704 | CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970582 | + | 0.000802212 | 0.9991978 |
Top |
Fusion Genomic Features for CTNNA1-MGME1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970581 | + | 0.002952364 | 0.9970476 |
| CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968808 | + | 2.49E-06 | 0.9999975 |
| CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17970581 | + | 0.002952364 | 0.9970476 |
| CTNNA1 | chr5 | 138163407 | + | MGME1 | chr20 | 17968808 | + | 2.49E-06 | 0.9999975 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
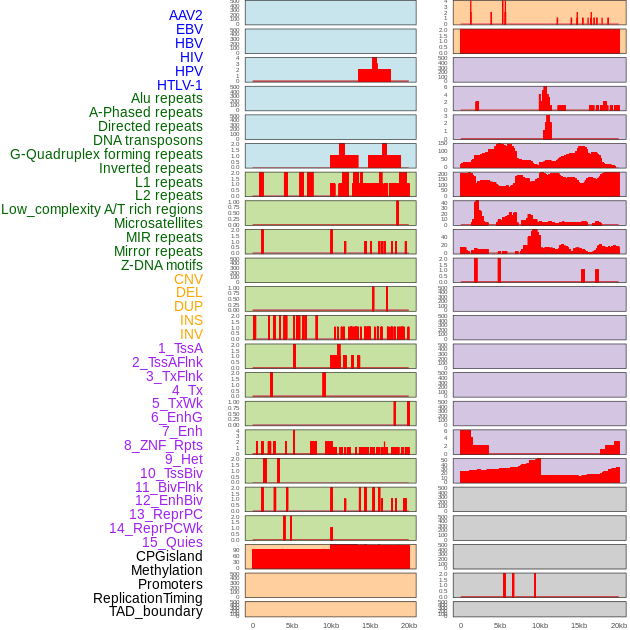 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
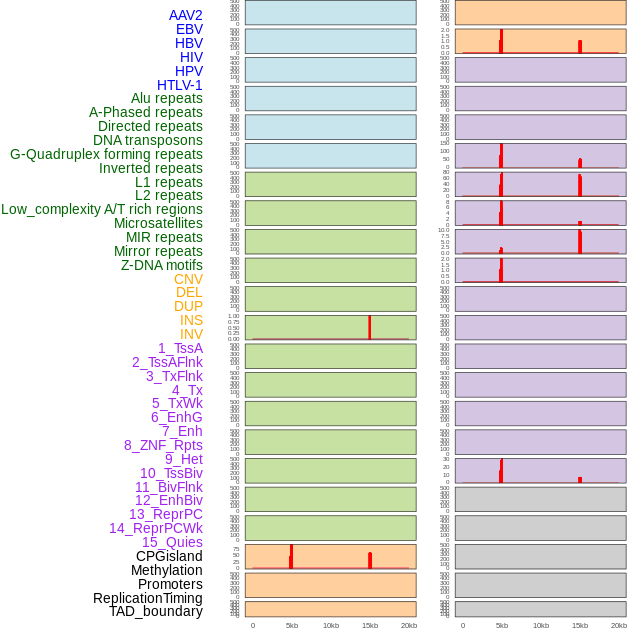 |
Top |
Fusion Protein Features for CTNNA1-MGME1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:138163407/chr20:17968808) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CTNNA1 | . |
| FUNCTION: Associates with the cytoplasmic domain of a variety of cadherins. The association of catenins to cadherins produces a complex which is linked to the actin filament network, and which seems to be of primary importance for cadherins cell-adhesion properties. Can associate with both E- and N-cadherins. Originally believed to be a stable component of E-cadherin/catenin adhesion complexes and to mediate the linkage of cadherins to the actin cytoskeleton at adherens junctions. In contrast, cortical actin was found to be much more dynamic than E-cadherin/catenin complexes and CTNNA1 was shown not to bind to F-actin when assembled in the complex suggesting a different linkage between actin and adherens junctions components. The homodimeric form may regulate actin filament assembly and inhibit actin branching by competing with the Arp2/3 complex for binding to actin filaments. Involved in the regulation of WWTR1/TAZ, YAP1 and TGFB1-dependent SMAD2 and SMAD3 nuclear accumulation (By similarity). May play a crucial role in cell differentiation. {ECO:0000250|UniProtKB:P26231, ECO:0000269|PubMed:25653389}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CTNNA1 | chr5:138163407 | chr20:17970582 | ENST00000302763 | + | 7 | 18 | 2_228 | 354 | 907.0 | Region | Note=Involved in homodimerization |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CTNNA1 | chr5:138163407 | chr20:17970582 | ENST00000540387 | + | 1 | 12 | 2_228 | 0 | 537.0 | Region | Note=Involved in homodimerization |
Top |
Fusion Gene Sequence for CTNNA1-MGME1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >20264_20264_1_CTNNA1-MGME1_CTNNA1_chr5_138163407_ENST00000302763_MGME1_chr20_17970582_ENST00000377704_length(transcript)=2336nt_BP=1152nt TCCTCCTCCTAGCCGGACTGGAGGGAGACAAAGCAGCGCCCGTCTGCTTCGGGCCTCTGGAATTTAGCGCTCGCCCAGCTAGCCGCAGAA ATGACTGCTGTCCATGCAGGCAACATAAACTTCAAGTGGGATCCTAAAAGTCTAGAGATCAGGACTCTGGCAGTTGAGAGACTGTTGGAG CCTCTTGTTACACAGGTTACAACCCTTGTAAACACCAATAGTAAAGGGCCCTCTAATAAGAAGAGAGGTCGTTCTAAGAAGGCCCATGTT TTGGCTGCATCTGTTGAACAAGCAACTGAGAATTTCTTGGAGAAGGGGGATAAAATTGCGAAGGAGAGCCAGTTTCTCAAGGAGGAGCTT GTGGCTGCTGTAGAAGATGTTCGAAAACAAGGTGATTTGATGAAGGCTGCTGCAGGAGAGTTCGCAGATGATCCCTGCTCTTCTGTGAAG CGAGGCAACATGGTTCGGGCAGCTCGAGCTTTGCTCTCTGCTGTTACCCGGTTGCTGATTTTGGCTGACATGGCAGATGTCTACAAATTA CTTGTTCAGCTGAAAGTTGTGGAAGATGGTATCTTGAAGTTGAGGAATGCTGGCAATGAACAAGACTTAGGAATCCAGTATAAAGCCCTA AAACCTGAAGTGGATAAGCTGAACATTATGGCAGCCAAAAGACAACAGGAATTGAAAGATGTTGGCCATCGTGATCAGATGGCTGCAGCT AGAGGAATCCTGCAGAAGAACGTTCCGATCCTCTATACTGCATCCCAGGCATGCCTACAGCACCCTGATGTCGCAGCCTATAAGGCCAAC AGGGACCTGATATACAAGCAGCTGCAGCAGGCGGTCACAGGCATTTCCAATGCAGCCCAGGCCACTGCCTCAGACGATGCCTCACAGCAC CAGGGTGGAGGAGGAGGAGAACTGGCATATGCACTCAATAACTTTGACAAACAAATCATTGTGGACCCCTTGAGCTTCAGCGAGGAGCGC TTTAGGCCTTCCCTGGAGGAGCGTCTGGAAAGCATCATTAGTGGGGCTGCCTTGATGGCCGACTCGTCCTGCACGCGTGATGACCGTCGT GAGCGAATTGTGGCAGAGTGTAATGCTGTCCGCCAGGCCCTGCAGGACCTGCTTTCGGAGTACATGGGCAATGTTCAATGTGGCTTAATT GTGGTGGCCTACAAAGATGGATCACCTGCCCACCCACATTTCATGGATGCAGAGCTCTGTTCCCAGTACTGGACCAAGTGGCTTCTTCGA CTAGAAGAATATACGGAAAAGAAAAAGAACCAGAATATTCAGAAACCAGAATATTCAGAATAGGGAGCAAGTTGCTATTTGGGAACATTC AGCACCTTCTCACAGTTTGGGAACATATATTGCTGTTTACTCCAGTGTAAAAATGAGGTGCCACTGGATCTGAGTGCTACACGAACACAA GTAGAAGTATTAATTTGTTGAAATGTGTTGTTACCAAAAAGACTGAAAAGCCCCAAAGTCTAGATATAAAGACCTAGACTTCGGCACGCG AAATCCCAGCTATGCTACCTCTTATTTACCTGAAAGGAGGACACGCAGGATGGGCAGTCATGCTGGTGACTCTTGTACTCCCTTGAGGGA CATTGGGGGGGGGGGGGCGTGGTCCCAGGCAGGATGCCCAGTCTTTGAGCTGAGATTGGAAGGCAGTGAGGCTGAGGGTGCCAAGATTTC CCCAGGGTTCACCCAGAGGGGAAGGGGCTACATGCCCCCAGCTGTGTGCAGGGAGGACACATCAGCCCACTACCGCTGCCAACACCAATG CCTAAAACTTGTTTCATACATTGGGGTTTTCTATATATTTCAGCTGGGAAAAGCTTACATTTAACCTTTTGAAAAAATAAATACGTGATT AGCCTCAACTAAACATTGCTGACTATAAAGACAGTATATTCACCATGTCGCTGGCAATATGTCATTGCGTAACACCAAATAACCCCCCAG AAGTAGCCAGAGGCCAGTTTGAACATCACAATTCTAAGTGTTTTAGTAACTATTTCTGGCGTGAGTCAACAGATCATGTAGATAGAGTCA ATTATTGTTTGTGGAGTTTTTCAGCTATAGGGGAGGGGAACTATTAAAATCCATTTGTTTCTATTCAATAGGTAATAAAAATTAGTTGTC CCTGGGTTTGGGAAACTTAAATGCCCATTACAGCCCTGGGGAAGGGTTTTCTGTCTTATGGAGTGAGTCTTAGCATTTAAGTTATACAGT TGCTGCCTTAAAATAGTAGCCTGCTACAATGACTTCTTTGGGTAGCCATTTTCATAAGAAATAAAATACAAGATATGAGTAATGAA >20264_20264_1_CTNNA1-MGME1_CTNNA1_chr5_138163407_ENST00000302763_MGME1_chr20_17970582_ENST00000377704_length(amino acids)=410AA_BP=354 MTAVHAGNINFKWDPKSLEIRTLAVERLLEPLVTQVTTLVNTNSKGPSNKKRGRSKKAHVLAASVEQATENFLEKGDKIAKESQFLKEEL VAAVEDVRKQGDLMKAAAGEFADDPCSSVKRGNMVRAARALLSAVTRLLILADMADVYKLLVQLKVVEDGILKLRNAGNEQDLGIQYKAL KPEVDKLNIMAAKRQQELKDVGHRDQMAAARGILQKNVPILYTASQACLQHPDVAAYKANRDLIYKQLQQAVTGISNAAQATASDDASQH QGGGGGELAYALNNFDKQIIVDPLSFSEERFRPSLEERLESIISGAALMADSSCTRDDRRERIVAECNAVRQALQDLLSEYMGNVQCGLI VVAYKDGSPAHPHFMDAELCSQYWTKWLLRLEEYTEKKKNQNIQKPEYSE -------------------------------------------------------------- >20264_20264_2_CTNNA1-MGME1_CTNNA1_chr5_138163407_ENST00000355078_MGME1_chr20_17970582_ENST00000377704_length(transcript)=2142nt_BP=958nt TTTCCTCCTCCTAGCCGGACTGGAGGGAGACAAAGCAGCGCCCGTCTGCTTCGGGCCTCTGGAATTTAGCGCTCGCCCAGCTAGCCGCAG AAATGACTGCTGTCCATGCAGGCAACATAAACTTCAAGTGGGATCCTAAAAGTCTAGAGATCAGGACTCTGGCAGTTGAGAGACTGTTGG AGCCTCTTGTTACACAGGTGATTTGATGAAGGCTGCTGCAGGAGAGTTCGCAGATGATCCCTGCTCTTCTGTGAAGCGAGGCAACATGGT TCGGGCAGCTCGAGCTTTGCTCTCTGCTGTTACCCGGTTGCTGATTTTGGCTGACATGGCAGATGTCTACAAATTACTTGTTCAGCTGAA AGTTGTGGAAGATGGTATCTTGAAGTTGAGGAATGCTGGCAATGAACAAGACTTAGGAATCCAGTATAAAGCCCTAAAACCTGAAGTGGA TAAGCTGAACATTATGGCAGCCAAAAGACAACAGGAATTGAAAGATGTTGGCCATCGTGATCAGATGGCTGCAGCTAGAGGAATCCTGCA GAAGAACGTTCCGATCCTCTATACTGCATCCCAGGCATGCCTACAGCACCCTGATGTCGCAGCCTATAAGGCCAACAGGGACCTGATATA CAAGCAGCTGCAGCAGGCGGTCACAGGCATTTCCAATGCAGCCCAGGCCACTGCCTCAGACGATGCCTCACAGCACCAGGGTGGAGGAGG AGGAGAACTGGCATATGCACTCAATAACTTTGACAAACAAATCATTGTGGACCCCTTGAGCTTCAGCGAGGAGCGCTTTAGGCCTTCCCT GGAGGAGCGTCTGGAAAGCATCATTAGTGGGGCTGCCTTGATGGCCGACTCGTCCTGCACGCGTGATGACCGTCGTGAGCGAATTGTGGC AGAGTGTAATGCTGTCCGCCAGGCCCTGCAGGACCTGCTTTCGGAGTACATGGGCAATGTTCAATGTGGCTTAATTGTGGTGGCCTACAA AGATGGATCACCTGCCCACCCACATTTCATGGATGCAGAGCTCTGTTCCCAGTACTGGACCAAGTGGCTTCTTCGACTAGAAGAATATAC GGAAAAGAAAAAGAACCAGAATATTCAGAAACCAGAATATTCAGAATAGGGAGCAAGTTGCTATTTGGGAACATTCAGCACCTTCTCACA GTTTGGGAACATATATTGCTGTTTACTCCAGTGTAAAAATGAGGTGCCACTGGATCTGAGTGCTACACGAACACAAGTAGAAGTATTAAT TTGTTGAAATGTGTTGTTACCAAAAAGACTGAAAAGCCCCAAAGTCTAGATATAAAGACCTAGACTTCGGCACGCGAAATCCCAGCTATG CTACCTCTTATTTACCTGAAAGGAGGACACGCAGGATGGGCAGTCATGCTGGTGACTCTTGTACTCCCTTGAGGGACATTGGGGGGGGGG GGGCGTGGTCCCAGGCAGGATGCCCAGTCTTTGAGCTGAGATTGGAAGGCAGTGAGGCTGAGGGTGCCAAGATTTCCCCAGGGTTCACCC AGAGGGGAAGGGGCTACATGCCCCCAGCTGTGTGCAGGGAGGACACATCAGCCCACTACCGCTGCCAACACCAATGCCTAAAACTTGTTT CATACATTGGGGTTTTCTATATATTTCAGCTGGGAAAAGCTTACATTTAACCTTTTGAAAAAATAAATACGTGATTAGCCTCAACTAAAC ATTGCTGACTATAAAGACAGTATATTCACCATGTCGCTGGCAATATGTCATTGCGTAACACCAAATAACCCCCCAGAAGTAGCCAGAGGC CAGTTTGAACATCACAATTCTAAGTGTTTTAGTAACTATTTCTGGCGTGAGTCAACAGATCATGTAGATAGAGTCAATTATTGTTTGTGG AGTTTTTCAGCTATAGGGGAGGGGAACTATTAAAATCCATTTGTTTCTATTCAATAGGTAATAAAAATTAGTTGTCCCTGGGTTTGGGAA ACTTAAATGCCCATTACAGCCCTGGGGAAGGGTTTTCTGTCTTATGGAGTGAGTCTTAGCATTTAAGTTATACAGTTGCTGCCTTAAAAT AGTAGCCTGCTACAATGACTTCTTTGGGTAGCCATTTTCATAAGAAATAAAATACAAGATATGAGTAATGAA >20264_20264_2_CTNNA1-MGME1_CTNNA1_chr5_138163407_ENST00000355078_MGME1_chr20_17970582_ENST00000377704_length(amino acids)=308AA_BP=252 MMKAAAGEFADDPCSSVKRGNMVRAARALLSAVTRLLILADMADVYKLLVQLKVVEDGILKLRNAGNEQDLGIQYKALKPEVDKLNIMAA KRQQELKDVGHRDQMAAARGILQKNVPILYTASQACLQHPDVAAYKANRDLIYKQLQQAVTGISNAAQATASDDASQHQGGGGGELAYAL NNFDKQIIVDPLSFSEERFRPSLEERLESIISGAALMADSSCTRDDRRERIVAECNAVRQALQDLLSEYMGNVQCGLIVVAYKDGSPAHP HFMDAELCSQYWTKWLLRLEEYTEKKKNQNIQKPEYSE -------------------------------------------------------------- >20264_20264_3_CTNNA1-MGME1_CTNNA1_chr5_138163407_ENST00000518825_MGME1_chr20_17970582_ENST00000377704_length(transcript)=2248nt_BP=1064nt AAATGACTGCTGTCCATGCAGGCAACATAAACTTCAAGTGGGATCCTAAAAGTCTAGAGATCAGGACTCTGGCAGTTGAGAGACTGTTGG AGCCTCTTGTTACACAGGTTACAACCCTTGTAAACACCAATAGTAAAGGGCCCTCTAATAAGAAGAGAGGTCGTTCTAAGAAGGCCCATG TTTTGGCTGCATCTGTTGAACAAGCAACTGAGAATTTCTTGGAGAAGGGGGATAAAATTGCGAAGGAGAGCCAGTTTCTCAAGGAGGAGC TTGTGGCTGCTGTAGAAGATGTTCGAAAACAAGGTGATTTGATGAAGGCTGCTGCAGGAGAGTTCGCAGATGATCCCTGCTCTTCTGTGA AGCGAGGCAACATGGTTCGGGCAGCTCGAGCTTTGCTCTCTGCTGTTACCCGGTTGCTGATTTTGGCTGACATGGCAGATGTCTACAAAT TACTTGTTCAGCTGAAAGTTGTGGAAGATGGTATCTTGAAGTTGAGGAATGCTGGCAATGAACAAGACTTAGGAATCCAGTATAAAGCCC TAAAACCTGAAGTGGATAAGCTGAACATTATGGCAGCCAAAAGACAACAGGAATTGAAAGATGTTGGCCATCGTGATCAGATGGCTGCAG CTAGAGGAATCCTGCAGAAGAACGTTCCGATCCTCTATACTGCATCCCAGGCATGCCTACAGCACCCTGATGTCGCAGCCTATAAGGCCA ACAGGGACCTGATATACAAGCAGCTGCAGCAGGCGGTCACAGGCATTTCCAATGCAGCCCAGGCCACTGCCTCAGACGATGCCTCACAGC ACCAGGGTGGAGGAGGAGGAGAACTGGCATATGCACTCAATAACTTTGACAAACAAATCATTGTGGACCCCTTGAGCTTCAGCGAGGAGC GCTTTAGGCCTTCCCTGGAGGAGCGTCTGGAAAGCATCATTAGTGGGGCTGCCTTGATGGCCGACTCGTCCTGCACGCGTGATGACCGTC GTGAGCGAATTGTGGCAGAGTGTAATGCTGTCCGCCAGGCCCTGCAGGACCTGCTTTCGGAGTACATGGGCAATGTTCAATGTGGCTTAA TTGTGGTGGCCTACAAAGATGGATCACCTGCCCACCCACATTTCATGGATGCAGAGCTCTGTTCCCAGTACTGGACCAAGTGGCTTCTTC GACTAGAAGAATATACGGAAAAGAAAAAGAACCAGAATATTCAGAAACCAGAATATTCAGAATAGGGAGCAAGTTGCTATTTGGGAACAT TCAGCACCTTCTCACAGTTTGGGAACATATATTGCTGTTTACTCCAGTGTAAAAATGAGGTGCCACTGGATCTGAGTGCTACACGAACAC AAGTAGAAGTATTAATTTGTTGAAATGTGTTGTTACCAAAAAGACTGAAAAGCCCCAAAGTCTAGATATAAAGACCTAGACTTCGGCACG CGAAATCCCAGCTATGCTACCTCTTATTTACCTGAAAGGAGGACACGCAGGATGGGCAGTCATGCTGGTGACTCTTGTACTCCCTTGAGG GACATTGGGGGGGGGGGGGCGTGGTCCCAGGCAGGATGCCCAGTCTTTGAGCTGAGATTGGAAGGCAGTGAGGCTGAGGGTGCCAAGATT TCCCCAGGGTTCACCCAGAGGGGAAGGGGCTACATGCCCCCAGCTGTGTGCAGGGAGGACACATCAGCCCACTACCGCTGCCAACACCAA TGCCTAAAACTTGTTTCATACATTGGGGTTTTCTATATATTTCAGCTGGGAAAAGCTTACATTTAACCTTTTGAAAAAATAAATACGTGA TTAGCCTCAACTAAACATTGCTGACTATAAAGACAGTATATTCACCATGTCGCTGGCAATATGTCATTGCGTAACACCAAATAACCCCCC AGAAGTAGCCAGAGGCCAGTTTGAACATCACAATTCTAAGTGTTTTAGTAACTATTTCTGGCGTGAGTCAACAGATCATGTAGATAGAGT CAATTATTGTTTGTGGAGTTTTTCAGCTATAGGGGAGGGGAACTATTAAAATCCATTTGTTTCTATTCAATAGGTAATAAAAATTAGTTG TCCCTGGGTTTGGGAAACTTAAATGCCCATTACAGCCCTGGGGAAGGGTTTTCTGTCTTATGGAGTGAGTCTTAGCATTTAAGTTATACA GTTGCTGCCTTAAAATAGTAGCCTGCTACAATGACTTCTTTGGGTAGCCATTTTCATAAGAAATAAAATACAAGATATGAGTAATGAA >20264_20264_3_CTNNA1-MGME1_CTNNA1_chr5_138163407_ENST00000518825_MGME1_chr20_17970582_ENST00000377704_length(amino acids)=410AA_BP=354 MTAVHAGNINFKWDPKSLEIRTLAVERLLEPLVTQVTTLVNTNSKGPSNKKRGRSKKAHVLAASVEQATENFLEKGDKIAKESQFLKEEL VAAVEDVRKQGDLMKAAAGEFADDPCSSVKRGNMVRAARALLSAVTRLLILADMADVYKLLVQLKVVEDGILKLRNAGNEQDLGIQYKAL KPEVDKLNIMAAKRQQELKDVGHRDQMAAARGILQKNVPILYTASQACLQHPDVAAYKANRDLIYKQLQQAVTGISNAAQATASDDASQH QGGGGGELAYALNNFDKQIIVDPLSFSEERFRPSLEERLESIISGAALMADSSCTRDDRRERIVAECNAVRQALQDLLSEYMGNVQCGLI VVAYKDGSPAHPHFMDAELCSQYWTKWLLRLEEYTEKKKNQNIQKPEYSE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CTNNA1-MGME1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | CTNNA1 | chr5:138163407 | chr20:17970582 | ENST00000302763 | + | 7 | 18 | 97_148 | 354.0 | 907.0 | JUP and CTNNB1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CTNNA1 | chr5:138163407 | chr20:17970582 | ENST00000302763 | + | 7 | 18 | 325_394 | 354.0 | 907.0 | alpha-actinin |
| Hgene | CTNNA1 | chr5:138163407 | chr20:17970582 | ENST00000540387 | + | 1 | 12 | 325_394 | 0 | 537.0 | alpha-actinin |
| Hgene | CTNNA1 | chr5:138163407 | chr20:17970582 | ENST00000540387 | + | 1 | 12 | 97_148 | 0 | 537.0 | JUP and CTNNB1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CTNNA1-MGME1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CTNNA1-MGME1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CTNNA1 | C1708349 | Hereditary Diffuse Gastric Cancer | 3 | ORPHANET |
| Hgene | CTNNA1 | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | CTNNA1 | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | CTNNA1 | C0238198 | Gastrointestinal Stromal Tumors | 1 | CTD_human |
| Hgene | CTNNA1 | C1837029 | Macular Dystrophy, Butterfly-Shaped Pigmentary, 2 | 1 | CTD_human;UNIPROT |
| Hgene | CTNNA1 | C1868569 | Patterned dystrophy of retinal pigment epithelium | 1 | CTD_human |
| Hgene | CTNNA1 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Hgene | CTNNA1 | C2713368 | Hematopoetic Myelodysplasia | 1 | CTD_human |
| Hgene | CTNNA1 | C3179349 | Gastrointestinal Stromal Sarcoma | 1 | CTD_human |
| Hgene | CTNNA1 | C3463824 | MYELODYSPLASTIC SYNDROME | 1 | CTD_human |
| Hgene | CTNNA1 | C4511237 | Butterfly-shaped pigmentary macular dystrophy | 1 | ORPHANET |
| Tgene | C0013911 | Emaciation | 1 | CTD_human | |
| Tgene | C0029089 | Ophthalmoplegia | 1 | CTD_human | |
| Tgene | C0035229 | Respiratory Insufficiency | 1 | CTD_human | |
| Tgene | C0162292 | External Ophthalmoplegia | 1 | CTD_human | |
| Tgene | C0235063 | Respiratory Depression | 1 | CTD_human | |
| Tgene | C0339693 | Internal Ophthalmoplegia | 1 | CTD_human | |
| Tgene | C0751401 | Ophthalmoparesis | 1 | CTD_human | |
| Tgene | C0751651 | Mitochondrial Diseases | 1 | CTD_human | |
| Tgene | C0949855 | Electron Transport Chain Deficiencies, Mitochondrial | 1 | CTD_human | |
| Tgene | C0949856 | Oxidative Phosphorylation Deficiencies | 1 | CTD_human | |
| Tgene | C0949857 | Mitochondrial Respiratory Chain Deficiencies | 1 | CTD_human | |
| Tgene | C1145670 | Respiratory Failure | 1 | CTD_human | |
| Tgene | C3554462 | MITOCHONDRIAL DNA DEPLETION SYNDROME 11 | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
