
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ADAMTS17-CERS3 (FusionGDB2 ID:HG170691TG204219) |
Fusion Gene Summary for ADAMTS17-CERS3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ADAMTS17-CERS3 | Fusion gene ID: hg170691tg204219 | Hgene | Tgene | Gene symbol | ADAMTS17 | CERS3 | Gene ID | 170691 | 204219 |
| Gene name | ADAM metallopeptidase with thrombospondin type 1 motif 17 | ceramide synthase 3 | |
| Synonyms | WMS4 | ARCI9|LASS3 | |
| Cytomap | ('ADAMTS17')('CERS3') 15q26.3 | 15q26.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | A disintegrin and metalloproteinase with thrombospondin motifs 17a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 17 | ceramide synthase 3LAG1 homolog, ceramide synthase 3LAG1 longevity assurance homolog 3dihydroceramide synthase 3sphingosine N-acyltransferase CERS3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8TE56 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000268070, ENST00000559976, | ||
| Fusion gene scores | * DoF score | 24 X 8 X 8=1536 | 8 X 7 X 7=392 |
| # samples | 14 | 11 | |
| ** MAII score | log2(14/1536*10)=-3.45567948377619 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/392*10)=-1.83335013059055 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ADAMTS17 [Title/Abstract] AND CERS3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ADAMTS17(100692817)-CERS3(100943070), # samples:2 ADAMTS17(100692817)-CERS3(100996251), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ADAMTS17-CERS3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ADAMTS17-CERS3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ADAMTS17-CERS3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ADAMTS17-CERS3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ADAMTS17-CERS3 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. ADAMTS17-CERS3 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ADAMTS17-CERS3 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CERS3 | GO:0046513 | ceramide biosynthetic process | 17977534|22038835 |
 Fusion gene breakpoints across ADAMTS17 (5'-gene) Fusion gene breakpoints across ADAMTS17 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
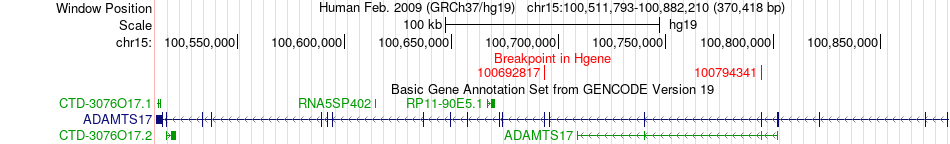 |
 Fusion gene breakpoints across CERS3 (3'-gene) Fusion gene breakpoints across CERS3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
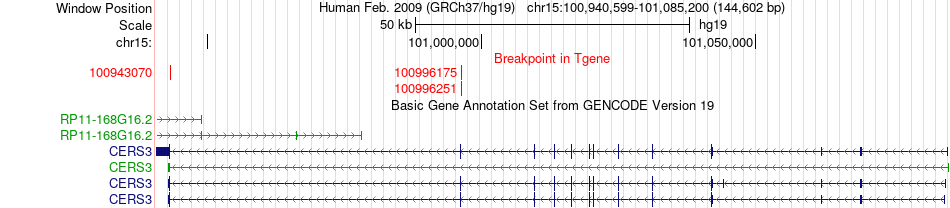 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-33-4532-01A | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| ChimerDB4 | LUSC | TCGA-33-4532-01A | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| ChimerDB4 | LUSC | TCGA-33-4532 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| ChimerDB4 | LUSC | TCGA-33-4532 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| ChimerDB4 | LUSC | TCGA-33-4532 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| ChimerDB4 | LUSC | TCGA-56-6545-01A | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
Top |
Fusion Gene ORF analysis for ADAMTS17-CERS3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000268070 | ENST00000560944 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| 5CDS-5UTR | ENST00000268070 | ENST00000560944 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| 5CDS-intron | ENST00000268070 | ENST00000560944 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| 5CDS-intron | ENST00000268070 | ENST00000560944 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| 5UTR-3CDS | ENST00000559976 | ENST00000284382 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| 5UTR-3CDS | ENST00000559976 | ENST00000394113 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| 5UTR-3CDS | ENST00000559976 | ENST00000538112 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| 5UTR-5UTR | ENST00000559976 | ENST00000560944 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000268070 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| Frame-shift | ENST00000268070 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| Frame-shift | ENST00000268070 | ENST00000284382 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000268070 | ENST00000394113 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000268070 | ENST00000394113 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| Frame-shift | ENST00000268070 | ENST00000394113 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| Frame-shift | ENST00000268070 | ENST00000538112 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000268070 | ENST00000538112 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| Frame-shift | ENST00000268070 | ENST00000538112 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| Frame-shift | ENST00000268070 | ENST00000538112 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000268070 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000268070 | ENST00000394113 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000559976 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000559976 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| intron-3CDS | ENST00000559976 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| intron-3CDS | ENST00000559976 | ENST00000394113 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000559976 | ENST00000394113 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| intron-3CDS | ENST00000559976 | ENST00000394113 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| intron-3CDS | ENST00000559976 | ENST00000538112 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000559976 | ENST00000538112 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| intron-3CDS | ENST00000559976 | ENST00000538112 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
| intron-5UTR | ENST00000559976 | ENST00000560944 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - |
| intron-intron | ENST00000559976 | ENST00000560944 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996175 | - |
| intron-intron | ENST00000559976 | ENST00000560944 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100996251 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000268070 | ADAMTS17 | chr15 | 100692817 | - | ENST00000284382 | CERS3 | chr15 | 100943070 | - | 4050 | 1579 | 37 | 1731 | 564 |
| ENST00000268070 | ADAMTS17 | chr15 | 100794341 | - | ENST00000394113 | CERS3 | chr15 | 100943070 | - | 1536 | 1181 | 37 | 1203 | 388 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000268070 | ENST00000284382 | ADAMTS17 | chr15 | 100692817 | - | CERS3 | chr15 | 100943070 | - | 0.007107494 | 0.9928925 |
| ENST00000268070 | ENST00000394113 | ADAMTS17 | chr15 | 100794341 | - | CERS3 | chr15 | 100943070 | - | 0.061279047 | 0.938721 |
Top |
Fusion Genomic Features for ADAMTS17-CERS3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
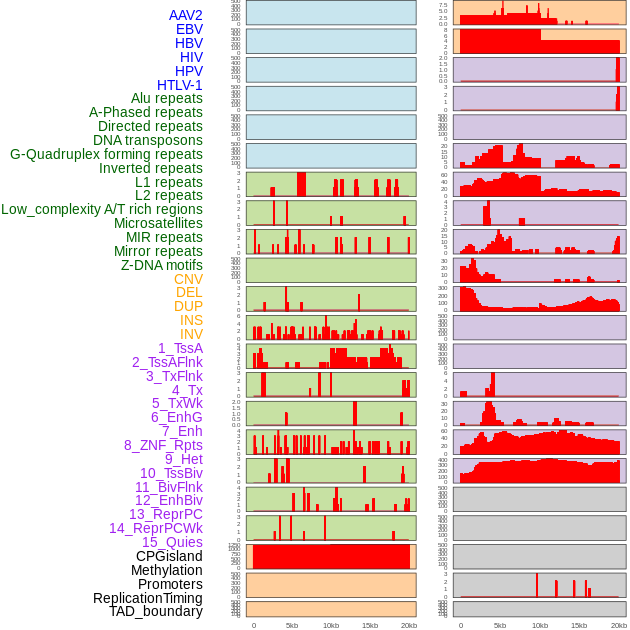 |
Top |
Fusion Protein Features for ADAMTS17-CERS3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:100692817/chr15:100943070) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ADAMTS17 | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 60_120 | 491 | 1096.0 | Compositional bias | Note=Arg-rich |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 60_120 | 358 | 1096.0 | Compositional bias | Note=Arg-rich |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 232_452 | 491 | 1096.0 | Domain | Peptidase M12B |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 199_206 | 491 | 1096.0 | Motif | Cysteine switch |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 199_206 | 358 | 1096.0 | Motif | Cysteine switch |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 599_701 | 491 | 1096.0 | Compositional bias | Note=Cys-rich |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 599_701 | 358 | 1096.0 | Compositional bias | Note=Cys-rich |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 1045_1084 | 491 | 1096.0 | Domain | PLAC |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 453_542 | 491 | 1096.0 | Domain | Note=Disintegrin |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 543_598 | 491 | 1096.0 | Domain | TSP type-1 1 |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 800_860 | 491 | 1096.0 | Domain | TSP type-1 2 |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 861_922 | 491 | 1096.0 | Domain | TSP type-1 3 |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 925_968 | 491 | 1096.0 | Domain | TSP type-1 4 |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 972_1029 | 491 | 1096.0 | Domain | TSP type-1 5 |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 1045_1084 | 358 | 1096.0 | Domain | PLAC |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 232_452 | 358 | 1096.0 | Domain | Peptidase M12B |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 453_542 | 358 | 1096.0 | Domain | Note=Disintegrin |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 543_598 | 358 | 1096.0 | Domain | TSP type-1 1 |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 800_860 | 358 | 1096.0 | Domain | TSP type-1 2 |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 861_922 | 358 | 1096.0 | Domain | TSP type-1 3 |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 925_968 | 358 | 1096.0 | Domain | TSP type-1 4 |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 972_1029 | 358 | 1096.0 | Domain | TSP type-1 5 |
| Hgene | ADAMTS17 | chr15:100692817 | chr15:100943070 | ENST00000268070 | - | 10 | 22 | 702_779 | 491 | 1096.0 | Region | Note=Spacer |
| Hgene | ADAMTS17 | chr15:100794341 | chr15:100943070 | ENST00000268070 | - | 7 | 22 | 702_779 | 358 | 1096.0 | Region | Note=Spacer |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000284382 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000394113 | 12 | 14 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100692817 | chr15:100943070 | ENST00000538112 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000284382 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000394113 | 12 | 14 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:100794341 | chr15:100943070 | ENST00000538112 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ADAMTS17-CERS3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2055_2055_1_ADAMTS17-CERS3_ADAMTS17_chr15_100692817_ENST00000268070_CERS3_chr15_100943070_ENST00000284382_length(transcript)=4050nt_BP=1579nt AATTACTTTTTCCAAAGGCTGGAGGGCTTCACTCCGGCTGGCGCCGCCGCCTAGCGCGCTCCTGCTTCGCCGCCACGGTCCGGGGGGGCT GCCGGTCCCGGGTACCATGTGTGACGGCGCCCTGCTGCCTCCGCTCGTCCTGCCCGTGCTGCTGCTGCTGGTTTGGGGACTGGACCCGGG CACAGCTGTCGGCGACGCGGCGGCCGACGTGGAGGTGGTGCTCCCGTGGCGGGTGCGCCCCGACGACGTGCACCTGCCGCCGCTGCCCGC AGCCCCCGGGCCCCGACGGCGGCGACGCCCCCGCACGCCCCCAGCCGCCCCGCGCGCCCGGCCCGGAGAGCGCGCCCTGCTGCTGCACCT GCCGGCCTTCGGGCGCGACCTGTACCTTCAGCTGCGCCGCGACCTGCGCTTCCTGTCCCGAGGCTTCGAGGTGGAGGAGGCGGGCGCGGC CCGGCGCCGCGGCCGCCCCGCCGAGCTGTGCTTCTACTCGGGCCGTGTGCTCGGCCACCCCGGCTCCCTCGTCTCGCTCAGCGCCTGCGG CGCCGCCGGCGGCCTGGTTGGCCTCATTCAGCTTGGGCAGGAGCAGGTGCTAATCCAGCCCCTCAACAACTCCCAGGGCCCATTCAGTGG ACGAGAACATCTGATCAGGCGCAAATGGTCCTTGACCCCCAGCCCTTCTGCTGAGGCCCAGAGACCTGAGCAGCTCTGCAAGGTTCTAAC AGAAAAGAAGAAGCCGACGTGGGGCAGGCCTTCGCGGGACTGGCGGGAGCGGAGGAACGCTATCCGGCTCACCAGCGAGCACACGGTGGA GACCCTGGTGGTGGCCGACGCCGACATGGTGCAGTACCACGGGGCCGAGGCCGCCCAGAGGTTCATCCTGACCGTCATGAACATGGTATA CAATATGTTTCAGCACCAGAGCCTGGGGATTAAAATTAACATTCAAGTGACCAAGCTTGTCCTGCTACGACAACGTCCCGCTAAGTTGTC CATTGGGCACCATGGTGAGCGGTCCCTGGAGAGCTTCTGTCACTGGCAGAACGAGGAGTATGGAGGAGCGCGATACCTCGGCAATAACCA GGTTCCCGGCGGGAAGGACGACCCGCCCCTGGTGGATGCTGCCGTGTTTGTGACCAGGACAGATTTCTGTGTACACAAGGATGAACCGTG TGACACTGTTGGAATTGCTTACTTAGGAGGTGTGTGCAGTGCTAAGAGGAAGTGTGTGCTTGCCGAAGACAATGGTCTCAATTTGGCCTT TACCATCGCCCATGAGCTGGGCCACAACTTGGGCATGAACCACGACGATGACCACTCATCTTGCGCTGGCAGGTCCCACATCATGTCAGG AGAGTGGGTGAAAGGCCGGAACCCAAGTGACCTCTCTTGGTCCTCCTGCAGCCGAGATGACCTTGAAAACTTCCTCAAGTCAAAAGTCAG CACCTGCTTGCTAGTCACGGACCCCAGAAGCCAGCACACAGTACGCCTCCCGCACAAGCTGCCGGGCATGCACTACAGTGCCAACGAGCA GTGCCAGATCCTGTTTGGCATGAATGCCACCTTCTGCAGAAACATGGAGAGCATCCAGGATGTGAGGAGTGATGACGAGGATTATGAAGA GGAAGAGGAAGAGGAAGAAGAAGAGGCTACCAAAGGCAAAGAGATGGATTGTTTAAAGAACGGCCTCAGGGCTGAGAGGCACCTCATTCC CAATGGCCAGCATGGCCATTAGCTGGAAGCCTACAGGACTCCCATGGCACAGCATGCTGCAAGTACTGTTGGCAGCCTGGCTTCCAGGCC CCACACCGACCCCACATTCTGCCCTTCCCTCTTTCTCACCACCGCCTTCCCTCCCACCTAAGATGTGTTTACCAAAATGTTGTTAACTTG TGTTAAAATGTTAAATATAAGCATGCCCATGGATTTTTACTGCAGTTAGGACTCAGACTGGTCAAAGATTTCAAAGATTTCTCCACAGAA CCGTCTCAGTTCTAATTGCACTCCCTCATGCATGTCACTTTCTCAGGGGCTCGCTTTGTTATAGACCCTTTCGCCTCGCCACCTTGCCTG TCCTCAGGACGCTTTCACAGGTGCTAAGTGATCTCATTTTTCCCAGGTGTGTTTGGCCACAAAGAGCAGCTTCTTTCTCAAAATGAGTTA GAAGTGGCAGTGGGACAGGAGCGGAAGGACCACACCAGGAGACACTTCCATCCTGAAGCTTAGGTGCCTCATCTCCACAGGGCGGTGGCA GTCCCTGCCTGCCACCCCACAGGGTTACAGCAAGGATTAAGTGAGATCACAGAAGTTTAAGTACACACTGTCAACCGTGGGGTCATCGTG AACCAACCCACTTTGCACTGTTTGGGAGACAGAAACCTGGCTAAACATGGCACTGCAATGGGCCTGAACAAAAGGCAGAATCTTTAAAAA TTCCTCTTAAATGACTCTGGAGATACCTGGAAATGAAAGTGCCAAGAAAGGTGTTCCATTTTATTTGCTAACTATTTATGCATTAGCTCC CCAAGGGTCATTCTCAGATTCTCCTGGAATTCTTCTCCCTCAGGACCCATGGTTCAAGGAAGGAAGCTTAGGCCCTGTTCCTTCCTGTCC TTTGCTGGTCTTTCCCTTTTTTCCTTTCTAGGAGGGAAGCTTCTGTGCTGCTGCCCTGAGCCCTTCCTTCAGGCCAGCACAGTACCTGGG GACCTCCACGGGGGAATGGGATCCAGGCCAGGTTGCTTGCTGAGCCTCATCACCCAGGAGGCCTGAGCCTCTGGGGAGGGCACGCATGTA CACTGCCAGACCCAGGGGAGATCTTGGGAACAGAGGATGCTACGTGATTTCCTCTGGCTTCCAACCCAATCAGCCTGCATCACAGTGAAA CACAACACAAAAGGCCATAAAAGGCATACCCCTGAGAAATGTTTAAGGGCAGGTGCTAGAGTCAGTGCCAGCTCCCTGGGAGGAGGGATA GGGACGGGCAGATCAGTAGCCAGCTTGTCCTCACCCTCCGGAAGGGAGCACCGGAGAAGACTTGCACACGTCCCTGCCTCCCTTGCGGCC TTGTCACCAGAGGGACACATCTCCTGGGCACAATGTGCAGGGCTGACCTGGGAGACTTCTCAGGTGGCTGCTGGCTGAGGAGAGGCCAGG CCTTCCCAAGGAAGACCCTGAGTAAAATCTGCATCTGTCCTCACCACCTGGCACAGTCTCTCATAACGGTCAGATTTTGTATGTTCATCC TATTTATTCAGGGGCTCGTACTAGAAAGCCACAGGAGAGGTCGGCTTGTAGGGACTGGAAAATCAGCCCCAAGCAGAGCAGCTGCAGGGC CCCTGGAGCCGGAAGGACACTGCACAGAACAGACTGCGTTATTGTTATTTTTAAATAAAAATATACATTTGAAACCTTAAGGCTAAACAA AAATAAACAAAAAATCCTTTAGAATTCTTTCCACAACAATATCTCTTTCTGAGAAATTGTTACAAACAAGGTCAGATTTTCTCTGTATAA CATTTGCTTTTATGAGGACAATATCATATGCATTATATGCATAATATGATATTATAAATCAAAATGCCTGCACCCACTTTAGGGTATAGC TATTGACTTATTATTAATATTATATTATTATTATTTTGCTGGAAGAAGGTCACACTAAGATATAATTTTTTATGTTTTCAGTTAACGGTA TGCTTTCTTCTTTGCTTATTTGGTTTTTGTCTCTGTACCAAATATCTTCTTGCTTAAGGTAGAAAAGTATTTGTTTACCTCTATCTCCAG TTTTTTTTCTTATTTGAATGTTGAAGGTAAAATTGATATACCAATTTTAACTATTTCTGATACAGCTGAAAGCACTAAACTACTTCATAA GAAGTAGATACTCATTTTTGTAACACTATTTAGGGCTTTTGTGGTTAATTTTAAAGGAAACCACTCTTTCTACAGGAAACAAGGGCTCAG GATTCTTCAGATGACCTTATAAAAATGCAGTCCACAGTGCTATCAATATTGTAACAGTAATGACTACAATAAAGCCAAAAGTCCAGTGTA >2055_2055_1_ADAMTS17-CERS3_ADAMTS17_chr15_100692817_ENST00000268070_CERS3_chr15_100943070_ENST00000284382_length(amino acids)=564AA_BP=514 MAPPPSALLLRRHGPGGLPVPGTMCDGALLPPLVLPVLLLLVWGLDPGTAVGDAAADVEVVLPWRVRPDDVHLPPLPAAPGPRRRRRPRT PPAAPRARPGERALLLHLPAFGRDLYLQLRRDLRFLSRGFEVEEAGAARRRGRPAELCFYSGRVLGHPGSLVSLSACGAAGGLVGLIQLG QEQVLIQPLNNSQGPFSGREHLIRRKWSLTPSPSAEAQRPEQLCKVLTEKKKPTWGRPSRDWRERRNAIRLTSEHTVETLVVADADMVQY HGAEAAQRFILTVMNMVYNMFQHQSLGIKINIQVTKLVLLRQRPAKLSIGHHGERSLESFCHWQNEEYGGARYLGNNQVPGGKDDPPLVD AAVFVTRTDFCVHKDEPCDTVGIAYLGGVCSAKRKCVLAEDNGLNLAFTIAHELGHNLGMNHDDDHSSCAGRSHIMSGEWVKGRNPSDLS WSSCSRDDLENFLKSKVSTCLLVTDPRSQHTVRLPHKLPGMHYSANEQCQILFGMNATFCRNMESIQDVRSDDEDYEEEEEEEEEEATKG KEMDCLKNGLRAERHLIPNGQHGH -------------------------------------------------------------- >2055_2055_2_ADAMTS17-CERS3_ADAMTS17_chr15_100794341_ENST00000268070_CERS3_chr15_100943070_ENST00000394113_length(transcript)=1536nt_BP=1181nt AATTACTTTTTCCAAAGGCTGGAGGGCTTCACTCCGGCTGGCGCCGCCGCCTAGCGCGCTCCTGCTTCGCCGCCACGGTCCGGGGGGGCT GCCGGTCCCGGGTACCATGTGTGACGGCGCCCTGCTGCCTCCGCTCGTCCTGCCCGTGCTGCTGCTGCTGGTTTGGGGACTGGACCCGGG CACAGCTGTCGGCGACGCGGCGGCCGACGTGGAGGTGGTGCTCCCGTGGCGGGTGCGCCCCGACGACGTGCACCTGCCGCCGCTGCCCGC AGCCCCCGGGCCCCGACGGCGGCGACGCCCCCGCACGCCCCCAGCCGCCCCGCGCGCCCGGCCCGGAGAGCGCGCCCTGCTGCTGCACCT GCCGGCCTTCGGGCGCGACCTGTACCTTCAGCTGCGCCGCGACCTGCGCTTCCTGTCCCGAGGCTTCGAGGTGGAGGAGGCGGGCGCGGC CCGGCGCCGCGGCCGCCCCGCCGAGCTGTGCTTCTACTCGGGCCGTGTGCTCGGCCACCCCGGCTCCCTCGTCTCGCTCAGCGCCTGCGG CGCCGCCGGCGGCCTGGTTGGCCTCATTCAGCTTGGGCAGGAGCAGGTGCTAATCCAGCCCCTCAACAACTCCCAGGGCCCATTCAGTGG ACGAGAACATCTGATCAGGCGCAAATGGTCCTTGACCCCCAGCCCTTCTGCTGAGGCCCAGAGACCTGAGCAGCTCTGCAAGGTTCTAAC AGAAAAGAAGAAGCCGACGTGGGGCAGGCCTTCGCGGGACTGGCGGGAGCGGAGGAACGCTATCCGGCTCACCAGCGAGCACACGGTGGA GACCCTGGTGGTGGCCGACGCCGACATGGTGCAGTACCACGGGGCCGAGGCCGCCCAGAGGTTCATCCTGACCGTCATGAACATGGTATA CAATATGTTTCAGCACCAGAGCCTGGGGATTAAAATTAACATTCAAGTGACCAAGCTTGTCCTGCTACGACAACGTCCCGCTAAGTTGTC CATTGGGCACCATGGTGAGCGGTCCCTGGAGAGCTTCTGTCACTGGCAGAACGAGGAGTATGGAGGAGCGCGATACCTCGGCAATAACCA GGTTCCCGGCGGGAAGGACGACCCGCCCCTGGTGGATGCTGCCGTGTTTGTGACCAGGACAGATTTCTGTGTACACAAGGATGAACCGTG TGACACTGTTGAGCATCCAGGATGTGAGGAGTGATGACGAGGATTATGAAGAGGAAGAGGAAGAGGAAGAAGAAGAGGCTACCAAAGGCA AAGAGATGGATTGTTTAAAGAACGGCCTCAGGGCTGAGAGGCACCTCATTCCCAATGGCCAGCATGGCCATTAGCTGGAAGCCTACAGGA CTCCCATGGCACAGCATGCTGCAAGTACTGTTGGCAGCCTGGCTTCCAGGCCCCACACCGACCCCACATTCTGCCCTTCCCTCTTTCTCA CCACCGCCTTCCCTCCCACCTAAGATGTGTTTACCAAAATGTTGTTAACTTGTGTTAAAATGTTAAATATAAGCATGCCCATGGATTTTT ACTGCA >2055_2055_2_ADAMTS17-CERS3_ADAMTS17_chr15_100794341_ENST00000268070_CERS3_chr15_100943070_ENST00000394113_length(amino acids)=388AA_BP= MAPPPSALLLRRHGPGGLPVPGTMCDGALLPPLVLPVLLLLVWGLDPGTAVGDAAADVEVVLPWRVRPDDVHLPPLPAAPGPRRRRRPRT PPAAPRARPGERALLLHLPAFGRDLYLQLRRDLRFLSRGFEVEEAGAARRRGRPAELCFYSGRVLGHPGSLVSLSACGAAGGLVGLIQLG QEQVLIQPLNNSQGPFSGREHLIRRKWSLTPSPSAEAQRPEQLCKVLTEKKKPTWGRPSRDWRERRNAIRLTSEHTVETLVVADADMVQY HGAEAAQRFILTVMNMVYNMFQHQSLGIKINIQVTKLVLLRQRPAKLSIGHHGERSLESFCHWQNEEYGGARYLGNNQVPGGKDDPPLVD AAVFVTRTDFCVHKDEPCDTVEHPGCEE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ADAMTS17-CERS3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ADAMTS17-CERS3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ADAMTS17-CERS3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ADAMTS17 | C2750787 | Weill-Marchesani-Like Syndrome | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | ADAMTS17 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Hgene | ADAMTS17 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | C3554349 | ICHTHYOSIS, CONGENITAL, AUTOSOMAL RECESSIVE 9 | 2 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C0079154 | Congenital Nonbullous Ichthyosiform Erythroderma | 1 | ORPHANET | |
| Tgene | C2750787 | Weill-Marchesani-Like Syndrome | 1 | ORPHANET |
