
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ECE1-PCP4 (FusionGDB2 ID:HG1889TG5121) |
Fusion Gene Summary for ECE1-PCP4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ECE1-PCP4 | Fusion gene ID: hg1889tg5121 | Hgene | Tgene | Gene symbol | ECE1 | PCP4 | Gene ID | 1889 | 5121 |
| Gene name | endothelin converting enzyme 1 | Purkinje cell protein 4 | |
| Synonyms | ECE | PEP-19 | |
| Cytomap | ('ECE1')('PCP4') 1p36.12 | 21q22.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | endothelin-converting enzyme 1ECE-1 | calmodulin regulator protein PCP4brain specific polypeptide PEP19brain-specific antigen PCP-4brain-specific polypeptide PEP-19 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P42892 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000264205, ENST00000357071, ENST00000374893, ENST00000415912, ENST00000436918, ENST00000528294, | ||
| Fusion gene scores | * DoF score | 16 X 15 X 9=2160 | 12 X 8 X 8=768 |
| # samples | 18 | 13 | |
| ** MAII score | log2(18/2160*10)=-3.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/768*10)=-2.5625946876927 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ECE1 [Title/Abstract] AND PCP4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ECE1(21571482)-PCP4(41270401), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ECE1-PCP4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ECE1-PCP4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ECE1-PCP4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ECE1-PCP4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ECE1 | GO:0010814 | substance P catabolic process | 18039931 |
| Hgene | ECE1 | GO:0010815 | bradykinin catabolic process | 18039931 |
| Hgene | ECE1 | GO:0010816 | calcitonin catabolic process | 18039931 |
| Hgene | ECE1 | GO:0016485 | protein processing | 7805846 |
| Hgene | ECE1 | GO:0016486 | peptide hormone processing | 7864876 |
| Hgene | ECE1 | GO:0034959 | endothelin maturation | 7805846 |
| Hgene | ECE1 | GO:0042447 | hormone catabolic process | 7864876 |
 Fusion gene breakpoints across ECE1 (5'-gene) Fusion gene breakpoints across ECE1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
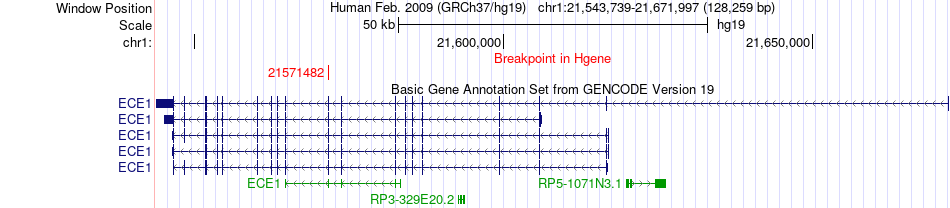 |
 Fusion gene breakpoints across PCP4 (3'-gene) Fusion gene breakpoints across PCP4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
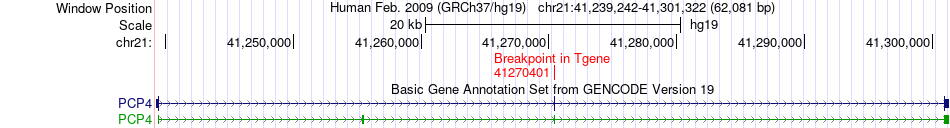 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-49-4501-01A | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
Top |
Fusion Gene ORF analysis for ECE1-PCP4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000264205 | ENST00000468717 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| 5CDS-3UTR | ENST00000357071 | ENST00000468717 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| 5CDS-3UTR | ENST00000374893 | ENST00000468717 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| 5CDS-3UTR | ENST00000415912 | ENST00000468717 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| 5CDS-3UTR | ENST00000436918 | ENST00000468717 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| 5UTR-3CDS | ENST00000528294 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| 5UTR-3UTR | ENST00000528294 | ENST00000468717 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| In-frame | ENST00000264205 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| In-frame | ENST00000357071 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| In-frame | ENST00000374893 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| In-frame | ENST00000415912 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
| In-frame | ENST00000436918 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000415912 | ECE1 | chr1 | 21571482 | - | ENST00000328619 | PCP4 | chr21 | 41270401 | + | 1822 | 1356 | 126 | 1535 | 469 |
| ENST00000357071 | ECE1 | chr1 | 21571482 | - | ENST00000328619 | PCP4 | chr21 | 41270401 | + | 1823 | 1357 | 46 | 1536 | 496 |
| ENST00000374893 | ECE1 | chr1 | 21571482 | - | ENST00000328619 | PCP4 | chr21 | 41270401 | + | 1819 | 1353 | 63 | 1532 | 489 |
| ENST00000436918 | ECE1 | chr1 | 21571482 | - | ENST00000328619 | PCP4 | chr21 | 41270401 | + | 1794 | 1328 | 38 | 1507 | 489 |
| ENST00000264205 | ECE1 | chr1 | 21571482 | - | ENST00000328619 | PCP4 | chr21 | 41270401 | + | 1794 | 1328 | 59 | 1507 | 482 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000415912 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + | 0.012173221 | 0.98782676 |
| ENST00000357071 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + | 0.007667148 | 0.9923328 |
| ENST00000374893 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + | 0.00381344 | 0.99618655 |
| ENST00000436918 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + | 0.003872473 | 0.99612755 |
| ENST00000264205 | ENST00000328619 | ECE1 | chr1 | 21571482 | - | PCP4 | chr21 | 41270401 | + | 0.003954306 | 0.99604565 |
Top |
Fusion Genomic Features for ECE1-PCP4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ECE1 | chr1 | 21571481 | - | PCP4 | chr21 | 41270400 | + | 2.80E-07 | 0.99999976 |
| ECE1 | chr1 | 21571481 | - | PCP4 | chr21 | 41270400 | + | 2.80E-07 | 0.99999976 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
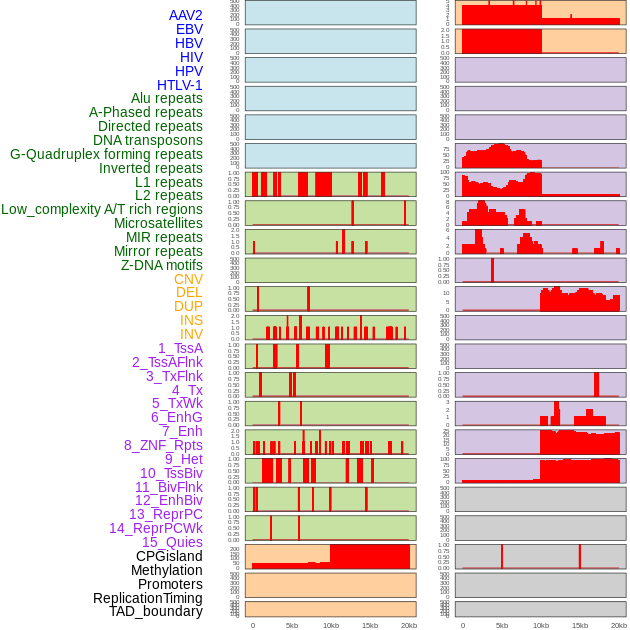 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
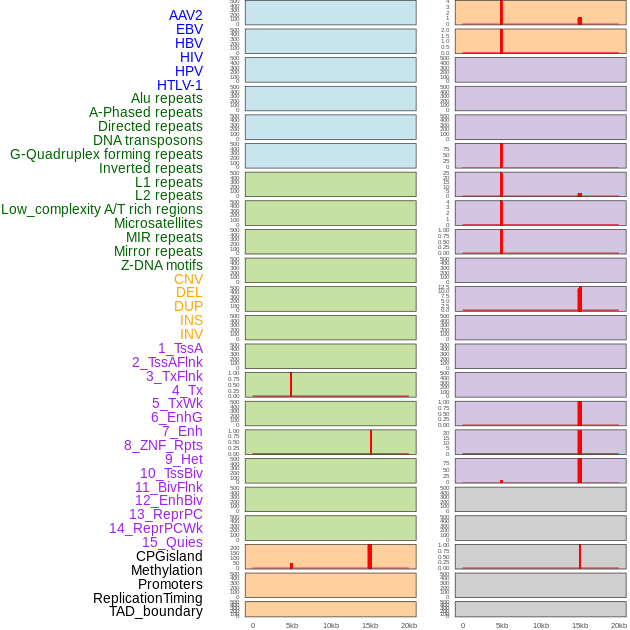 |
Top |
Fusion Protein Features for ECE1-PCP4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:21571482/chr21:41270401) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ECE1 | . |
| FUNCTION: Converts big endothelin-1 to endothelin-1. {ECO:0000269|PubMed:9396733}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000264205 | - | 9 | 18 | 1_68 | 423 | 768.0 | Topological domain | Cytoplasmic |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000357071 | - | 8 | 17 | 1_68 | 414 | 759.0 | Topological domain | Cytoplasmic |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000374893 | - | 10 | 19 | 1_68 | 426 | 771.0 | Topological domain | Cytoplasmic |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000415912 | - | 10 | 19 | 1_68 | 410 | 755.0 | Topological domain | Cytoplasmic |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000264205 | - | 9 | 18 | 69_89 | 423 | 768.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000357071 | - | 8 | 17 | 69_89 | 414 | 759.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000374893 | - | 10 | 19 | 69_89 | 426 | 771.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000415912 | - | 10 | 19 | 69_89 | 410 | 755.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | PCP4 | chr1:21571482 | chr21:41270401 | ENST00000328619 | 0 | 3 | 39_62 | 3 | 63.0 | Domain | IQ | |
| Tgene | PCP4 | chr1:21571482 | chr21:41270401 | ENST00000328619 | 0 | 3 | 28_40 | 3 | 63.0 | Region | Acidic%3B binds calcium and is required for modulating the calcium-binding kinetics of calmodulin |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000264205 | - | 9 | 18 | 98_770 | 423 | 768.0 | Domain | Peptidase M13 |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000357071 | - | 8 | 17 | 98_770 | 414 | 759.0 | Domain | Peptidase M13 |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000374893 | - | 10 | 19 | 98_770 | 426 | 771.0 | Domain | Peptidase M13 |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000415912 | - | 10 | 19 | 98_770 | 410 | 755.0 | Domain | Peptidase M13 |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000264205 | - | 9 | 18 | 90_770 | 423 | 768.0 | Topological domain | Extracellular |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000357071 | - | 8 | 17 | 90_770 | 414 | 759.0 | Topological domain | Extracellular |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000374893 | - | 10 | 19 | 90_770 | 426 | 771.0 | Topological domain | Extracellular |
| Hgene | ECE1 | chr1:21571482 | chr21:41270401 | ENST00000415912 | - | 10 | 19 | 90_770 | 410 | 755.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for ECE1-PCP4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >24856_24856_1_ECE1-PCP4_ECE1_chr1_21571482_ENST00000264205_PCP4_chr21_41270401_ENST00000328619_length(transcript)=1794nt_BP=1328nt GGGGCGCGGCGGCGGCGGCGCCAGGGTCGGGGCCGCTTCCCCATTCGGGCGCGAGAGCCATGGAGGCGCTGAGGGAGTCCGTGCTGCATT TGGCCTTGCAGATGTCGACGTACAAGCGGGCCACGCTGGACGAGGAGGACCTGGTGGACTCGCTCTCCGAGGGCGACGCATACCCCAACG GCCTGCAGGTGAACTTCCACAGCCCCCGGAGTGGCCAGAGGTGCTGGGCTGCACGGACCCAGGTGGAGAAGCGGCTGGTGGTGTTGGTGG TACTTCTGGCGGCAGGACTGGTGGCCTGCTTGGCAGCACTGGGCATCCAGTACCAGACAAGATCCCCCTCTGTGTGCCTGAGCGAAGCTT GTGTCTCAGTGACCAGCTCCATCTTGAGCTCCATGGACCCCACAGTGGACCCCTGCCATGACTTCTTCAGCTACGCCTGTGGGGGCTGGA TCAAGGCCAACCCAGTCCCTGATGGCCACTCACGCTGGGGGACCTTCAGCAACCTCTGGGAACACAACCAAGCAATCATCAAGCACCTCC TCGAAAACTCCACGGCCAGCGTGAGCGAGGCAGAGAGAAAGGCGCAAGTATACTACCGTGCGTGCATGAACGAGACCAGGATCGAGGAGC TCAGGGCCAAACCTCTAATGGAGTTGATTGAGAGGCTCGGGGGCTGGAACATCACAGGTCCCTGGGCCAAGGACAACTTCCAGGACACCC TGCAGGTGGTCACCGCCCACTACCGCACCTCACCCTTCTTCTCTGTCTATGTCAGTGCCGATTCCAAGAACTCCAACAGCAACGTGATCC AGGTGGACCAGTCTGGCCTGGGCTTGCCCTCGAGAGACTATTACCTGAACAAAACTGAAAACGAGAAGGTGCTGACCGGATATCTGAACT ACATGGTCCAGCTGGGGAAGCTGCTGGGCGGCGGGGACGAGGAGGCCATCCGGCCCCAGATGCAGCAGATCTTGGACTTTGAGACGGCAC TGGCCAACATCACCATCCCACAGGAGAAGCGCCGTGATGAGGAGCTCATCTACCACAAAGTGACGGCAGCCGAGCTGCAGACCTTGGCAC CCGCCATCAACTGGTTGCCTTTTCTCAACACCATCTTCTACCCCGTGGAGATCAATGAATCCGAGCCTATTGTGGTCTATGACAAGGAAT ACCTTGAGCAGATCTCCACTCTCATCAACACCACCGACAGATGCCTGCTCAACAACTACATGATCTGGAACCTGGTGCGGAAAACAAGCT CCTTCCTTGACCAGCGCTTTCAGGACGCCGATGAGAAGTTCATGGAAGTCATGTACGGGACCAAGAAGCGACAAGGTGCTGGGGCAACCA ATGGAAAAGACAAGACATCTGGTGAAAATGATGGACAGAAGAAAGTTCAAGAAGAATTTGACATTGACATGGATGCACCAGAGACAGAAC GTGCAGCGGTGGCCATTCAGTCTCAGTTCAGAAAATTCCAGAAGAAGAAGGCTGGGTCTCAGTCCTAGTGGGAGAACCCCCTCCTAGTCC ACCTGAAAACACCAAATTCAACCATCATCTGTCAAGAAATTAAAAGAACAACACCCTAGAGAGAAGTCATCCACACACAATCCACACACG CATAGCAAACCTCCAATGCATGTACAGAAACCTGTGATATTTATACCCTTGTAGGAAGGTATAGACAATGGAATTGTGAGTAGCTTAATC TCTATGTTTCTCTCCATTTTCATTCCTCCTGCAACTATTTTCCTTGATGTTGTAATAAAATGAAGTTACGATGAGTGAATTCAA >24856_24856_1_ECE1-PCP4_ECE1_chr1_21571482_ENST00000264205_PCP4_chr21_41270401_ENST00000328619_length(amino acids)=482AA_BP=423 MEALRESVLHLALQMSTYKRATLDEEDLVDSLSEGDAYPNGLQVNFHSPRSGQRCWAARTQVEKRLVVLVVLLAAGLVACLAALGIQYQT RSPSVCLSEACVSVTSSILSSMDPTVDPCHDFFSYACGGWIKANPVPDGHSRWGTFSNLWEHNQAIIKHLLENSTASVSEAERKAQVYYR ACMNETRIEELRAKPLMELIERLGGWNITGPWAKDNFQDTLQVVTAHYRTSPFFSVYVSADSKNSNSNVIQVDQSGLGLPSRDYYLNKTE NEKVLTGYLNYMVQLGKLLGGGDEEAIRPQMQQILDFETALANITIPQEKRRDEELIYHKVTAAELQTLAPAINWLPFLNTIFYPVEINE SEPIVVYDKEYLEQISTLINTTDRCLLNNYMIWNLVRKTSSFLDQRFQDADEKFMEVMYGTKKRQGAGATNGKDKTSGENDGQKKVQEEF DIDMDAPETERAAVAIQSQFRKFQKKKAGSQS -------------------------------------------------------------- >24856_24856_2_ECE1-PCP4_ECE1_chr1_21571482_ENST00000357071_PCP4_chr21_41270401_ENST00000328619_length(transcript)=1823nt_BP=1357nt GACTCTCTGATGTTTGGGAGCAGATCCGAGCAGCTGAGCAGGGTGGCTGTTCCTTTCCTGGATTAGGGCTGAATCTGTGGGAACCAGACC ACCCCTGAGACAGGAGGCAGCCCTGATGCCTCTCCAGGGCCTGGGCCTGCAGCGGAACCCCTTCCTCCAAGGGAAGCGGGGCCCGGGGCT CACGTCTTCCCCGCCCCTCCTGCCTCCTTCCCTGCAGGTGAACTTCCACAGCCCCCGGAGTGGCCAGAGGTGCTGGGCTGCACGGACCCA GGTGGAGAAGCGGCTGGTGGTGTTGGTGGTACTTCTGGCGGCAGGACTGGTGGCCTGCTTGGCAGCACTGGGCATCCAGTACCAGACAAG ATCCCCCTCTGTGTGCCTGAGCGAAGCTTGTGTCTCAGTGACCAGCTCCATCTTGAGCTCCATGGACCCCACAGTGGACCCCTGCCATGA CTTCTTCAGCTACGCCTGTGGGGGCTGGATCAAGGCCAACCCAGTCCCTGATGGCCACTCACGCTGGGGGACCTTCAGCAACCTCTGGGA ACACAACCAAGCAATCATCAAGCACCTCCTCGAAAACTCCACGGCCAGCGTGAGCGAGGCAGAGAGAAAGGCGCAAGTATACTACCGTGC GTGCATGAACGAGACCAGGATCGAGGAGCTCAGGGCCAAACCTCTAATGGAGTTGATTGAGAGGCTCGGGGGCTGGAACATCACAGGTCC CTGGGCCAAGGACAACTTCCAGGACACCCTGCAGGTGGTCACCGCCCACTACCGCACCTCACCCTTCTTCTCTGTCTATGTCAGTGCCGA TTCCAAGAACTCCAACAGCAACGTGATCCAGGTGGACCAGTCTGGCCTGGGCTTGCCCTCGAGAGACTATTACCTGAACAAAACTGAAAA CGAGAAGGTGCTGACCGGATATCTGAACTACATGGTCCAGCTGGGGAAGCTGCTGGGCGGCGGGGACGAGGAGGCCATCCGGCCCCAGAT GCAGCAGATCTTGGACTTTGAGACGGCACTGGCCAACATCACCATCCCACAGGAGAAGCGCCGTGATGAGGAGCTCATCTACCACAAAGT GACGGCAGCCGAGCTGCAGACCTTGGCACCCGCCATCAACTGGTTGCCTTTTCTCAACACCATCTTCTACCCCGTGGAGATCAATGAATC CGAGCCTATTGTGGTCTATGACAAGGAATACCTTGAGCAGATCTCCACTCTCATCAACACCACCGACAGATGCCTGCTCAACAACTACAT GATCTGGAACCTGGTGCGGAAAACAAGCTCCTTCCTTGACCAGCGCTTTCAGGACGCCGATGAGAAGTTCATGGAAGTCATGTACGGGAC CAAGAAGCGACAAGGTGCTGGGGCAACCAATGGAAAAGACAAGACATCTGGTGAAAATGATGGACAGAAGAAAGTTCAAGAAGAATTTGA CATTGACATGGATGCACCAGAGACAGAACGTGCAGCGGTGGCCATTCAGTCTCAGTTCAGAAAATTCCAGAAGAAGAAGGCTGGGTCTCA GTCCTAGTGGGAGAACCCCCTCCTAGTCCACCTGAAAACACCAAATTCAACCATCATCTGTCAAGAAATTAAAAGAACAACACCCTAGAG AGAAGTCATCCACACACAATCCACACACGCATAGCAAACCTCCAATGCATGTACAGAAACCTGTGATATTTATACCCTTGTAGGAAGGTA TAGACAATGGAATTGTGAGTAGCTTAATCTCTATGTTTCTCTCCATTTTCATTCCTCCTGCAACTATTTTCCTTGATGTTGTAATAAAAT GAAGTTACGATGAGTGAATTCAA >24856_24856_2_ECE1-PCP4_ECE1_chr1_21571482_ENST00000357071_PCP4_chr21_41270401_ENST00000328619_length(amino acids)=496AA_BP=437 MFLSWIRAESVGTRPPLRQEAALMPLQGLGLQRNPFLQGKRGPGLTSSPPLLPPSLQVNFHSPRSGQRCWAARTQVEKRLVVLVVLLAAG LVACLAALGIQYQTRSPSVCLSEACVSVTSSILSSMDPTVDPCHDFFSYACGGWIKANPVPDGHSRWGTFSNLWEHNQAIIKHLLENSTA SVSEAERKAQVYYRACMNETRIEELRAKPLMELIERLGGWNITGPWAKDNFQDTLQVVTAHYRTSPFFSVYVSADSKNSNSNVIQVDQSG LGLPSRDYYLNKTENEKVLTGYLNYMVQLGKLLGGGDEEAIRPQMQQILDFETALANITIPQEKRRDEELIYHKVTAAELQTLAPAINWL PFLNTIFYPVEINESEPIVVYDKEYLEQISTLINTTDRCLLNNYMIWNLVRKTSSFLDQRFQDADEKFMEVMYGTKKRQGAGATNGKDKT SGENDGQKKVQEEFDIDMDAPETERAAVAIQSQFRKFQKKKAGSQS -------------------------------------------------------------- >24856_24856_3_ECE1-PCP4_ECE1_chr1_21571482_ENST00000374893_PCP4_chr21_41270401_ENST00000328619_length(transcript)=1819nt_BP=1353nt AGGTGGTGCAACGCCTGGCCCGGCCCATCCCATCCCGGCCACCCGGGCAGCGGGACCAGGCGTCTGGGGCACAGCATGCGGGGCGTGTGG CCGCCCCCGGTGTCCGCCCTGCTGTCGGCGCTGGGGATGTCGACGTACAAGCGGGCCACGCTGGACGAGGAGGACCTGGTGGACTCGCTC TCCGAGGGCGACGCATACCCCAACGGCCTGCAGGTGAACTTCCACAGCCCCCGGAGTGGCCAGAGGTGCTGGGCTGCACGGACCCAGGTG GAGAAGCGGCTGGTGGTGTTGGTGGTACTTCTGGCGGCAGGACTGGTGGCCTGCTTGGCAGCACTGGGCATCCAGTACCAGACAAGATCC CCCTCTGTGTGCCTGAGCGAAGCTTGTGTCTCAGTGACCAGCTCCATCTTGAGCTCCATGGACCCCACAGTGGACCCCTGCCATGACTTC TTCAGCTACGCCTGTGGGGGCTGGATCAAGGCCAACCCAGTCCCTGATGGCCACTCACGCTGGGGGACCTTCAGCAACCTCTGGGAACAC AACCAAGCAATCATCAAGCACCTCCTCGAAAACTCCACGGCCAGCGTGAGCGAGGCAGAGAGAAAGGCGCAAGTATACTACCGTGCGTGC ATGAACGAGACCAGGATCGAGGAGCTCAGGGCCAAACCTCTAATGGAGTTGATTGAGAGGCTCGGGGGCTGGAACATCACAGGTCCCTGG GCCAAGGACAACTTCCAGGACACCCTGCAGGTGGTCACCGCCCACTACCGCACCTCACCCTTCTTCTCTGTCTATGTCAGTGCCGATTCC AAGAACTCCAACAGCAACGTGATCCAGGTGGACCAGTCTGGCCTGGGCTTGCCCTCGAGAGACTATTACCTGAACAAAACTGAAAACGAG AAGGTGCTGACCGGATATCTGAACTACATGGTCCAGCTGGGGAAGCTGCTGGGCGGCGGGGACGAGGAGGCCATCCGGCCCCAGATGCAG CAGATCTTGGACTTTGAGACGGCACTGGCCAACATCACCATCCCACAGGAGAAGCGCCGTGATGAGGAGCTCATCTACCACAAAGTGACG GCAGCCGAGCTGCAGACCTTGGCACCCGCCATCAACTGGTTGCCTTTTCTCAACACCATCTTCTACCCCGTGGAGATCAATGAATCCGAG CCTATTGTGGTCTATGACAAGGAATACCTTGAGCAGATCTCCACTCTCATCAACACCACCGACAGATGCCTGCTCAACAACTACATGATC TGGAACCTGGTGCGGAAAACAAGCTCCTTCCTTGACCAGCGCTTTCAGGACGCCGATGAGAAGTTCATGGAAGTCATGTACGGGACCAAG AAGCGACAAGGTGCTGGGGCAACCAATGGAAAAGACAAGACATCTGGTGAAAATGATGGACAGAAGAAAGTTCAAGAAGAATTTGACATT GACATGGATGCACCAGAGACAGAACGTGCAGCGGTGGCCATTCAGTCTCAGTTCAGAAAATTCCAGAAGAAGAAGGCTGGGTCTCAGTCC TAGTGGGAGAACCCCCTCCTAGTCCACCTGAAAACACCAAATTCAACCATCATCTGTCAAGAAATTAAAAGAACAACACCCTAGAGAGAA GTCATCCACACACAATCCACACACGCATAGCAAACCTCCAATGCATGTACAGAAACCTGTGATATTTATACCCTTGTAGGAAGGTATAGA CAATGGAATTGTGAGTAGCTTAATCTCTATGTTTCTCTCCATTTTCATTCCTCCTGCAACTATTTTCCTTGATGTTGTAATAAAATGAAG TTACGATGAGTGAATTCAA >24856_24856_3_ECE1-PCP4_ECE1_chr1_21571482_ENST00000374893_PCP4_chr21_41270401_ENST00000328619_length(amino acids)=489AA_BP=430 MGHSMRGVWPPPVSALLSALGMSTYKRATLDEEDLVDSLSEGDAYPNGLQVNFHSPRSGQRCWAARTQVEKRLVVLVVLLAAGLVACLAA LGIQYQTRSPSVCLSEACVSVTSSILSSMDPTVDPCHDFFSYACGGWIKANPVPDGHSRWGTFSNLWEHNQAIIKHLLENSTASVSEAER KAQVYYRACMNETRIEELRAKPLMELIERLGGWNITGPWAKDNFQDTLQVVTAHYRTSPFFSVYVSADSKNSNSNVIQVDQSGLGLPSRD YYLNKTENEKVLTGYLNYMVQLGKLLGGGDEEAIRPQMQQILDFETALANITIPQEKRRDEELIYHKVTAAELQTLAPAINWLPFLNTIF YPVEINESEPIVVYDKEYLEQISTLINTTDRCLLNNYMIWNLVRKTSSFLDQRFQDADEKFMEVMYGTKKRQGAGATNGKDKTSGENDGQ KKVQEEFDIDMDAPETERAAVAIQSQFRKFQKKKAGSQS -------------------------------------------------------------- >24856_24856_4_ECE1-PCP4_ECE1_chr1_21571482_ENST00000415912_PCP4_chr21_41270401_ENST00000328619_length(transcript)=1822nt_BP=1356nt GAGCGCGCCGCCTGGGCCAGGCAGCCGAGCCGTCCGAGCAGCTGGGCTGGGAGCAGGGAACCCGGAGCTGGGAATCGGGAGCCGGGCGCG GGGAGCTGCGCGAAGCCGGGGCGGAGCACGCGAGCTATGATGTCGACGTACAAGCGGGCCACGCTGGACGAGGAGGACCTGGTGGACTCG CTCTCCGAGGGCGACGCATACCCCAACGGCCTGCAGGTGAACTTCCACAGCCCCCGGAGTGGCCAGAGGTGCTGGGCTGCACGGACCCAG GTGGAGAAGCGGCTGGTGGTGTTGGTGGTACTTCTGGCGGCAGGACTGGTGGCCTGCTTGGCAGCACTGGGCATCCAGTACCAGACAAGA TCCCCCTCTGTGTGCCTGAGCGAAGCTTGTGTCTCAGTGACCAGCTCCATCTTGAGCTCCATGGACCCCACAGTGGACCCCTGCCATGAC TTCTTCAGCTACGCCTGTGGGGGCTGGATCAAGGCCAACCCAGTCCCTGATGGCCACTCACGCTGGGGGACCTTCAGCAACCTCTGGGAA CACAACCAAGCAATCATCAAGCACCTCCTCGAAAACTCCACGGCCAGCGTGAGCGAGGCAGAGAGAAAGGCGCAAGTATACTACCGTGCG TGCATGAACGAGACCAGGATCGAGGAGCTCAGGGCCAAACCTCTAATGGAGTTGATTGAGAGGCTCGGGGGCTGGAACATCACAGGTCCC TGGGCCAAGGACAACTTCCAGGACACCCTGCAGGTGGTCACCGCCCACTACCGCACCTCACCCTTCTTCTCTGTCTATGTCAGTGCCGAT TCCAAGAACTCCAACAGCAACGTGATCCAGGTGGACCAGTCTGGCCTGGGCTTGCCCTCGAGAGACTATTACCTGAACAAAACTGAAAAC GAGAAGGTGCTGACCGGATATCTGAACTACATGGTCCAGCTGGGGAAGCTGCTGGGCGGCGGGGACGAGGAGGCCATCCGGCCCCAGATG CAGCAGATCTTGGACTTTGAGACGGCACTGGCCAACATCACCATCCCACAGGAGAAGCGCCGTGATGAGGAGCTCATCTACCACAAAGTG ACGGCAGCCGAGCTGCAGACCTTGGCACCCGCCATCAACTGGTTGCCTTTTCTCAACACCATCTTCTACCCCGTGGAGATCAATGAATCC GAGCCTATTGTGGTCTATGACAAGGAATACCTTGAGCAGATCTCCACTCTCATCAACACCACCGACAGATGCCTGCTCAACAACTACATG ATCTGGAACCTGGTGCGGAAAACAAGCTCCTTCCTTGACCAGCGCTTTCAGGACGCCGATGAGAAGTTCATGGAAGTCATGTACGGGACC AAGAAGCGACAAGGTGCTGGGGCAACCAATGGAAAAGACAAGACATCTGGTGAAAATGATGGACAGAAGAAAGTTCAAGAAGAATTTGAC ATTGACATGGATGCACCAGAGACAGAACGTGCAGCGGTGGCCATTCAGTCTCAGTTCAGAAAATTCCAGAAGAAGAAGGCTGGGTCTCAG TCCTAGTGGGAGAACCCCCTCCTAGTCCACCTGAAAACACCAAATTCAACCATCATCTGTCAAGAAATTAAAAGAACAACACCCTAGAGA GAAGTCATCCACACACAATCCACACACGCATAGCAAACCTCCAATGCATGTACAGAAACCTGTGATATTTATACCCTTGTAGGAAGGTAT AGACAATGGAATTGTGAGTAGCTTAATCTCTATGTTTCTCTCCATTTTCATTCCTCCTGCAACTATTTTCCTTGATGTTGTAATAAAATG AAGTTACGATGAGTGAATTCAA >24856_24856_4_ECE1-PCP4_ECE1_chr1_21571482_ENST00000415912_PCP4_chr21_41270401_ENST00000328619_length(amino acids)=469AA_BP=410 MMSTYKRATLDEEDLVDSLSEGDAYPNGLQVNFHSPRSGQRCWAARTQVEKRLVVLVVLLAAGLVACLAALGIQYQTRSPSVCLSEACVS VTSSILSSMDPTVDPCHDFFSYACGGWIKANPVPDGHSRWGTFSNLWEHNQAIIKHLLENSTASVSEAERKAQVYYRACMNETRIEELRA KPLMELIERLGGWNITGPWAKDNFQDTLQVVTAHYRTSPFFSVYVSADSKNSNSNVIQVDQSGLGLPSRDYYLNKTENEKVLTGYLNYMV QLGKLLGGGDEEAIRPQMQQILDFETALANITIPQEKRRDEELIYHKVTAAELQTLAPAINWLPFLNTIFYPVEINESEPIVVYDKEYLE QISTLINTTDRCLLNNYMIWNLVRKTSSFLDQRFQDADEKFMEVMYGTKKRQGAGATNGKDKTSGENDGQKKVQEEFDIDMDAPETERAA VAIQSQFRKFQKKKAGSQS -------------------------------------------------------------- >24856_24856_5_ECE1-PCP4_ECE1_chr1_21571482_ENST00000436918_PCP4_chr21_41270401_ENST00000328619_length(transcript)=1794nt_BP=1328nt CATCCCATCCCGGCCACCCGGGCAGCGGGACCAGGCGTCTGGGGCACAGCATGCGGGGCGTGTGGCCGCCCCCGGTGTCCGCCCTGCTGT CGGCGCTGGGGATGTCGACGTACAAGCGGGCCACGCTGGACGAGGAGGACCTGGTGGACTCGCTCTCCGAGGGCGACGCATACCCCAACG GCCTGCAGGTGAACTTCCACAGCCCCCGGAGTGGCCAGAGGTGCTGGGCTGCACGGACCCAGGTGGAGAAGCGGCTGGTGGTGTTGGTGG TACTTCTGGCGGCAGGACTGGTGGCCTGCTTGGCAGCACTGGGCATCCAGTACCAGACAAGATCCCCCTCTGTGTGCCTGAGCGAAGCTT GTGTCTCAGTGACCAGCTCCATCTTGAGCTCCATGGACCCCACAGTGGACCCCTGCCATGACTTCTTCAGCTACGCCTGTGGGGGCTGGA TCAAGGCCAACCCAGTCCCTGATGGCCACTCACGCTGGGGGACCTTCAGCAACCTCTGGGAACACAACCAAGCAATCATCAAGCACCTCC TCGAAAACTCCACGGCCAGCGTGAGCGAGGCAGAGAGAAAGGCGCAAGTATACTACCGTGCGTGCATGAACGAGACCAGGATCGAGGAGC TCAGGGCCAAACCTCTAATGGAGTTGATTGAGAGGCTCGGGGGCTGGAACATCACAGGTCCCTGGGCCAAGGACAACTTCCAGGACACCC TGCAGGTGGTCACCGCCCACTACCGCACCTCACCCTTCTTCTCTGTCTATGTCAGTGCCGATTCCAAGAACTCCAACAGCAACGTGATCC AGGTGGACCAGTCTGGCCTGGGCTTGCCCTCGAGAGACTATTACCTGAACAAAACTGAAAACGAGAAGGTGCTGACCGGATATCTGAACT ACATGGTCCAGCTGGGGAAGCTGCTGGGCGGCGGGGACGAGGAGGCCATCCGGCCCCAGATGCAGCAGATCTTGGACTTTGAGACGGCAC TGGCCAACATCACCATCCCACAGGAGAAGCGCCGTGATGAGGAGCTCATCTACCACAAAGTGACGGCAGCCGAGCTGCAGACCTTGGCAC CCGCCATCAACTGGTTGCCTTTTCTCAACACCATCTTCTACCCCGTGGAGATCAATGAATCCGAGCCTATTGTGGTCTATGACAAGGAAT ACCTTGAGCAGATCTCCACTCTCATCAACACCACCGACAGATGCCTGCTCAACAACTACATGATCTGGAACCTGGTGCGGAAAACAAGCT CCTTCCTTGACCAGCGCTTTCAGGACGCCGATGAGAAGTTCATGGAAGTCATGTACGGGACCAAGAAGCGACAAGGTGCTGGGGCAACCA ATGGAAAAGACAAGACATCTGGTGAAAATGATGGACAGAAGAAAGTTCAAGAAGAATTTGACATTGACATGGATGCACCAGAGACAGAAC GTGCAGCGGTGGCCATTCAGTCTCAGTTCAGAAAATTCCAGAAGAAGAAGGCTGGGTCTCAGTCCTAGTGGGAGAACCCCCTCCTAGTCC ACCTGAAAACACCAAATTCAACCATCATCTGTCAAGAAATTAAAAGAACAACACCCTAGAGAGAAGTCATCCACACACAATCCACACACG CATAGCAAACCTCCAATGCATGTACAGAAACCTGTGATATTTATACCCTTGTAGGAAGGTATAGACAATGGAATTGTGAGTAGCTTAATC TCTATGTTTCTCTCCATTTTCATTCCTCCTGCAACTATTTTCCTTGATGTTGTAATAAAATGAAGTTACGATGAGTGAATTCAA >24856_24856_5_ECE1-PCP4_ECE1_chr1_21571482_ENST00000436918_PCP4_chr21_41270401_ENST00000328619_length(amino acids)=489AA_BP=430 MGHSMRGVWPPPVSALLSALGMSTYKRATLDEEDLVDSLSEGDAYPNGLQVNFHSPRSGQRCWAARTQVEKRLVVLVVLLAAGLVACLAA LGIQYQTRSPSVCLSEACVSVTSSILSSMDPTVDPCHDFFSYACGGWIKANPVPDGHSRWGTFSNLWEHNQAIIKHLLENSTASVSEAER KAQVYYRACMNETRIEELRAKPLMELIERLGGWNITGPWAKDNFQDTLQVVTAHYRTSPFFSVYVSADSKNSNSNVIQVDQSGLGLPSRD YYLNKTENEKVLTGYLNYMVQLGKLLGGGDEEAIRPQMQQILDFETALANITIPQEKRRDEELIYHKVTAAELQTLAPAINWLPFLNTIF YPVEINESEPIVVYDKEYLEQISTLINTTDRCLLNNYMIWNLVRKTSSFLDQRFQDADEKFMEVMYGTKKRQGAGATNGKDKTSGENDGQ KKVQEEFDIDMDAPETERAAVAIQSQFRKFQKKKAGSQS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ECE1-PCP4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ECE1-PCP4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ECE1-PCP4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ECE1 | C0018798 | Congenital Heart Defects | 2 | CTD_human |
| Hgene | ECE1 | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Hgene | ECE1 | C0019569 | Hirschsprung Disease | 1 | CTD_human |
| Hgene | ECE1 | C0020538 | Hypertensive disease | 1 | CTD_human |
| Hgene | ECE1 | C0085758 | Aganglionosis, Colonic | 1 | CTD_human |
| Hgene | ECE1 | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Hgene | ECE1 | C0393912 | Segmental Autonomic Dysfunction | 1 | CTD_human |
| Hgene | ECE1 | C0750944 | Peripheral Autonomic Nervous System Diseases | 1 | CTD_human |
| Hgene | ECE1 | C0750945 | Nervous System Diseases, Parasympathetic | 1 | CTD_human |
| Hgene | ECE1 | C0750946 | Nervous System Diseases, Sympathetic | 1 | CTD_human |
| Hgene | ECE1 | C1145628 | Autonomic nervous system disorders | 1 | CTD_human |
| Hgene | ECE1 | C1257840 | Aganglionosis, Rectosigmoid Colon | 1 | CTD_human |
| Hgene | ECE1 | C3151237 | Hirschsprung Disease, Cardiac Defects, and Autonomic Dysfunction | 1 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | ECE1 | C3661523 | Congenital Intestinal Aganglionosis | 1 | CTD_human |
| Tgene | C0007102 | Malignant tumor of colon | 1 | CTD_human | |
| Tgene | C0009375 | Colonic Neoplasms | 1 | CTD_human |
