
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EGFR-INSL4 (FusionGDB2 ID:HG1956TG3641) |
Fusion Gene Summary for EGFR-INSL4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EGFR-INSL4 | Fusion gene ID: hg1956tg3641 | Hgene | Tgene | Gene symbol | EGFR | INSL4 | Gene ID | 1956 | 3641 |
| Gene name | epidermal growth factor receptor | insulin like 4 | |
| Synonyms | ERBB|ERBB1|HER1|NISBD2|PIG61|mENA | EPIL|PLACENTIN | |
| Cytomap | ('EGFR')('INSL4') 7p11.2 | 9p24.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | epidermal growth factor receptoravian erythroblastic leukemia viral (v-erb-b) oncogene homologcell growth inhibiting protein 40cell proliferation-inducing protein 61epidermal growth factor receptor tyrosine kinase domainerb-b2 receptor tyrosine kinas | early placenta insulin-like peptideearly placenta insulin-like peptide (EPIL)insulin-like 4 (placenta)insulin-like peptide 4 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000463948, ENST00000275493, ENST00000342916, ENST00000344576, ENST00000420316, ENST00000442591, ENST00000455089, ENST00000454757, | ||
| Fusion gene scores | * DoF score | 41 X 25 X 14=14350 | 3 X 2 X 3=18 |
| # samples | 53 | 4 | |
| ** MAII score | log2(53/14350*10)=-4.75891456699985 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/18*10)=1.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: EGFR [Title/Abstract] AND INSL4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EGFR(55087058)-INSL4(5233654), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | EGFR-INSL4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EGFR-INSL4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EGFR | GO:0001934 | positive regulation of protein phosphorylation | 20551055 |
| Hgene | EGFR | GO:0007165 | signal transduction | 10572067 |
| Hgene | EGFR | GO:0007166 | cell surface receptor signaling pathway | 7736574 |
| Hgene | EGFR | GO:0007173 | epidermal growth factor receptor signaling pathway | 7736574|12435727 |
| Hgene | EGFR | GO:0008283 | cell proliferation | 17115032 |
| Hgene | EGFR | GO:0008284 | positive regulation of cell proliferation | 7736574 |
| Hgene | EGFR | GO:0010750 | positive regulation of nitric oxide mediated signal transduction | 12828935 |
| Hgene | EGFR | GO:0018108 | peptidyl-tyrosine phosphorylation | 22732145 |
| Hgene | EGFR | GO:0030307 | positive regulation of cell growth | 15467833 |
| Hgene | EGFR | GO:0042177 | negative regulation of protein catabolic process | 17115032 |
| Hgene | EGFR | GO:0042327 | positive regulation of phosphorylation | 15082764 |
| Hgene | EGFR | GO:0043406 | positive regulation of MAP kinase activity | 10572067 |
| Hgene | EGFR | GO:0045739 | positive regulation of DNA repair | 17115032 |
| Hgene | EGFR | GO:0045740 | positive regulation of DNA replication | 17115032 |
| Hgene | EGFR | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20551055 |
| Hgene | EGFR | GO:0050679 | positive regulation of epithelial cell proliferation | 10572067 |
| Hgene | EGFR | GO:0050999 | regulation of nitric-oxide synthase activity | 12828935 |
| Hgene | EGFR | GO:0070141 | response to UV-A | 18483258 |
| Hgene | EGFR | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 20551055 |
| Hgene | EGFR | GO:0071392 | cellular response to estradiol stimulus | 20551055 |
| Hgene | EGFR | GO:1900020 | positive regulation of protein kinase C activity | 22732145 |
| Hgene | EGFR | GO:1903078 | positive regulation of protein localization to plasma membrane | 22732145 |
 Fusion gene breakpoints across EGFR (5'-gene) Fusion gene breakpoints across EGFR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
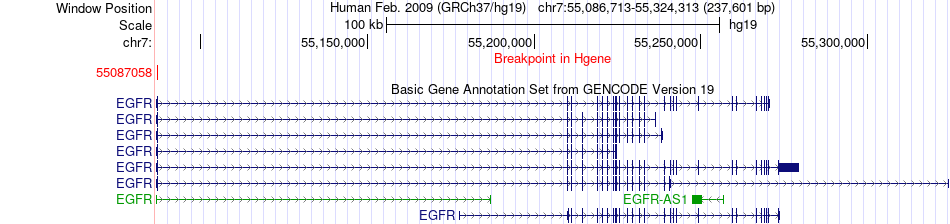 |
 Fusion gene breakpoints across INSL4 (3'-gene) Fusion gene breakpoints across INSL4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
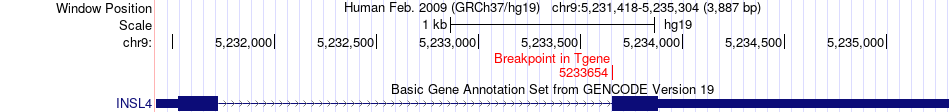 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-6706-01A | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
Top |
Fusion Gene ORF analysis for EGFR-INSL4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000463948 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| In-frame | ENST00000275493 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| In-frame | ENST00000342916 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| In-frame | ENST00000344576 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| In-frame | ENST00000420316 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| In-frame | ENST00000442591 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| In-frame | ENST00000455089 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
| intron-3CDS | ENST00000454757 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000455089 | EGFR | chr7 | 55087058 | + | ENST00000239316 | INSL4 | chr9 | 5233654 | + | 1996 | 345 | 398 | 0 | 133 |
| ENST00000342916 | EGFR | chr7 | 55087058 | + | ENST00000239316 | INSL4 | chr9 | 5233654 | + | 1985 | 334 | 472 | 83 | 129 |
| ENST00000344576 | EGFR | chr7 | 55087058 | + | ENST00000239316 | INSL4 | chr9 | 5233654 | + | 1984 | 333 | 471 | 82 | 129 |
| ENST00000420316 | EGFR | chr7 | 55087058 | + | ENST00000239316 | INSL4 | chr9 | 5233654 | + | 1983 | 332 | 470 | 81 | 129 |
| ENST00000275493 | EGFR | chr7 | 55087058 | + | ENST00000239316 | INSL4 | chr9 | 5233654 | + | 1916 | 265 | 403 | 14 | 129 |
| ENST00000442591 | EGFR | chr7 | 55087058 | + | ENST00000239316 | INSL4 | chr9 | 5233654 | + | 1899 | 248 | 386 | 0 | 129 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000455089 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + | 0.044436697 | 0.95556325 |
| ENST00000342916 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + | 0.040829748 | 0.9591703 |
| ENST00000344576 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + | 0.044101942 | 0.9558981 |
| ENST00000420316 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + | 0.036607247 | 0.9633928 |
| ENST00000275493 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + | 0.025662336 | 0.9743377 |
| ENST00000442591 | ENST00000239316 | EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233654 | + | 0.021379769 | 0.9786202 |
Top |
Fusion Genomic Features for EGFR-INSL4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233653 | + | 0.000744206 | 0.9992557 |
| EGFR | chr7 | 55087058 | + | INSL4 | chr9 | 5233653 | + | 0.000744206 | 0.9992557 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
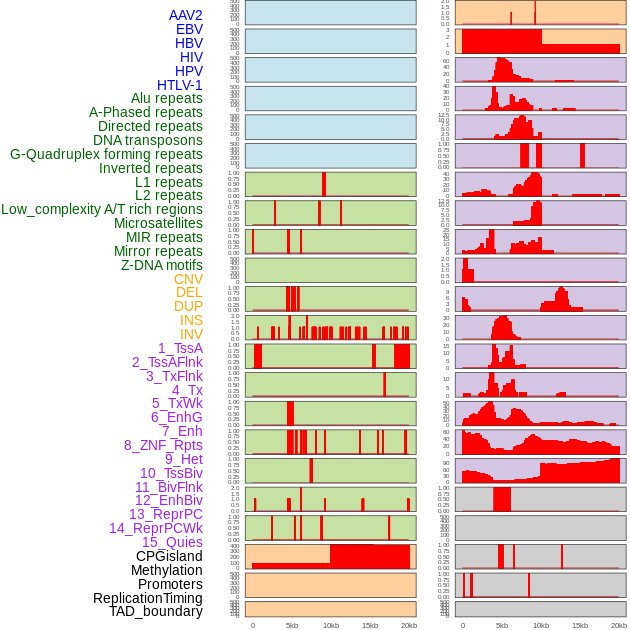 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
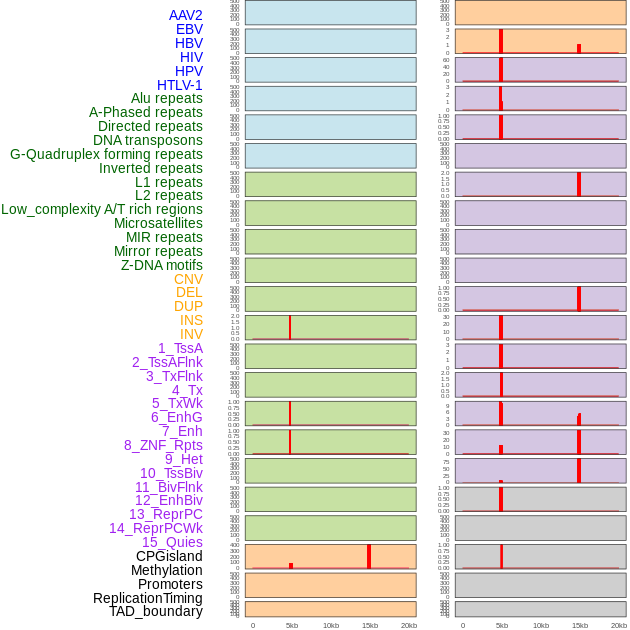 |
Top |
Fusion Protein Features for EGFR-INSL4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:55087058/chr9:5233654) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 712_979 | 29 | 1211.0 | Domain | Protein kinase |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 712_979 | 29 | 629.0 | Domain | Protein kinase |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 712_979 | 29 | 706.0 | Domain | Protein kinase |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 712_979 | 29 | 406.0 | Domain | Protein kinase |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 718_726 | 29 | 1211.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 790_791 | 29 | 1211.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 718_726 | 29 | 629.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 790_791 | 29 | 629.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 718_726 | 29 | 706.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 790_791 | 29 | 706.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 718_726 | 29 | 406.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 790_791 | 29 | 406.0 | Nucleotide binding | ATP |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 688_704 | 29 | 1211.0 | Region | Note=Important for dimerization%2C phosphorylation and activation |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 688_704 | 29 | 629.0 | Region | Note=Important for dimerization%2C phosphorylation and activation |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 688_704 | 29 | 706.0 | Region | Note=Important for dimerization%2C phosphorylation and activation |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 688_704 | 29 | 406.0 | Region | Note=Important for dimerization%2C phosphorylation and activation |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 390_600 | 29 | 1211.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 75_300 | 29 | 1211.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 390_600 | 29 | 629.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 75_300 | 29 | 629.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 390_600 | 29 | 706.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 75_300 | 29 | 706.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 390_600 | 29 | 406.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 75_300 | 29 | 406.0 | Repeat | Note=Approximate |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 25_645 | 29 | 1211.0 | Topological domain | Extracellular |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 669_1210 | 29 | 1211.0 | Topological domain | Cytoplasmic |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 25_645 | 29 | 629.0 | Topological domain | Extracellular |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 669_1210 | 29 | 629.0 | Topological domain | Cytoplasmic |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 25_645 | 29 | 706.0 | Topological domain | Extracellular |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 669_1210 | 29 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 25_645 | 29 | 406.0 | Topological domain | Extracellular |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 669_1210 | 29 | 406.0 | Topological domain | Cytoplasmic |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000275493 | + | 1 | 28 | 646_668 | 29 | 1211.0 | Transmembrane | Helical |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000342916 | + | 1 | 16 | 646_668 | 29 | 629.0 | Transmembrane | Helical |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000344576 | + | 1 | 16 | 646_668 | 29 | 706.0 | Transmembrane | Helical |
| Hgene | EGFR | chr7:55087058 | chr9:5233654 | ENST00000420316 | + | 1 | 10 | 646_668 | 29 | 406.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for EGFR-INSL4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25460_25460_1_EGFR-INSL4_EGFR_chr7_55087058_ENST00000275493_INSL4_chr9_5233654_ENST00000239316_length(transcript)=1916nt_BP=265nt GCCGGAGTCCCGAGCTAGCCCCGGCGGCCGCCGCCGCCCAGACCGGACGACAGGCCACCTCGTCGGCGTCCGCCCGAGTCCCCGCCTCGC CGCCAACGCCACAACCACCGCGCACGGCCCCCTGACTCCGTCCAGTATTGATCGGGAGAGCCGGAGCGAGCTCTTCGGGGAGCAGCGATG CGACCCTCCGGGACGGCCGGGGCAGCGCTCCTGGCGCTGCTGGCTGCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGAAATG GTGTCAACCTCCAACAACAAAGATGGACAAGCCTTAGGTACGACATCAGAATTCATTCCTAATTTGTCACCAGAGCTGAAGAAACCACTG TCTGAAGGGCAGCCATCATTGAAGAAAATAATACTTTCCCGCAAAAAGAGAAGTGGACGTCACAGATTTGATCCATTCTGTTGTGAAGTA ATTTGTGACGATGGAACTTCAGTTAAATTATGTACATAGTAGAGTAATCATGGACTGGACATCTCATCCATTCTCATATGTATTCTCAAT GACAAATTCACTGATGCCCAATTAAATGATTGCTGTTTATTAGAACATGAGAATCATTATTAGTATTCACATGTTTCACTTGCTCTGTAC TAATATAGTCTCTCCAAATGGAGGAAGTTAGATAGATGGAATAAGTTCTAGTAGTTGATAGTACAATGGGGAAATTATACTTAACAATAA TTCACTGTATATTCCAAAATAGCTAGAAGAGAATTGTAATGATTCCAACACAAAAAAGATGAATGTTAGTTTGGATGCATATCCCAATTA CCCTGATTTGATCATTACACATTATATACACGTATCAAAATATCACATGTACTACAAAAATGTGTATAACTGTAATATGTGAACTACAAA CTTACTGGAAATAAATTTTTAAAGTTTTACACACATAAACACATACGTTCACACACACTCACTCAAGTCTCTTCATTTTTCCTCTTTTCA CACTAGAAGAAATGTAAGAGATAAAGGATCTAACCATCTCTAACATCAGCTCATAGTACGGAGGAAAAGCAGTGAAGGAATGAACTGGTG GGAGAGGTATTAGGCATGTTCACCCTAGGGTAAACCCAGAGACCTCCCTAGAACTTTGCCCAATGCATATCCTCCACCTGCCTCTCAGCA CGCTGCCTTTAGACCTTCAATCATTCTATAAACTCCTCTTTTGTCCCTGGCATTACTCCACATCCCCTCATCAAAGTGGTGAATTGGCAC AAATACTGGCATTCCTCTGGCTACACATACCTCCTCCCTATGTGTCTCTAAATCCTAAATCAAAGAGTACATTTTATACTCTGTCAGTGA GGTTCTACTAGTTGTGTTTGTTTGTTTGCAGGCTAACTGAAGCTGCTACTACCTCATTATCAGGTCCTATAAACTTAATTCAAGTGGCAT GATTTGAGTAGCCCCCTGTGCCTGCCCATAAAGTGCTCTGAATCTAGTGTGGGACATGGAGAAGCAAAGGATAATAAGTGCAATGATACT TGTAGGTCATCATAGAGACTGTAATGATCCACAGGAATCAGGGTCGTGTTTAATGCCAACATGTGAAAGAATAAATATACAGGGAACAGT GTAGGTAAGAGGAGAGAGCAAAATTTCCCTAAGGAAACTGATGGCTGATCTGCATCTTGAAAGACATCTAAGCAGAAGAGAGTGCAAAAA GGGACAAGTAGAAGCCCAGGAAAATGCACAAAGGCCTGGAAGCATAGCATGTGTGGAAACAGCCAGAGTCAGCAGGGCAGGAGGTGGTGT GGAGAAAAGGAATTGTCAAGATCTGGTTGGAATGGAGACAGGAGAGAGGATCTCAAACGATTCCACACAATGCAGCTATGGGGTTTAGGC TGCATCCTGGAGTAAGGAAGAACTGC >25460_25460_1_EGFR-INSL4_EGFR_chr7_55087058_ENST00000275493_INSL4_chr9_5233654_ENST00000239316_length(amino acids)=129AA_BP=1 MRESIIFFNDGCPSDSGFFSSGDKLGMNSDVVPKACPSLLLEVDTISFFSSRARLAGQSAASSARSAAPAVPEGRIAAPRRARSGSPDQY WTESGGRARWLWRWRRGGDSGGRRRGGLSSGLGGGGRRG -------------------------------------------------------------- >25460_25460_2_EGFR-INSL4_EGFR_chr7_55087058_ENST00000342916_INSL4_chr9_5233654_ENST00000239316_length(transcript)=1985nt_BP=334nt CCCCGGCGCAGCGCGGCCGCAGCAGCCTCCGCCCCCCGCACGGTGTGAGCGCCCGACGCGGCCGAGGCGGCCGGAGTCCCGAGCTAGCCC CGGCGGCCGCCGCCGCCCAGACCGGACGACAGGCCACCTCGTCGGCGTCCGCCCGAGTCCCCGCCTCGCCGCCAACGCCACAACCACCGC GCACGGCCCCCTGACTCCGTCCAGTATTGATCGGGAGAGCCGGAGCGAGCTCTTCGGGGAGCAGCGATGCGACCCTCCGGGACGGCCGGG GCAGCGCTCCTGGCGCTGCTGGCTGCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGAAATGGTGTCAACCTCCAACAACAAA GATGGACAAGCCTTAGGTACGACATCAGAATTCATTCCTAATTTGTCACCAGAGCTGAAGAAACCACTGTCTGAAGGGCAGCCATCATTG AAGAAAATAATACTTTCCCGCAAAAAGAGAAGTGGACGTCACAGATTTGATCCATTCTGTTGTGAAGTAATTTGTGACGATGGAACTTCA GTTAAATTATGTACATAGTAGAGTAATCATGGACTGGACATCTCATCCATTCTCATATGTATTCTCAATGACAAATTCACTGATGCCCAA TTAAATGATTGCTGTTTATTAGAACATGAGAATCATTATTAGTATTCACATGTTTCACTTGCTCTGTACTAATATAGTCTCTCCAAATGG AGGAAGTTAGATAGATGGAATAAGTTCTAGTAGTTGATAGTACAATGGGGAAATTATACTTAACAATAATTCACTGTATATTCCAAAATA GCTAGAAGAGAATTGTAATGATTCCAACACAAAAAAGATGAATGTTAGTTTGGATGCATATCCCAATTACCCTGATTTGATCATTACACA TTATATACACGTATCAAAATATCACATGTACTACAAAAATGTGTATAACTGTAATATGTGAACTACAAACTTACTGGAAATAAATTTTTA AAGTTTTACACACATAAACACATACGTTCACACACACTCACTCAAGTCTCTTCATTTTTCCTCTTTTCACACTAGAAGAAATGTAAGAGA TAAAGGATCTAACCATCTCTAACATCAGCTCATAGTACGGAGGAAAAGCAGTGAAGGAATGAACTGGTGGGAGAGGTATTAGGCATGTTC ACCCTAGGGTAAACCCAGAGACCTCCCTAGAACTTTGCCCAATGCATATCCTCCACCTGCCTCTCAGCACGCTGCCTTTAGACCTTCAAT CATTCTATAAACTCCTCTTTTGTCCCTGGCATTACTCCACATCCCCTCATCAAAGTGGTGAATTGGCACAAATACTGGCATTCCTCTGGC TACACATACCTCCTCCCTATGTGTCTCTAAATCCTAAATCAAAGAGTACATTTTATACTCTGTCAGTGAGGTTCTACTAGTTGTGTTTGT TTGTTTGCAGGCTAACTGAAGCTGCTACTACCTCATTATCAGGTCCTATAAACTTAATTCAAGTGGCATGATTTGAGTAGCCCCCTGTGC CTGCCCATAAAGTGCTCTGAATCTAGTGTGGGACATGGAGAAGCAAAGGATAATAAGTGCAATGATACTTGTAGGTCATCATAGAGACTG TAATGATCCACAGGAATCAGGGTCGTGTTTAATGCCAACATGTGAAAGAATAAATATACAGGGAACAGTGTAGGTAAGAGGAGAGAGCAA AATTTCCCTAAGGAAACTGATGGCTGATCTGCATCTTGAAAGACATCTAAGCAGAAGAGAGTGCAAAAAGGGACAAGTAGAAGCCCAGGA AAATGCACAAAGGCCTGGAAGCATAGCATGTGTGGAAACAGCCAGAGTCAGCAGGGCAGGAGGTGGTGTGGAGAAAAGGAATTGTCAAGA TCTGGTTGGAATGGAGACAGGAGAGAGGATCTCAAACGATTCCACACAATGCAGCTATGGGGTTTAGGCTGCATCCTGGAGTAAGGAAGA ACTGC >25460_25460_2_EGFR-INSL4_EGFR_chr7_55087058_ENST00000342916_INSL4_chr9_5233654_ENST00000239316_length(amino acids)=129AA_BP=1 MRESIIFFNDGCPSDSGFFSSGDKLGMNSDVVPKACPSLLLEVDTISFFSSRARLAGQSAASSARSAAPAVPEGRIAAPRRARSGSPDQY WTESGGRARWLWRWRRGGDSGGRRRGGLSSGLGGGGRRG -------------------------------------------------------------- >25460_25460_3_EGFR-INSL4_EGFR_chr7_55087058_ENST00000344576_INSL4_chr9_5233654_ENST00000239316_length(transcript)=1984nt_BP=333nt CCCGGCGCAGCGCGGCCGCAGCAGCCTCCGCCCCCCGCACGGTGTGAGCGCCCGACGCGGCCGAGGCGGCCGGAGTCCCGAGCTAGCCCC GGCGGCCGCCGCCGCCCAGACCGGACGACAGGCCACCTCGTCGGCGTCCGCCCGAGTCCCCGCCTCGCCGCCAACGCCACAACCACCGCG CACGGCCCCCTGACTCCGTCCAGTATTGATCGGGAGAGCCGGAGCGAGCTCTTCGGGGAGCAGCGATGCGACCCTCCGGGACGGCCGGGG CAGCGCTCCTGGCGCTGCTGGCTGCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGAAATGGTGTCAACCTCCAACAACAAAG ATGGACAAGCCTTAGGTACGACATCAGAATTCATTCCTAATTTGTCACCAGAGCTGAAGAAACCACTGTCTGAAGGGCAGCCATCATTGA AGAAAATAATACTTTCCCGCAAAAAGAGAAGTGGACGTCACAGATTTGATCCATTCTGTTGTGAAGTAATTTGTGACGATGGAACTTCAG TTAAATTATGTACATAGTAGAGTAATCATGGACTGGACATCTCATCCATTCTCATATGTATTCTCAATGACAAATTCACTGATGCCCAAT TAAATGATTGCTGTTTATTAGAACATGAGAATCATTATTAGTATTCACATGTTTCACTTGCTCTGTACTAATATAGTCTCTCCAAATGGA GGAAGTTAGATAGATGGAATAAGTTCTAGTAGTTGATAGTACAATGGGGAAATTATACTTAACAATAATTCACTGTATATTCCAAAATAG CTAGAAGAGAATTGTAATGATTCCAACACAAAAAAGATGAATGTTAGTTTGGATGCATATCCCAATTACCCTGATTTGATCATTACACAT TATATACACGTATCAAAATATCACATGTACTACAAAAATGTGTATAACTGTAATATGTGAACTACAAACTTACTGGAAATAAATTTTTAA AGTTTTACACACATAAACACATACGTTCACACACACTCACTCAAGTCTCTTCATTTTTCCTCTTTTCACACTAGAAGAAATGTAAGAGAT AAAGGATCTAACCATCTCTAACATCAGCTCATAGTACGGAGGAAAAGCAGTGAAGGAATGAACTGGTGGGAGAGGTATTAGGCATGTTCA CCCTAGGGTAAACCCAGAGACCTCCCTAGAACTTTGCCCAATGCATATCCTCCACCTGCCTCTCAGCACGCTGCCTTTAGACCTTCAATC ATTCTATAAACTCCTCTTTTGTCCCTGGCATTACTCCACATCCCCTCATCAAAGTGGTGAATTGGCACAAATACTGGCATTCCTCTGGCT ACACATACCTCCTCCCTATGTGTCTCTAAATCCTAAATCAAAGAGTACATTTTATACTCTGTCAGTGAGGTTCTACTAGTTGTGTTTGTT TGTTTGCAGGCTAACTGAAGCTGCTACTACCTCATTATCAGGTCCTATAAACTTAATTCAAGTGGCATGATTTGAGTAGCCCCCTGTGCC TGCCCATAAAGTGCTCTGAATCTAGTGTGGGACATGGAGAAGCAAAGGATAATAAGTGCAATGATACTTGTAGGTCATCATAGAGACTGT AATGATCCACAGGAATCAGGGTCGTGTTTAATGCCAACATGTGAAAGAATAAATATACAGGGAACAGTGTAGGTAAGAGGAGAGAGCAAA ATTTCCCTAAGGAAACTGATGGCTGATCTGCATCTTGAAAGACATCTAAGCAGAAGAGAGTGCAAAAAGGGACAAGTAGAAGCCCAGGAA AATGCACAAAGGCCTGGAAGCATAGCATGTGTGGAAACAGCCAGAGTCAGCAGGGCAGGAGGTGGTGTGGAGAAAAGGAATTGTCAAGAT CTGGTTGGAATGGAGACAGGAGAGAGGATCTCAAACGATTCCACACAATGCAGCTATGGGGTTTAGGCTGCATCCTGGAGTAAGGAAGAA CTGC >25460_25460_3_EGFR-INSL4_EGFR_chr7_55087058_ENST00000344576_INSL4_chr9_5233654_ENST00000239316_length(amino acids)=129AA_BP=1 MRESIIFFNDGCPSDSGFFSSGDKLGMNSDVVPKACPSLLLEVDTISFFSSRARLAGQSAASSARSAAPAVPEGRIAAPRRARSGSPDQY WTESGGRARWLWRWRRGGDSGGRRRGGLSSGLGGGGRRG -------------------------------------------------------------- >25460_25460_4_EGFR-INSL4_EGFR_chr7_55087058_ENST00000420316_INSL4_chr9_5233654_ENST00000239316_length(transcript)=1983nt_BP=332nt CCGGCGCAGCGCGGCCGCAGCAGCCTCCGCCCCCCGCACGGTGTGAGCGCCCGACGCGGCCGAGGCGGCCGGAGTCCCGAGCTAGCCCCG GCGGCCGCCGCCGCCCAGACCGGACGACAGGCCACCTCGTCGGCGTCCGCCCGAGTCCCCGCCTCGCCGCCAACGCCACAACCACCGCGC ACGGCCCCCTGACTCCGTCCAGTATTGATCGGGAGAGCCGGAGCGAGCTCTTCGGGGAGCAGCGATGCGACCCTCCGGGACGGCCGGGGC AGCGCTCCTGGCGCTGCTGGCTGCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGAAATGGTGTCAACCTCCAACAACAAAGA TGGACAAGCCTTAGGTACGACATCAGAATTCATTCCTAATTTGTCACCAGAGCTGAAGAAACCACTGTCTGAAGGGCAGCCATCATTGAA GAAAATAATACTTTCCCGCAAAAAGAGAAGTGGACGTCACAGATTTGATCCATTCTGTTGTGAAGTAATTTGTGACGATGGAACTTCAGT TAAATTATGTACATAGTAGAGTAATCATGGACTGGACATCTCATCCATTCTCATATGTATTCTCAATGACAAATTCACTGATGCCCAATT AAATGATTGCTGTTTATTAGAACATGAGAATCATTATTAGTATTCACATGTTTCACTTGCTCTGTACTAATATAGTCTCTCCAAATGGAG GAAGTTAGATAGATGGAATAAGTTCTAGTAGTTGATAGTACAATGGGGAAATTATACTTAACAATAATTCACTGTATATTCCAAAATAGC TAGAAGAGAATTGTAATGATTCCAACACAAAAAAGATGAATGTTAGTTTGGATGCATATCCCAATTACCCTGATTTGATCATTACACATT ATATACACGTATCAAAATATCACATGTACTACAAAAATGTGTATAACTGTAATATGTGAACTACAAACTTACTGGAAATAAATTTTTAAA GTTTTACACACATAAACACATACGTTCACACACACTCACTCAAGTCTCTTCATTTTTCCTCTTTTCACACTAGAAGAAATGTAAGAGATA AAGGATCTAACCATCTCTAACATCAGCTCATAGTACGGAGGAAAAGCAGTGAAGGAATGAACTGGTGGGAGAGGTATTAGGCATGTTCAC CCTAGGGTAAACCCAGAGACCTCCCTAGAACTTTGCCCAATGCATATCCTCCACCTGCCTCTCAGCACGCTGCCTTTAGACCTTCAATCA TTCTATAAACTCCTCTTTTGTCCCTGGCATTACTCCACATCCCCTCATCAAAGTGGTGAATTGGCACAAATACTGGCATTCCTCTGGCTA CACATACCTCCTCCCTATGTGTCTCTAAATCCTAAATCAAAGAGTACATTTTATACTCTGTCAGTGAGGTTCTACTAGTTGTGTTTGTTT GTTTGCAGGCTAACTGAAGCTGCTACTACCTCATTATCAGGTCCTATAAACTTAATTCAAGTGGCATGATTTGAGTAGCCCCCTGTGCCT GCCCATAAAGTGCTCTGAATCTAGTGTGGGACATGGAGAAGCAAAGGATAATAAGTGCAATGATACTTGTAGGTCATCATAGAGACTGTA ATGATCCACAGGAATCAGGGTCGTGTTTAATGCCAACATGTGAAAGAATAAATATACAGGGAACAGTGTAGGTAAGAGGAGAGAGCAAAA TTTCCCTAAGGAAACTGATGGCTGATCTGCATCTTGAAAGACATCTAAGCAGAAGAGAGTGCAAAAAGGGACAAGTAGAAGCCCAGGAAA ATGCACAAAGGCCTGGAAGCATAGCATGTGTGGAAACAGCCAGAGTCAGCAGGGCAGGAGGTGGTGTGGAGAAAAGGAATTGTCAAGATC TGGTTGGAATGGAGACAGGAGAGAGGATCTCAAACGATTCCACACAATGCAGCTATGGGGTTTAGGCTGCATCCTGGAGTAAGGAAGAAC TGC >25460_25460_4_EGFR-INSL4_EGFR_chr7_55087058_ENST00000420316_INSL4_chr9_5233654_ENST00000239316_length(amino acids)=129AA_BP=1 MRESIIFFNDGCPSDSGFFSSGDKLGMNSDVVPKACPSLLLEVDTISFFSSRARLAGQSAASSARSAAPAVPEGRIAAPRRARSGSPDQY WTESGGRARWLWRWRRGGDSGGRRRGGLSSGLGGGGRRG -------------------------------------------------------------- >25460_25460_5_EGFR-INSL4_EGFR_chr7_55087058_ENST00000442591_INSL4_chr9_5233654_ENST00000239316_length(transcript)=1899nt_BP=248nt GCCCCGGCGGCCGCCGCCGCCCAGACCGGACGACAGGCCACCTCGTCGGCGTCCGCCCGAGTCCCCGCCTCGCCGCCAACGCCACAACCA CCGCGCACGGCCCCCTGACTCCGTCCAGTATTGATCGGGAGAGCCGGAGCGAGCTCTTCGGGGAGCAGCGATGCGACCCTCCGGGACGGC CGGGGCAGCGCTCCTGGCGCTGCTGGCTGCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGAAATGGTGTCAACCTCCAACAA CAAAGATGGACAAGCCTTAGGTACGACATCAGAATTCATTCCTAATTTGTCACCAGAGCTGAAGAAACCACTGTCTGAAGGGCAGCCATC ATTGAAGAAAATAATACTTTCCCGCAAAAAGAGAAGTGGACGTCACAGATTTGATCCATTCTGTTGTGAAGTAATTTGTGACGATGGAAC TTCAGTTAAATTATGTACATAGTAGAGTAATCATGGACTGGACATCTCATCCATTCTCATATGTATTCTCAATGACAAATTCACTGATGC CCAATTAAATGATTGCTGTTTATTAGAACATGAGAATCATTATTAGTATTCACATGTTTCACTTGCTCTGTACTAATATAGTCTCTCCAA ATGGAGGAAGTTAGATAGATGGAATAAGTTCTAGTAGTTGATAGTACAATGGGGAAATTATACTTAACAATAATTCACTGTATATTCCAA AATAGCTAGAAGAGAATTGTAATGATTCCAACACAAAAAAGATGAATGTTAGTTTGGATGCATATCCCAATTACCCTGATTTGATCATTA CACATTATATACACGTATCAAAATATCACATGTACTACAAAAATGTGTATAACTGTAATATGTGAACTACAAACTTACTGGAAATAAATT TTTAAAGTTTTACACACATAAACACATACGTTCACACACACTCACTCAAGTCTCTTCATTTTTCCTCTTTTCACACTAGAAGAAATGTAA GAGATAAAGGATCTAACCATCTCTAACATCAGCTCATAGTACGGAGGAAAAGCAGTGAAGGAATGAACTGGTGGGAGAGGTATTAGGCAT GTTCACCCTAGGGTAAACCCAGAGACCTCCCTAGAACTTTGCCCAATGCATATCCTCCACCTGCCTCTCAGCACGCTGCCTTTAGACCTT CAATCATTCTATAAACTCCTCTTTTGTCCCTGGCATTACTCCACATCCCCTCATCAAAGTGGTGAATTGGCACAAATACTGGCATTCCTC TGGCTACACATACCTCCTCCCTATGTGTCTCTAAATCCTAAATCAAAGAGTACATTTTATACTCTGTCAGTGAGGTTCTACTAGTTGTGT TTGTTTGTTTGCAGGCTAACTGAAGCTGCTACTACCTCATTATCAGGTCCTATAAACTTAATTCAAGTGGCATGATTTGAGTAGCCCCCT GTGCCTGCCCATAAAGTGCTCTGAATCTAGTGTGGGACATGGAGAAGCAAAGGATAATAAGTGCAATGATACTTGTAGGTCATCATAGAG ACTGTAATGATCCACAGGAATCAGGGTCGTGTTTAATGCCAACATGTGAAAGAATAAATATACAGGGAACAGTGTAGGTAAGAGGAGAGA GCAAAATTTCCCTAAGGAAACTGATGGCTGATCTGCATCTTGAAAGACATCTAAGCAGAAGAGAGTGCAAAAAGGGACAAGTAGAAGCCC AGGAAAATGCACAAAGGCCTGGAAGCATAGCATGTGTGGAAACAGCCAGAGTCAGCAGGGCAGGAGGTGGTGTGGAGAAAAGGAATTGTC AAGATCTGGTTGGAATGGAGACAGGAGAGAGGATCTCAAACGATTCCACACAATGCAGCTATGGGGTTTAGGCTGCATCCTGGAGTAAGG AAGAACTGC >25460_25460_5_EGFR-INSL4_EGFR_chr7_55087058_ENST00000442591_INSL4_chr9_5233654_ENST00000239316_length(amino acids)=129AA_BP=1 MRESIIFFNDGCPSDSGFFSSGDKLGMNSDVVPKACPSLLLEVDTISFFSSRARLAGQSAASSARSAAPAVPEGRIAAPRRARSGSPDQY WTESGGRARWLWRWRRGGDSGGRRRGGLSSGLGGGGRRG -------------------------------------------------------------- >25460_25460_6_EGFR-INSL4_EGFR_chr7_55087058_ENST00000455089_INSL4_chr9_5233654_ENST00000239316_length(transcript)=1996nt_BP=345nt GTCCGGGCAGCCCCCGGCGCAGCGCGGCCGCAGCAGCCTCCGCCCCCCGCACGGTGTGAGCGCCCGACGCGGCCGAGGCGGCCGGAGTCC CGAGCTAGCCCCGGCGGCCGCCGCCGCCCAGACCGGACGACAGGCCACCTCGTCGGCGTCCGCCCGAGTCCCCGCCTCGCCGCCAACGCC ACAACCACCGCGCACGGCCCCCTGACTCCGTCCAGTATTGATCGGGAGAGCCGGAGCGAGCTCTTCGGGGAGCAGCGATGCGACCCTCCG GGACGGCCGGGGCAGCGCTCCTGGCGCTGCTGGCTGCGCTCTGCCCGGCGAGTCGGGCTCTGGAGGAAAAGAAAGAAATGGTGTCAACCT CCAACAACAAAGATGGACAAGCCTTAGGTACGACATCAGAATTCATTCCTAATTTGTCACCAGAGCTGAAGAAACCACTGTCTGAAGGGC AGCCATCATTGAAGAAAATAATACTTTCCCGCAAAAAGAGAAGTGGACGTCACAGATTTGATCCATTCTGTTGTGAAGTAATTTGTGACG ATGGAACTTCAGTTAAATTATGTACATAGTAGAGTAATCATGGACTGGACATCTCATCCATTCTCATATGTATTCTCAATGACAAATTCA CTGATGCCCAATTAAATGATTGCTGTTTATTAGAACATGAGAATCATTATTAGTATTCACATGTTTCACTTGCTCTGTACTAATATAGTC TCTCCAAATGGAGGAAGTTAGATAGATGGAATAAGTTCTAGTAGTTGATAGTACAATGGGGAAATTATACTTAACAATAATTCACTGTAT ATTCCAAAATAGCTAGAAGAGAATTGTAATGATTCCAACACAAAAAAGATGAATGTTAGTTTGGATGCATATCCCAATTACCCTGATTTG ATCATTACACATTATATACACGTATCAAAATATCACATGTACTACAAAAATGTGTATAACTGTAATATGTGAACTACAAACTTACTGGAA ATAAATTTTTAAAGTTTTACACACATAAACACATACGTTCACACACACTCACTCAAGTCTCTTCATTTTTCCTCTTTTCACACTAGAAGA AATGTAAGAGATAAAGGATCTAACCATCTCTAACATCAGCTCATAGTACGGAGGAAAAGCAGTGAAGGAATGAACTGGTGGGAGAGGTAT TAGGCATGTTCACCCTAGGGTAAACCCAGAGACCTCCCTAGAACTTTGCCCAATGCATATCCTCCACCTGCCTCTCAGCACGCTGCCTTT AGACCTTCAATCATTCTATAAACTCCTCTTTTGTCCCTGGCATTACTCCACATCCCCTCATCAAAGTGGTGAATTGGCACAAATACTGGC ATTCCTCTGGCTACACATACCTCCTCCCTATGTGTCTCTAAATCCTAAATCAAAGAGTACATTTTATACTCTGTCAGTGAGGTTCTACTA GTTGTGTTTGTTTGTTTGCAGGCTAACTGAAGCTGCTACTACCTCATTATCAGGTCCTATAAACTTAATTCAAGTGGCATGATTTGAGTA GCCCCCTGTGCCTGCCCATAAAGTGCTCTGAATCTAGTGTGGGACATGGAGAAGCAAAGGATAATAAGTGCAATGATACTTGTAGGTCAT CATAGAGACTGTAATGATCCACAGGAATCAGGGTCGTGTTTAATGCCAACATGTGAAAGAATAAATATACAGGGAACAGTGTAGGTAAGA GGAGAGAGCAAAATTTCCCTAAGGAAACTGATGGCTGATCTGCATCTTGAAAGACATCTAAGCAGAAGAGAGTGCAAAAAGGGACAAGTA GAAGCCCAGGAAAATGCACAAAGGCCTGGAAGCATAGCATGTGTGGAAACAGCCAGAGTCAGCAGGGCAGGAGGTGGTGTGGAGAAAAGG AATTGTCAAGATCTGGTTGGAATGGAGACAGGAGAGAGGATCTCAAACGATTCCACACAATGCAGCTATGGGGTTTAGGCTGCATCCTGG AGTAAGGAAGAACTGC >25460_25460_6_EGFR-INSL4_EGFR_chr7_55087058_ENST00000455089_INSL4_chr9_5233654_ENST00000239316_length(amino acids)=133AA_BP=1 MMSYLRLVHLCCWRLTPFLSFPPEPDSPGRAQPAAPGALPRPSRRVASLLPEELAPALPINTGRSQGAVRGGCGVGGEAGTRADADEVAC RPVWAAAAAGASSGLRPPRPRRALTPCGGRRLLRPRCAGGCPD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EGFR-INSL4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EGFR-INSL4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EGFR-INSL4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | EGFR | C0007131 | Non-Small Cell Lung Carcinoma | 16 | CGI;CTD_human |
| Hgene | EGFR | C0024121 | Lung Neoplasms | 7 | CGI;CTD_human |
| Hgene | EGFR | C0242379 | Malignant neoplasm of lung | 7 | CGI;CTD_human |
| Hgene | EGFR | C0006142 | Malignant neoplasm of breast | 6 | CTD_human |
| Hgene | EGFR | C0678222 | Breast Carcinoma | 6 | CTD_human |
| Hgene | EGFR | C1257931 | Mammary Neoplasms, Human | 6 | CTD_human |
| Hgene | EGFR | C1458155 | Mammary Neoplasms | 6 | CTD_human |
| Hgene | EGFR | C4704874 | Mammary Carcinoma, Human | 6 | CTD_human |
| Hgene | EGFR | C0001418 | Adenocarcinoma | 5 | CTD_human |
| Hgene | EGFR | C0205641 | Adenocarcinoma, Basal Cell | 5 | CTD_human |
| Hgene | EGFR | C0205642 | Adenocarcinoma, Oxyphilic | 5 | CTD_human |
| Hgene | EGFR | C0205643 | Carcinoma, Cribriform | 5 | CTD_human |
| Hgene | EGFR | C0205644 | Carcinoma, Granular Cell | 5 | CTD_human |
| Hgene | EGFR | C0205645 | Adenocarcinoma, Tubular | 5 | CTD_human |
| Hgene | EGFR | C0334588 | Giant Cell Glioblastoma | 5 | CTD_human;ORPHANET |
| Hgene | EGFR | C0007137 | Squamous cell carcinoma | 3 | CTD_human |
| Hgene | EGFR | C0007873 | Uterine Cervical Neoplasm | 3 | CTD_human |
| Hgene | EGFR | C0014859 | Esophageal Neoplasms | 3 | CGI;CTD_human |
| Hgene | EGFR | C0017636 | Glioblastoma | 3 | CGI;CTD_human |
| Hgene | EGFR | C0018671 | Head and Neck Neoplasms | 3 | CGI;CTD_human |
| Hgene | EGFR | C0018675 | Head Neoplasms | 3 | CTD_human |
| Hgene | EGFR | C0024623 | Malignant neoplasm of stomach | 3 | CTD_human |
| Hgene | EGFR | C0027533 | Neck Neoplasms | 3 | CTD_human |
| Hgene | EGFR | C0027627 | Neoplasm Metastasis | 3 | CTD_human |
| Hgene | EGFR | C0033578 | Prostatic Neoplasms | 3 | CTD_human |
| Hgene | EGFR | C0038356 | Stomach Neoplasms | 3 | CTD_human |
| Hgene | EGFR | C0278996 | Malignant Head and Neck Neoplasm | 3 | CGI;CTD_human |
| Hgene | EGFR | C0376358 | Malignant neoplasm of prostate | 3 | CTD_human |
| Hgene | EGFR | C0546837 | Malignant neoplasm of esophagus | 3 | CGI;CTD_human |
| Hgene | EGFR | C0746787 | Cancer of Neck | 3 | CTD_human |
| Hgene | EGFR | C0751177 | Cancer of Head | 3 | CTD_human |
| Hgene | EGFR | C0887900 | Upper Aerodigestive Tract Neoplasms | 3 | CTD_human |
| Hgene | EGFR | C1621958 | Glioblastoma Multiforme | 3 | CTD_human |
| Hgene | EGFR | C1708349 | Hereditary Diffuse Gastric Cancer | 3 | CTD_human |
| Hgene | EGFR | C4048328 | cervical cancer | 3 | CTD_human |
| Hgene | EGFR | C0005684 | Malignant neoplasm of urinary bladder | 2 | CTD_human |
| Hgene | EGFR | C0005695 | Bladder Neoplasm | 2 | CTD_human |
| Hgene | EGFR | C0007102 | Malignant tumor of colon | 2 | CTD_human |
| Hgene | EGFR | C0009375 | Colonic Neoplasms | 2 | CTD_human |
| Hgene | EGFR | C0009402 | Colorectal Carcinoma | 2 | CTD_human |
| Hgene | EGFR | C0009404 | Colorectal Neoplasms | 2 | CTD_human |
| Hgene | EGFR | C0024668 | Mammary Neoplasms, Experimental | 2 | CTD_human |
| Hgene | EGFR | C0027626 | Neoplasm Invasiveness | 2 | CTD_human |
| Hgene | EGFR | C0027643 | Neoplasm Recurrence, Local | 2 | CTD_human |
| Hgene | EGFR | C0152013 | Adenocarcinoma of lung (disorder) | 2 | CGI;CTD_human |
| Hgene | EGFR | C0206726 | gliosarcoma | 2 | ORPHANET |
| Hgene | EGFR | C0919267 | ovarian neoplasm | 2 | CTD_human |
| Hgene | EGFR | C1140680 | Malignant neoplasm of ovary | 2 | CTD_human |
| Hgene | EGFR | C4015130 | INFLAMMATORY SKIN AND BOWEL DISEASE, NEONATAL, 2 | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | EGFR | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
| Hgene | EGFR | C0003865 | Arthritis, Adjuvant-Induced | 1 | CTD_human |
| Hgene | EGFR | C0005396 | Bile Duct Neoplasms | 1 | CTD_human |
| Hgene | EGFR | C0007097 | Carcinoma | 1 | CTD_human |
| Hgene | EGFR | C0007113 | Rectal Carcinoma | 1 | CTD_human |
| Hgene | EGFR | C0007193 | Cardiomyopathy, Dilated | 1 | CTD_human |
| Hgene | EGFR | C0011603 | Dermatitis | 1 | CTD_human |
| Hgene | EGFR | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Hgene | EGFR | C0014175 | Endometriosis | 1 | CTD_human |
| Hgene | EGFR | C0016978 | gallbladder neoplasm | 1 | CTD_human |
| Hgene | EGFR | C0021655 | Insulin Resistance | 1 | CTD_human |
| Hgene | EGFR | C0022660 | Kidney Failure, Acute | 1 | CTD_human |
| Hgene | EGFR | C0024667 | Animal Mammary Neoplasms | 1 | CTD_human |
| Hgene | EGFR | C0024809 | Marijuana Abuse | 1 | PSYGENET |
| Hgene | EGFR | C0025500 | Mesothelioma | 1 | CTD_human |
| Hgene | EGFR | C0027439 | Nasopharyngeal Neoplasms | 1 | CTD_human |
| Hgene | EGFR | C0029463 | Osteosarcoma | 1 | CTD_human |
| Hgene | EGFR | C0030297 | Pancreatic Neoplasm | 1 | CTD_human |
| Hgene | EGFR | C0030354 | Papilloma | 1 | CTD_human |
| Hgene | EGFR | C0032580 | Adenomatous Polyposis Coli | 1 | CTD_human |
| Hgene | EGFR | C0034885 | Rectal Neoplasms | 1 | CTD_human |
| Hgene | EGFR | C0041696 | Unipolar Depression | 1 | PSYGENET |
| Hgene | EGFR | C0085548 | Autosomal Recessive Polycystic Kidney Disease | 1 | CTD_human |
| Hgene | EGFR | C0085762 | Alcohol abuse | 1 | PSYGENET |
| Hgene | EGFR | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Hgene | EGFR | C0153452 | Malignant neoplasm of gallbladder | 1 | CTD_human |
| Hgene | EGFR | C0205696 | Anaplastic carcinoma | 1 | CTD_human |
| Hgene | EGFR | C0205697 | Carcinoma, Spindle-Cell | 1 | CTD_human |
| Hgene | EGFR | C0205698 | Undifferentiated carcinoma | 1 | CTD_human |
| Hgene | EGFR | C0205699 | Carcinomatosis | 1 | CTD_human |
| Hgene | EGFR | C0205874 | Papilloma, Squamous Cell | 1 | CTD_human |
| Hgene | EGFR | C0205875 | Papillomatosis | 1 | CTD_human |
| Hgene | EGFR | C0206686 | Adrenocortical carcinoma | 1 | CTD_human |
| Hgene | EGFR | C0206698 | Cholangiocarcinoma | 1 | CTD_human |
| Hgene | EGFR | C0235874 | Disease Exacerbation | 1 | CTD_human |
| Hgene | EGFR | C0238301 | Cancer of Nasopharynx | 1 | CTD_human |
| Hgene | EGFR | C0263454 | Chloracne | 1 | CTD_human |
| Hgene | EGFR | C0269102 | Endometrioma | 1 | CTD_human |
| Hgene | EGFR | C0279626 | Squamous cell carcinoma of esophagus | 1 | CGI;CTD_human |
| Hgene | EGFR | C0345905 | Intrahepatic Cholangiocarcinoma | 1 | CTD_human |
| Hgene | EGFR | C0345967 | Malignant mesothelioma | 1 | CTD_human |
| Hgene | EGFR | C0346647 | Malignant neoplasm of pancreas | 1 | CTD_human |
| Hgene | EGFR | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Hgene | EGFR | C0740277 | Bile duct carcinoma | 1 | CTD_human |
| Hgene | EGFR | C0920563 | Insulin Sensitivity | 1 | CTD_human |
| Hgene | EGFR | C0971858 | Arthritis, Collagen-Induced | 1 | CTD_human |
| Hgene | EGFR | C0993582 | Arthritis, Experimental | 1 | CTD_human |
| Hgene | EGFR | C1257925 | Mammary Carcinoma, Animal | 1 | CTD_human |
| Hgene | EGFR | C1269683 | Major Depressive Disorder | 1 | PSYGENET |
| Hgene | EGFR | C1449563 | Cardiomyopathy, Familial Idiopathic | 1 | CTD_human |
| Hgene | EGFR | C1565662 | Acute Kidney Insufficiency | 1 | CTD_human |
| Hgene | EGFR | C2239176 | Liver carcinoma | 1 | CTD_human |
| Hgene | EGFR | C2609414 | Acute kidney injury | 1 | CTD_human |
| Hgene | EGFR | C2713442 | Polyposis, Adenomatous Intestinal | 1 | CTD_human |
| Hgene | EGFR | C2713443 | Familial Intestinal Polyposis | 1 | CTD_human |
| Hgene | EGFR | C3805278 | Extrahepatic Cholangiocarcinoma | 1 | CTD_human |
| Hgene | EGFR | C4751120 | Neonatal inflammatory skin and bowel disease | 1 | ORPHANET |
