
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ERG-APP (FusionGDB2 ID:HG2078TG351) |
Fusion Gene Summary for ERG-APP |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ERG-APP | Fusion gene ID: hg2078tg351 | Hgene | Tgene | Gene symbol | ERG | APP | Gene ID | 2078 | 351 |
| Gene name | ETS transcription factor ERG | amyloid beta precursor protein | |
| Synonyms | erg-3|p55 | AAA|ABETA|ABPP|AD1|APPI|CTFgamma|CVAP|PN-II|PN2|preA4 | |
| Cytomap | ('ERG')('APP') 21q22.2 | 21q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcriptional regulator ERGERG, ETS transcription factorFUS/ERG fusion proteinTMPRSS2/ERG fusionerythroblast transformation-specific transcription factor ERG variant 10ets-relatedtranscriptional regulator ERG (transforming protein ERG)v-ets avian | amyloid-beta precursor proteinalzheimer disease amyloid proteinamyloid beta (A4) precursor proteinamyloid beta A4 proteinamyloid precursor proteinbeta-amyloid peptidebeta-amyloid peptide(1-40)beta-amyloid peptide(1-42)beta-amyloid precursor protei | |
| Modification date | 20200320 | 20200329 | |
| UniProtAcc | P11308 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000288319, ENST00000398905, ENST00000398907, ENST00000429727, ENST00000398910, ENST00000417133, ENST00000442448, ENST00000453032, ENST00000485493, ENST00000398897, ENST00000398911, ENST00000398919, | ||
| Fusion gene scores | * DoF score | 19 X 16 X 5=1520 | 25 X 18 X 10=4500 |
| # samples | 31 | 27 | |
| ** MAII score | log2(31/1520*10)=-2.29373120305671 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/4500*10)=-4.05889368905357 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ERG [Title/Abstract] AND APP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ERG(39870286)-APP(27484463), # samples:1 ERG(40033582)-APP(27484463), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ERG-APP seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ERG-APP seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ERG-APP seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ERG-APP seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ERG | GO:0045944 | positive regulation of transcription by RNA polymerase II | 18195090|23913826 |
| Tgene | APP | GO:0001934 | positive regulation of protein phosphorylation | 11404397 |
| Tgene | APP | GO:0008285 | negative regulation of cell proliferation | 22944668 |
| Tgene | APP | GO:1905606 | regulation of presynapse assembly | 19726636 |
 Fusion gene breakpoints across ERG (5'-gene) Fusion gene breakpoints across ERG (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
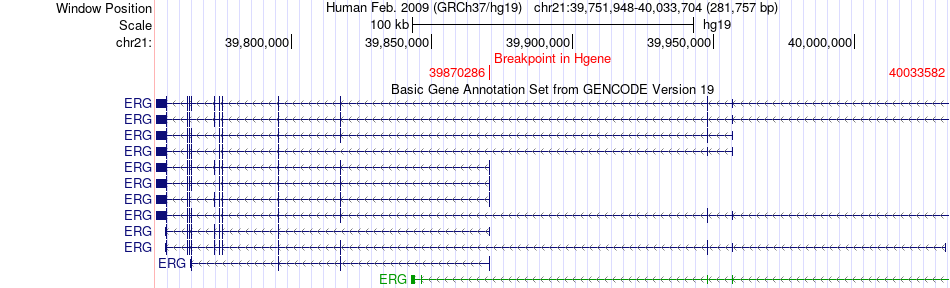 |
 Fusion gene breakpoints across APP (3'-gene) Fusion gene breakpoints across APP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
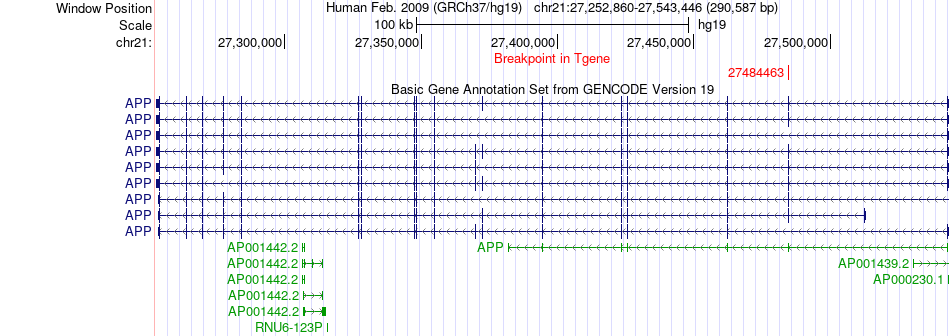 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-8530-01A | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| ChimerDB4 | STAD | TCGA-D7-A4Z0 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
Top |
Fusion Gene ORF analysis for ERG-APP |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000288319 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000288319 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000288319 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000398905 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000398905 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000398905 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000398907 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000398907 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000398907 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000429727 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000429727 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-5UTR | ENST00000429727 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000288319 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000288319 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000288319 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000288319 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000288319 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000288319 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398905 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398905 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398905 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398905 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398905 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398905 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398907 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398907 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398907 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398907 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398907 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000398907 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000429727 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000429727 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000429727 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000429727 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000429727 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5CDS-intron | ENST00000429727 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-3CDS | ENST00000398910 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-3CDS | ENST00000417133 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-3CDS | ENST00000442448 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-3CDS | ENST00000453032 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-3CDS | ENST00000485493 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000398910 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000398910 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000398910 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000417133 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000417133 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000417133 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000442448 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000442448 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000442448 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000453032 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000453032 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000453032 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000485493 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000485493 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-5UTR | ENST00000485493 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000398910 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000398910 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000398910 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000398910 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000398910 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000398910 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000417133 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000417133 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000417133 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000417133 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000417133 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000417133 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000442448 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000442448 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000442448 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000442448 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000442448 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000442448 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000453032 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000453032 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000453032 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000453032 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000453032 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000453032 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000485493 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000485493 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000485493 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000485493 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000485493 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| 5UTR-intron | ENST00000485493 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| In-frame | ENST00000288319 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| In-frame | ENST00000398905 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| In-frame | ENST00000398907 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| In-frame | ENST00000429727 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000288319 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398897 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398897 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398905 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398907 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398910 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398911 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398911 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398919 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000398919 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000417133 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000429727 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000442448 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000453032 | ENST00000346798 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-3CDS | ENST00000485493 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000288319 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000288319 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000288319 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398897 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398897 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398897 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398897 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398897 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398897 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398905 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398905 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398905 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398907 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398907 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398907 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398910 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398910 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398910 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398911 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398911 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398911 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398911 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398911 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398911 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398919 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398919 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398919 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398919 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398919 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000398919 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000417133 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000417133 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000417133 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000429727 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000429727 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000429727 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000442448 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000442448 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000442448 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000453032 | ENST00000440126 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000453032 | ENST00000448388 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000453032 | ENST00000474136 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000485493 | ENST00000440126 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000485493 | ENST00000448388 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-5UTR | ENST00000485493 | ENST00000474136 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000288319 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000288319 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000288319 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000288319 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000288319 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000288319 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398897 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398905 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398905 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398905 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398905 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398905 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398905 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398907 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398907 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398907 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398907 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398907 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398907 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398910 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398910 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398910 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398910 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398910 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398910 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398911 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000398919 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000417133 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000417133 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000417133 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000417133 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000417133 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000417133 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000429727 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000429727 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000429727 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000429727 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000429727 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000429727 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000442448 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000442448 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000442448 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000442448 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000442448 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000442448 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000453032 | ENST00000348990 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000453032 | ENST00000354192 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000453032 | ENST00000357903 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000453032 | ENST00000358918 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000453032 | ENST00000359726 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000453032 | ENST00000439274 | ERG | chr21 | 40033582 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000485493 | ENST00000348990 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000485493 | ENST00000354192 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000485493 | ENST00000357903 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000485493 | ENST00000358918 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000485493 | ENST00000359726 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
| intron-intron | ENST00000485493 | ENST00000439274 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000398907 | ERG | chr21 | 39870286 | - | ENST00000346798 | APP | chr21 | 27484463 | - | 3456 | 80 | 62 | 2335 | 757 |
| ENST00000398905 | ERG | chr21 | 39870286 | - | ENST00000346798 | APP | chr21 | 27484463 | - | 3456 | 80 | 62 | 2335 | 757 |
| ENST00000288319 | ERG | chr21 | 39870286 | - | ENST00000346798 | APP | chr21 | 27484463 | - | 3497 | 121 | 103 | 2376 | 757 |
| ENST00000429727 | ERG | chr21 | 39870286 | - | ENST00000346798 | APP | chr21 | 27484463 | - | 3435 | 59 | 41 | 2314 | 757 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000398907 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - | 0.002401826 | 0.99759823 |
| ENST00000398905 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - | 0.002401826 | 0.99759823 |
| ENST00000288319 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - | 0.002449989 | 0.9975501 |
| ENST00000429727 | ENST00000346798 | ERG | chr21 | 39870286 | - | APP | chr21 | 27484463 | - | 0.002389516 | 0.99761045 |
Top |
Fusion Genomic Features for ERG-APP |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
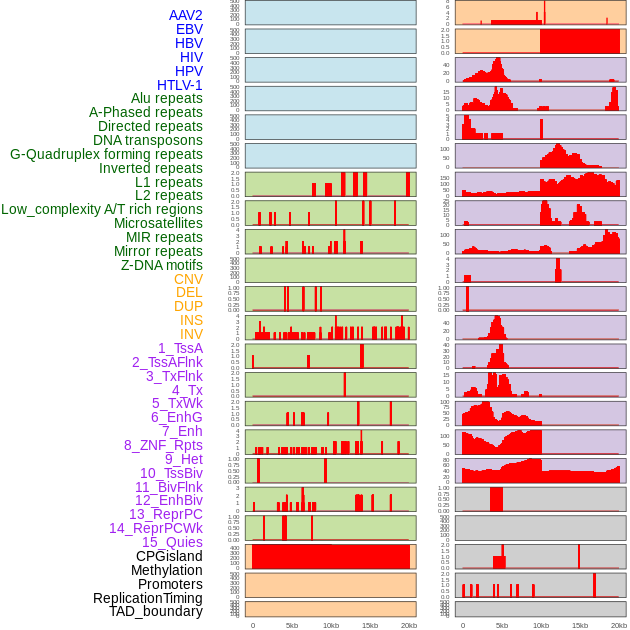 |
Top |
Fusion Protein Features for ERG-APP |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr21:39870286/chr21:27484463) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ERG | . |
| FUNCTION: Transcriptional regulator. May participate in transcriptional regulation through the recruitment of SETDB1 histone methyltransferase and subsequent modification of local chromatin structure. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 230_260 | 19 | 771.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 274_280 | 19 | 771.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 230_260 | 19 | 696.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 274_280 | 19 | 696.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 230_260 | 0 | 640.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 274_280 | 0 | 640.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 230_260 | 19 | 752.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 274_280 | 19 | 752.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 230_260 | 19 | 753.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 274_280 | 19 | 753.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 230_260 | 19 | 715.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 274_280 | 19 | 715.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 230_260 | 14 | 747.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 274_280 | 14 | 747.0 | Compositional bias | Note=Poly-Thr | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 28_189 | 19 | 771.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 374_565 | 19 | 771.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 28_189 | 19 | 696.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 374_565 | 19 | 696.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 28_189 | 0 | 640.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 374_565 | 0 | 640.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 28_189 | 19 | 752.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 374_565 | 19 | 752.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 28_189 | 19 | 753.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 374_565 | 19 | 753.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 28_189 | 19 | 715.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 374_565 | 19 | 715.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 28_189 | 14 | 747.0 | Domain | E1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 374_565 | 14 | 747.0 | Domain | E2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 344_365 | 19 | 771.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 724_734 | 19 | 771.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 344_365 | 19 | 696.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 724_734 | 19 | 696.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 344_365 | 0 | 640.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 724_734 | 0 | 640.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 344_365 | 19 | 752.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 724_734 | 19 | 752.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 344_365 | 19 | 753.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 724_734 | 19 | 753.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 344_365 | 19 | 715.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 724_734 | 19 | 715.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 344_365 | 14 | 747.0 | Motif | OX-2 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 724_734 | 14 | 747.0 | Motif | Note=Basolateral sorting signal | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 131_189 | 19 | 771.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 181_188 | 19 | 771.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 28_123 | 19 | 771.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 391_423 | 19 | 771.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 491_522 | 19 | 771.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 523_540 | 19 | 771.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 96_110 | 19 | 771.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 131_189 | 19 | 696.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 181_188 | 19 | 696.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 28_123 | 19 | 696.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 391_423 | 19 | 696.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 491_522 | 19 | 696.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 523_540 | 19 | 696.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 96_110 | 19 | 696.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 131_189 | 0 | 640.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 181_188 | 0 | 640.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 28_123 | 0 | 640.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 391_423 | 0 | 640.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 491_522 | 0 | 640.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 523_540 | 0 | 640.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 96_110 | 0 | 640.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 131_189 | 19 | 752.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 181_188 | 19 | 752.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 28_123 | 19 | 752.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 391_423 | 19 | 752.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 491_522 | 19 | 752.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 523_540 | 19 | 752.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 96_110 | 19 | 752.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 131_189 | 19 | 753.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 181_188 | 19 | 753.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 28_123 | 19 | 753.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 391_423 | 19 | 753.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 491_522 | 19 | 753.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 523_540 | 19 | 753.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 96_110 | 19 | 753.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 131_189 | 19 | 715.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 181_188 | 19 | 715.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 28_123 | 19 | 715.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 391_423 | 19 | 715.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 491_522 | 19 | 715.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 523_540 | 19 | 715.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 96_110 | 19 | 715.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 131_189 | 14 | 747.0 | Region | CuBD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 181_188 | 14 | 747.0 | Region | Note=Zinc-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 28_123 | 14 | 747.0 | Region | GFLD subdomain | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 391_423 | 14 | 747.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 491_522 | 14 | 747.0 | Region | Note=Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 523_540 | 14 | 747.0 | Region | Collagen-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 96_110 | 14 | 747.0 | Region | Heparin-binding | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 18_701 | 19 | 771.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 723_770 | 19 | 771.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 18_701 | 19 | 696.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 723_770 | 19 | 696.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 18_701 | 0 | 640.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 723_770 | 0 | 640.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 18_701 | 19 | 752.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 723_770 | 19 | 752.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 18_701 | 19 | 753.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 723_770 | 19 | 753.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 18_701 | 19 | 715.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 723_770 | 19 | 715.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 18_701 | 14 | 747.0 | Topological domain | Extracellular | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 723_770 | 14 | 747.0 | Topological domain | Cytoplasmic | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 702_722 | 19 | 771.0 | Transmembrane | Helical | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 702_722 | 19 | 696.0 | Transmembrane | Helical | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 702_722 | 0 | 640.0 | Transmembrane | Helical | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 702_722 | 19 | 752.0 | Transmembrane | Helical | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 702_722 | 19 | 753.0 | Transmembrane | Helical | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 702_722 | 19 | 715.0 | Transmembrane | Helical | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 702_722 | 14 | 747.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000288319 | - | 1 | 10 | 311_391 | 6 | 480.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000398897 | - | 1 | 9 | 311_391 | 0 | 364.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000398911 | - | 1 | 10 | 311_391 | 0 | 463.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000398919 | - | 1 | 12 | 311_391 | 0 | 487.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000417133 | - | 1 | 12 | 311_391 | 0 | 487.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000442448 | - | 1 | 11 | 311_391 | 0 | 463.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000453032 | - | 1 | 9 | 311_391 | 0 | 388.0 | DNA binding | ETS |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000288319 | - | 1 | 10 | 113_199 | 6 | 480.0 | Domain | PNT |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000398897 | - | 1 | 9 | 113_199 | 0 | 364.0 | Domain | PNT |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000398911 | - | 1 | 10 | 113_199 | 0 | 463.0 | Domain | PNT |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000398919 | - | 1 | 12 | 113_199 | 0 | 487.0 | Domain | PNT |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000417133 | - | 1 | 12 | 113_199 | 0 | 487.0 | Domain | PNT |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000442448 | - | 1 | 11 | 113_199 | 0 | 463.0 | Domain | PNT |
| Hgene | ERG | chr21:39870286 | chr21:27484463 | ENST00000453032 | - | 1 | 9 | 113_199 | 0 | 388.0 | Domain | PNT |
Top |
Fusion Gene Sequence for ERG-APP |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27334_27334_1_ERG-APP_ERG_chr21_39870286_ENST00000288319_APP_chr21_27484463_ENST00000346798_length(transcript)=3497nt_BP=121nt AATCTCATCCGCTCTAAACAACCTCATCAAAACTACTTTCTGGTCAGAGAGAAGCAATAATTATTATTAACATTTATTAACGATCAATAA ACTTGATCGCATTATGGCCAGCACTATTAAGGTACCCACTGATGGTAATGCTGGCCTGCTGGCTGAACCCCAGATTGCCATGTTCTGTGG CAGACTGAACATGCACATGAATGTCCAGAATGGGAAGTGGGATTCAGATCCATCAGGGACCAAAACCTGCATTGATACCAAGGAAGGCAT CCTGCAGTATTGCCAAGAAGTCTACCCTGAACTGCAGATCACCAATGTGGTAGAAGCCAACCAACCAGTGACCATCCAGAACTGGTGCAA GCGGGGCCGCAAGCAGTGCAAGACCCATCCCCACTTTGTGATTCCCTACCGCTGCTTAGTTGGTGAGTTTGTAAGTGATGCCCTTCTCGT TCCTGACAAGTGCAAATTCTTACACCAGGAGAGGATGGATGTTTGCGAAACTCATCTTCACTGGCACACCGTCGCCAAAGAGACATGCAG TGAGAAGAGTACCAACTTGCATGACTACGGCATGTTGCTGCCCTGCGGAATTGACAAGTTCCGAGGGGTAGAGTTTGTGTGTTGCCCACT GGCTGAAGAAAGTGACAATGTGGATTCTGCTGATGCGGAGGAGGATGACTCGGATGTCTGGTGGGGCGGAGCAGACACAGACTATGCAGA TGGGAGTGAAGACAAAGTAGTAGAAGTAGCAGAGGAGGAAGAAGTGGCTGAGGTGGAAGAAGAAGAAGCCGATGATGACGAGGACGATGA GGATGGTGATGAGGTAGAGGAAGAGGCTGAGGAACCCTACGAAGAAGCCACAGAGAGAACCACCAGCATTGCCACCACCACCACCACCAC CACAGAGTCTGTGGAAGAGGTGGTTCGAGAGGTGTGCTCTGAACAAGCCGAGACGGGGCCGTGCCGAGCAATGATCTCCCGCTGGTACTT TGATGTGACTGAAGGGAAGTGTGCCCCATTCTTTTACGGCGGATGTGGCGGCAACCGGAACAACTTTGACACAGAAGAGTACTGCATGGC CGTGTGTGGCAGCGCCATGTCCCAAAGTTTACTCAAGACTACCCAGGAACCTCTTGCCCGAGATCCTGTTAAACTTCCTACAACAGCAGC CAGTACCCCTGATGCCGTTGACAAGTATCTCGAGACACCTGGGGATGAGAATGAACATGCCCATTTCCAGAAAGCCAAAGAGAGGCTTGA GGCCAAGCACCGAGAGAGAATGTCCCAGGTCATGAGAGAATGGGAAGAGGCAGAACGTCAAGCAAAGAACTTGCCTAAAGCTGATAAGAA GGCAGTTATCCAGCATTTCCAGGAGAAAGTGGAATCTTTGGAACAGGAAGCAGCCAACGAGAGACAGCAGCTGGTGGAGACACACATGGC CAGAGTGGAAGCCATGCTCAATGACCGCCGCCGCCTGGCCCTGGAGAACTACATCACCGCTCTGCAGGCTGTTCCTCCTCGGCCTCGTCA CGTGTTCAATATGCTAAAGAAGTATGTCCGCGCAGAACAGAAGGACAGACAGCACACCCTAAAGCATTTCGAGCATGTGCGCATGGTGGA TCCCAAGAAAGCCGCTCAGATCCGGTCCCAGGTTATGACACACCTCCGTGTGATTTATGAGCGCATGAATCAGTCTCTCTCCCTGCTCTA CAACGTGCCTGCAGTGGCCGAGGAGATTCAGGATGAAGTTGATGAGCTGCTTCAGAAAGAGCAAAACTATTCAGATGACGTCTTGGCCAA CATGATTAGTGAACCAAGGATCAGTTACGGAAACGATGCTCTCATGCCATCTTTGACCGAAACGAAAACCACCGTGGAGCTCCTTCCCGT GAATGGAGAGTTCAGCCTGGACGATCTCCAGCCGTGGCATTCTTTTGGGGCTGACTCTGTGCCAGCCAACACAGAAAACGAAGTTGAGCC TGTTGATGCCCGCCCTGCTGCCGACCGAGGACTGACCACTCGACCAGGTTCTGGGTTGACAAATATCAAGACGGAGGAGATCTCTGAAGT GAAGATGGATGCAGAATTCCGACATGACTCAGGATATGAAGTTCATCATCAAAAATTGGTGTTCTTTGCAGAAGATGTGGGTTCAAACAA AGGTGCAATCATTGGACTCATGGTGGGCGGTGTTGTCATAGCGACAGTGATCGTCATCACCTTGGTGATGCTGAAGAAGAAACAGTACAC ATCCATTCATCATGGTGTGGTGGAGGTTGACGCCGCTGTCACCCCAGAGGAGCGCCACCTGTCCAAGATGCAGCAGAACGGCTACGAAAA TCCAACCTACAAGTTCTTTGAGCAGATGCAGAACTAGACCCCCGCCACAGCAGCCTCTGAAGTTGGACAGCAAAACCATTGCTTCACTAC CCATCGGTGTCCATTTATAGAATAATGTGGGAAGAAACAAACCCGTTTTATGATTTACTCATTATCGCCTTTTGACAGCTGTGCTGTAAC ACAAGTAGATGCCTGAACTTGAATTAATCCACACATCAGTAATGTATTCTATCTCTCTTTACATTTTGGTCTCTATACTACATTATTAAT GGGTTTTGTGTACTGTAAAGAATTTAGCTGTATCAAACTAGTGCATGAATAGATTCTCTCCTGATTATTTATCACATAGCCCCTTAGCCA GTTGTATATTATTCTTGTGGTTTGTGACCCAATTAAGTCCTACTTTACATATGCTTTAAGAATCGATGGGGGATGCTTCATGTGAACGTG GGAGTTCAGCTGCTTCTCTTGCCTAAGTATTCCTTTCCTGATCACTATGCATTTTAAAGTTAAACATTTTTAAGTATTTCAGATGCTTTA GAGAGATTTTTTTTCCATGACTGCATTTTACTGTACAGATTGCTGCTTCTGCTATATTTGTGATATAGGAATTAAGAGGATACACACGTT TGTTTCTTCGTGCCTGTTTTATGTGCACACATTAGGCATTGAGACTTCAAGCTTTTCTTTTTTTGTCCACGTATCTTTGGGTCTTTGATA AAGAAAAGAATCCCTGTTCATTGTAAGCACTTTTACGGGGCGGGTGGGGAGGGGTGCTCTGCTGGTCTTCAATTACCAAGAATTCTCCAA AACAATTTTCTGCAGGATGATTGTACAGAATCATTGCTTATGACATGATCGCTTTCTACACTGTATTACATAAATAAATTAAATAAAATA ACCCCGGGCAAGACTTTTCTTTGAAGGATGACTACAGACATTAAATAATCGAAGTAATTTTGGGTGGGGAGAAGAGGCAGATTCAATTTT CTTTAACCAGTCTGAAGTTTCATTTATGATACAAAAGAAGATGAAAATGGAAGTGGCAATATAAGGGGATGAGGAAGGCATGCCTGGACA AACCCTTCTTTTAAGATGTGTCTTCAATTTGTATAAAATGGTGTTTTCATGTAAATAAATACATTCTTGGAGGAGCA >27334_27334_1_ERG-APP_ERG_chr21_39870286_ENST00000288319_APP_chr21_27484463_ENST00000346798_length(amino acids)=757AA_BP=6 MASTIKVPTDGNAGLLAEPQIAMFCGRLNMHMNVQNGKWDSDPSGTKTCIDTKEGILQYCQEVYPELQITNVVEANQPVTIQNWCKRGRK QCKTHPHFVIPYRCLVGEFVSDALLVPDKCKFLHQERMDVCETHLHWHTVAKETCSEKSTNLHDYGMLLPCGIDKFRGVEFVCCPLAEES DNVDSADAEEDDSDVWWGGADTDYADGSEDKVVEVAEEEEVAEVEEEEADDDEDDEDGDEVEEEAEEPYEEATERTTSIATTTTTTTESV EEVVREVCSEQAETGPCRAMISRWYFDVTEGKCAPFFYGGCGGNRNNFDTEEYCMAVCGSAMSQSLLKTTQEPLARDPVKLPTTAASTPD AVDKYLETPGDENEHAHFQKAKERLEAKHRERMSQVMREWEEAERQAKNLPKADKKAVIQHFQEKVESLEQEAANERQQLVETHMARVEA MLNDRRRLALENYITALQAVPPRPRHVFNMLKKYVRAEQKDRQHTLKHFEHVRMVDPKKAAQIRSQVMTHLRVIYERMNQSLSLLYNVPA VAEEIQDEVDELLQKEQNYSDDVLANMISEPRISYGNDALMPSLTETKTTVELLPVNGEFSLDDLQPWHSFGADSVPANTENEVEPVDAR PAADRGLTTRPGSGLTNIKTEEISEVKMDAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIATVIVITLVMLKKKQYTSIHH GVVEVDAAVTPEERHLSKMQQNGYENPTYKFFEQMQN -------------------------------------------------------------- >27334_27334_2_ERG-APP_ERG_chr21_39870286_ENST00000398905_APP_chr21_27484463_ENST00000346798_length(transcript)=3456nt_BP=80nt GGTCAGAGAGAAGCAATAATTATTATTAACATTTATTAACGATCAATAAACTTGATCGCATTATGGCCAGCACTATTAAGGTACCCACTG ATGGTAATGCTGGCCTGCTGGCTGAACCCCAGATTGCCATGTTCTGTGGCAGACTGAACATGCACATGAATGTCCAGAATGGGAAGTGGG ATTCAGATCCATCAGGGACCAAAACCTGCATTGATACCAAGGAAGGCATCCTGCAGTATTGCCAAGAAGTCTACCCTGAACTGCAGATCA CCAATGTGGTAGAAGCCAACCAACCAGTGACCATCCAGAACTGGTGCAAGCGGGGCCGCAAGCAGTGCAAGACCCATCCCCACTTTGTGA TTCCCTACCGCTGCTTAGTTGGTGAGTTTGTAAGTGATGCCCTTCTCGTTCCTGACAAGTGCAAATTCTTACACCAGGAGAGGATGGATG TTTGCGAAACTCATCTTCACTGGCACACCGTCGCCAAAGAGACATGCAGTGAGAAGAGTACCAACTTGCATGACTACGGCATGTTGCTGC CCTGCGGAATTGACAAGTTCCGAGGGGTAGAGTTTGTGTGTTGCCCACTGGCTGAAGAAAGTGACAATGTGGATTCTGCTGATGCGGAGG AGGATGACTCGGATGTCTGGTGGGGCGGAGCAGACACAGACTATGCAGATGGGAGTGAAGACAAAGTAGTAGAAGTAGCAGAGGAGGAAG AAGTGGCTGAGGTGGAAGAAGAAGAAGCCGATGATGACGAGGACGATGAGGATGGTGATGAGGTAGAGGAAGAGGCTGAGGAACCCTACG AAGAAGCCACAGAGAGAACCACCAGCATTGCCACCACCACCACCACCACCACAGAGTCTGTGGAAGAGGTGGTTCGAGAGGTGTGCTCTG AACAAGCCGAGACGGGGCCGTGCCGAGCAATGATCTCCCGCTGGTACTTTGATGTGACTGAAGGGAAGTGTGCCCCATTCTTTTACGGCG GATGTGGCGGCAACCGGAACAACTTTGACACAGAAGAGTACTGCATGGCCGTGTGTGGCAGCGCCATGTCCCAAAGTTTACTCAAGACTA CCCAGGAACCTCTTGCCCGAGATCCTGTTAAACTTCCTACAACAGCAGCCAGTACCCCTGATGCCGTTGACAAGTATCTCGAGACACCTG GGGATGAGAATGAACATGCCCATTTCCAGAAAGCCAAAGAGAGGCTTGAGGCCAAGCACCGAGAGAGAATGTCCCAGGTCATGAGAGAAT GGGAAGAGGCAGAACGTCAAGCAAAGAACTTGCCTAAAGCTGATAAGAAGGCAGTTATCCAGCATTTCCAGGAGAAAGTGGAATCTTTGG AACAGGAAGCAGCCAACGAGAGACAGCAGCTGGTGGAGACACACATGGCCAGAGTGGAAGCCATGCTCAATGACCGCCGCCGCCTGGCCC TGGAGAACTACATCACCGCTCTGCAGGCTGTTCCTCCTCGGCCTCGTCACGTGTTCAATATGCTAAAGAAGTATGTCCGCGCAGAACAGA AGGACAGACAGCACACCCTAAAGCATTTCGAGCATGTGCGCATGGTGGATCCCAAGAAAGCCGCTCAGATCCGGTCCCAGGTTATGACAC ACCTCCGTGTGATTTATGAGCGCATGAATCAGTCTCTCTCCCTGCTCTACAACGTGCCTGCAGTGGCCGAGGAGATTCAGGATGAAGTTG ATGAGCTGCTTCAGAAAGAGCAAAACTATTCAGATGACGTCTTGGCCAACATGATTAGTGAACCAAGGATCAGTTACGGAAACGATGCTC TCATGCCATCTTTGACCGAAACGAAAACCACCGTGGAGCTCCTTCCCGTGAATGGAGAGTTCAGCCTGGACGATCTCCAGCCGTGGCATT CTTTTGGGGCTGACTCTGTGCCAGCCAACACAGAAAACGAAGTTGAGCCTGTTGATGCCCGCCCTGCTGCCGACCGAGGACTGACCACTC GACCAGGTTCTGGGTTGACAAATATCAAGACGGAGGAGATCTCTGAAGTGAAGATGGATGCAGAATTCCGACATGACTCAGGATATGAAG TTCATCATCAAAAATTGGTGTTCTTTGCAGAAGATGTGGGTTCAAACAAAGGTGCAATCATTGGACTCATGGTGGGCGGTGTTGTCATAG CGACAGTGATCGTCATCACCTTGGTGATGCTGAAGAAGAAACAGTACACATCCATTCATCATGGTGTGGTGGAGGTTGACGCCGCTGTCA CCCCAGAGGAGCGCCACCTGTCCAAGATGCAGCAGAACGGCTACGAAAATCCAACCTACAAGTTCTTTGAGCAGATGCAGAACTAGACCC CCGCCACAGCAGCCTCTGAAGTTGGACAGCAAAACCATTGCTTCACTACCCATCGGTGTCCATTTATAGAATAATGTGGGAAGAAACAAA CCCGTTTTATGATTTACTCATTATCGCCTTTTGACAGCTGTGCTGTAACACAAGTAGATGCCTGAACTTGAATTAATCCACACATCAGTA ATGTATTCTATCTCTCTTTACATTTTGGTCTCTATACTACATTATTAATGGGTTTTGTGTACTGTAAAGAATTTAGCTGTATCAAACTAG TGCATGAATAGATTCTCTCCTGATTATTTATCACATAGCCCCTTAGCCAGTTGTATATTATTCTTGTGGTTTGTGACCCAATTAAGTCCT ACTTTACATATGCTTTAAGAATCGATGGGGGATGCTTCATGTGAACGTGGGAGTTCAGCTGCTTCTCTTGCCTAAGTATTCCTTTCCTGA TCACTATGCATTTTAAAGTTAAACATTTTTAAGTATTTCAGATGCTTTAGAGAGATTTTTTTTCCATGACTGCATTTTACTGTACAGATT GCTGCTTCTGCTATATTTGTGATATAGGAATTAAGAGGATACACACGTTTGTTTCTTCGTGCCTGTTTTATGTGCACACATTAGGCATTG AGACTTCAAGCTTTTCTTTTTTTGTCCACGTATCTTTGGGTCTTTGATAAAGAAAAGAATCCCTGTTCATTGTAAGCACTTTTACGGGGC GGGTGGGGAGGGGTGCTCTGCTGGTCTTCAATTACCAAGAATTCTCCAAAACAATTTTCTGCAGGATGATTGTACAGAATCATTGCTTAT GACATGATCGCTTTCTACACTGTATTACATAAATAAATTAAATAAAATAACCCCGGGCAAGACTTTTCTTTGAAGGATGACTACAGACAT TAAATAATCGAAGTAATTTTGGGTGGGGAGAAGAGGCAGATTCAATTTTCTTTAACCAGTCTGAAGTTTCATTTATGATACAAAAGAAGA TGAAAATGGAAGTGGCAATATAAGGGGATGAGGAAGGCATGCCTGGACAAACCCTTCTTTTAAGATGTGTCTTCAATTTGTATAAAATGG TGTTTTCATGTAAATAAATACATTCTTGGAGGAGCA >27334_27334_2_ERG-APP_ERG_chr21_39870286_ENST00000398905_APP_chr21_27484463_ENST00000346798_length(amino acids)=757AA_BP=6 MASTIKVPTDGNAGLLAEPQIAMFCGRLNMHMNVQNGKWDSDPSGTKTCIDTKEGILQYCQEVYPELQITNVVEANQPVTIQNWCKRGRK QCKTHPHFVIPYRCLVGEFVSDALLVPDKCKFLHQERMDVCETHLHWHTVAKETCSEKSTNLHDYGMLLPCGIDKFRGVEFVCCPLAEES DNVDSADAEEDDSDVWWGGADTDYADGSEDKVVEVAEEEEVAEVEEEEADDDEDDEDGDEVEEEAEEPYEEATERTTSIATTTTTTTESV EEVVREVCSEQAETGPCRAMISRWYFDVTEGKCAPFFYGGCGGNRNNFDTEEYCMAVCGSAMSQSLLKTTQEPLARDPVKLPTTAASTPD AVDKYLETPGDENEHAHFQKAKERLEAKHRERMSQVMREWEEAERQAKNLPKADKKAVIQHFQEKVESLEQEAANERQQLVETHMARVEA MLNDRRRLALENYITALQAVPPRPRHVFNMLKKYVRAEQKDRQHTLKHFEHVRMVDPKKAAQIRSQVMTHLRVIYERMNQSLSLLYNVPA VAEEIQDEVDELLQKEQNYSDDVLANMISEPRISYGNDALMPSLTETKTTVELLPVNGEFSLDDLQPWHSFGADSVPANTENEVEPVDAR PAADRGLTTRPGSGLTNIKTEEISEVKMDAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIATVIVITLVMLKKKQYTSIHH GVVEVDAAVTPEERHLSKMQQNGYENPTYKFFEQMQN -------------------------------------------------------------- >27334_27334_3_ERG-APP_ERG_chr21_39870286_ENST00000398907_APP_chr21_27484463_ENST00000346798_length(transcript)=3456nt_BP=80nt GGTCAGAGAGAAGCAATAATTATTATTAACATTTATTAACGATCAATAAACTTGATCGCATTATGGCCAGCACTATTAAGGTACCCACTG ATGGTAATGCTGGCCTGCTGGCTGAACCCCAGATTGCCATGTTCTGTGGCAGACTGAACATGCACATGAATGTCCAGAATGGGAAGTGGG ATTCAGATCCATCAGGGACCAAAACCTGCATTGATACCAAGGAAGGCATCCTGCAGTATTGCCAAGAAGTCTACCCTGAACTGCAGATCA CCAATGTGGTAGAAGCCAACCAACCAGTGACCATCCAGAACTGGTGCAAGCGGGGCCGCAAGCAGTGCAAGACCCATCCCCACTTTGTGA TTCCCTACCGCTGCTTAGTTGGTGAGTTTGTAAGTGATGCCCTTCTCGTTCCTGACAAGTGCAAATTCTTACACCAGGAGAGGATGGATG TTTGCGAAACTCATCTTCACTGGCACACCGTCGCCAAAGAGACATGCAGTGAGAAGAGTACCAACTTGCATGACTACGGCATGTTGCTGC CCTGCGGAATTGACAAGTTCCGAGGGGTAGAGTTTGTGTGTTGCCCACTGGCTGAAGAAAGTGACAATGTGGATTCTGCTGATGCGGAGG AGGATGACTCGGATGTCTGGTGGGGCGGAGCAGACACAGACTATGCAGATGGGAGTGAAGACAAAGTAGTAGAAGTAGCAGAGGAGGAAG AAGTGGCTGAGGTGGAAGAAGAAGAAGCCGATGATGACGAGGACGATGAGGATGGTGATGAGGTAGAGGAAGAGGCTGAGGAACCCTACG AAGAAGCCACAGAGAGAACCACCAGCATTGCCACCACCACCACCACCACCACAGAGTCTGTGGAAGAGGTGGTTCGAGAGGTGTGCTCTG AACAAGCCGAGACGGGGCCGTGCCGAGCAATGATCTCCCGCTGGTACTTTGATGTGACTGAAGGGAAGTGTGCCCCATTCTTTTACGGCG GATGTGGCGGCAACCGGAACAACTTTGACACAGAAGAGTACTGCATGGCCGTGTGTGGCAGCGCCATGTCCCAAAGTTTACTCAAGACTA CCCAGGAACCTCTTGCCCGAGATCCTGTTAAACTTCCTACAACAGCAGCCAGTACCCCTGATGCCGTTGACAAGTATCTCGAGACACCTG GGGATGAGAATGAACATGCCCATTTCCAGAAAGCCAAAGAGAGGCTTGAGGCCAAGCACCGAGAGAGAATGTCCCAGGTCATGAGAGAAT GGGAAGAGGCAGAACGTCAAGCAAAGAACTTGCCTAAAGCTGATAAGAAGGCAGTTATCCAGCATTTCCAGGAGAAAGTGGAATCTTTGG AACAGGAAGCAGCCAACGAGAGACAGCAGCTGGTGGAGACACACATGGCCAGAGTGGAAGCCATGCTCAATGACCGCCGCCGCCTGGCCC TGGAGAACTACATCACCGCTCTGCAGGCTGTTCCTCCTCGGCCTCGTCACGTGTTCAATATGCTAAAGAAGTATGTCCGCGCAGAACAGA AGGACAGACAGCACACCCTAAAGCATTTCGAGCATGTGCGCATGGTGGATCCCAAGAAAGCCGCTCAGATCCGGTCCCAGGTTATGACAC ACCTCCGTGTGATTTATGAGCGCATGAATCAGTCTCTCTCCCTGCTCTACAACGTGCCTGCAGTGGCCGAGGAGATTCAGGATGAAGTTG ATGAGCTGCTTCAGAAAGAGCAAAACTATTCAGATGACGTCTTGGCCAACATGATTAGTGAACCAAGGATCAGTTACGGAAACGATGCTC TCATGCCATCTTTGACCGAAACGAAAACCACCGTGGAGCTCCTTCCCGTGAATGGAGAGTTCAGCCTGGACGATCTCCAGCCGTGGCATT CTTTTGGGGCTGACTCTGTGCCAGCCAACACAGAAAACGAAGTTGAGCCTGTTGATGCCCGCCCTGCTGCCGACCGAGGACTGACCACTC GACCAGGTTCTGGGTTGACAAATATCAAGACGGAGGAGATCTCTGAAGTGAAGATGGATGCAGAATTCCGACATGACTCAGGATATGAAG TTCATCATCAAAAATTGGTGTTCTTTGCAGAAGATGTGGGTTCAAACAAAGGTGCAATCATTGGACTCATGGTGGGCGGTGTTGTCATAG CGACAGTGATCGTCATCACCTTGGTGATGCTGAAGAAGAAACAGTACACATCCATTCATCATGGTGTGGTGGAGGTTGACGCCGCTGTCA CCCCAGAGGAGCGCCACCTGTCCAAGATGCAGCAGAACGGCTACGAAAATCCAACCTACAAGTTCTTTGAGCAGATGCAGAACTAGACCC CCGCCACAGCAGCCTCTGAAGTTGGACAGCAAAACCATTGCTTCACTACCCATCGGTGTCCATTTATAGAATAATGTGGGAAGAAACAAA CCCGTTTTATGATTTACTCATTATCGCCTTTTGACAGCTGTGCTGTAACACAAGTAGATGCCTGAACTTGAATTAATCCACACATCAGTA ATGTATTCTATCTCTCTTTACATTTTGGTCTCTATACTACATTATTAATGGGTTTTGTGTACTGTAAAGAATTTAGCTGTATCAAACTAG TGCATGAATAGATTCTCTCCTGATTATTTATCACATAGCCCCTTAGCCAGTTGTATATTATTCTTGTGGTTTGTGACCCAATTAAGTCCT ACTTTACATATGCTTTAAGAATCGATGGGGGATGCTTCATGTGAACGTGGGAGTTCAGCTGCTTCTCTTGCCTAAGTATTCCTTTCCTGA TCACTATGCATTTTAAAGTTAAACATTTTTAAGTATTTCAGATGCTTTAGAGAGATTTTTTTTCCATGACTGCATTTTACTGTACAGATT GCTGCTTCTGCTATATTTGTGATATAGGAATTAAGAGGATACACACGTTTGTTTCTTCGTGCCTGTTTTATGTGCACACATTAGGCATTG AGACTTCAAGCTTTTCTTTTTTTGTCCACGTATCTTTGGGTCTTTGATAAAGAAAAGAATCCCTGTTCATTGTAAGCACTTTTACGGGGC GGGTGGGGAGGGGTGCTCTGCTGGTCTTCAATTACCAAGAATTCTCCAAAACAATTTTCTGCAGGATGATTGTACAGAATCATTGCTTAT GACATGATCGCTTTCTACACTGTATTACATAAATAAATTAAATAAAATAACCCCGGGCAAGACTTTTCTTTGAAGGATGACTACAGACAT TAAATAATCGAAGTAATTTTGGGTGGGGAGAAGAGGCAGATTCAATTTTCTTTAACCAGTCTGAAGTTTCATTTATGATACAAAAGAAGA TGAAAATGGAAGTGGCAATATAAGGGGATGAGGAAGGCATGCCTGGACAAACCCTTCTTTTAAGATGTGTCTTCAATTTGTATAAAATGG TGTTTTCATGTAAATAAATACATTCTTGGAGGAGCA >27334_27334_3_ERG-APP_ERG_chr21_39870286_ENST00000398907_APP_chr21_27484463_ENST00000346798_length(amino acids)=757AA_BP=6 MASTIKVPTDGNAGLLAEPQIAMFCGRLNMHMNVQNGKWDSDPSGTKTCIDTKEGILQYCQEVYPELQITNVVEANQPVTIQNWCKRGRK QCKTHPHFVIPYRCLVGEFVSDALLVPDKCKFLHQERMDVCETHLHWHTVAKETCSEKSTNLHDYGMLLPCGIDKFRGVEFVCCPLAEES DNVDSADAEEDDSDVWWGGADTDYADGSEDKVVEVAEEEEVAEVEEEEADDDEDDEDGDEVEEEAEEPYEEATERTTSIATTTTTTTESV EEVVREVCSEQAETGPCRAMISRWYFDVTEGKCAPFFYGGCGGNRNNFDTEEYCMAVCGSAMSQSLLKTTQEPLARDPVKLPTTAASTPD AVDKYLETPGDENEHAHFQKAKERLEAKHRERMSQVMREWEEAERQAKNLPKADKKAVIQHFQEKVESLEQEAANERQQLVETHMARVEA MLNDRRRLALENYITALQAVPPRPRHVFNMLKKYVRAEQKDRQHTLKHFEHVRMVDPKKAAQIRSQVMTHLRVIYERMNQSLSLLYNVPA VAEEIQDEVDELLQKEQNYSDDVLANMISEPRISYGNDALMPSLTETKTTVELLPVNGEFSLDDLQPWHSFGADSVPANTENEVEPVDAR PAADRGLTTRPGSGLTNIKTEEISEVKMDAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIATVIVITLVMLKKKQYTSIHH GVVEVDAAVTPEERHLSKMQQNGYENPTYKFFEQMQN -------------------------------------------------------------- >27334_27334_4_ERG-APP_ERG_chr21_39870286_ENST00000429727_APP_chr21_27484463_ENST00000346798_length(transcript)=3435nt_BP=59nt ATTATTAACATTTATTAACGATCAATAAACTTGATCGCATTATGGCCAGCACTATTAAGGTACCCACTGATGGTAATGCTGGCCTGCTGG CTGAACCCCAGATTGCCATGTTCTGTGGCAGACTGAACATGCACATGAATGTCCAGAATGGGAAGTGGGATTCAGATCCATCAGGGACCA AAACCTGCATTGATACCAAGGAAGGCATCCTGCAGTATTGCCAAGAAGTCTACCCTGAACTGCAGATCACCAATGTGGTAGAAGCCAACC AACCAGTGACCATCCAGAACTGGTGCAAGCGGGGCCGCAAGCAGTGCAAGACCCATCCCCACTTTGTGATTCCCTACCGCTGCTTAGTTG GTGAGTTTGTAAGTGATGCCCTTCTCGTTCCTGACAAGTGCAAATTCTTACACCAGGAGAGGATGGATGTTTGCGAAACTCATCTTCACT GGCACACCGTCGCCAAAGAGACATGCAGTGAGAAGAGTACCAACTTGCATGACTACGGCATGTTGCTGCCCTGCGGAATTGACAAGTTCC GAGGGGTAGAGTTTGTGTGTTGCCCACTGGCTGAAGAAAGTGACAATGTGGATTCTGCTGATGCGGAGGAGGATGACTCGGATGTCTGGT GGGGCGGAGCAGACACAGACTATGCAGATGGGAGTGAAGACAAAGTAGTAGAAGTAGCAGAGGAGGAAGAAGTGGCTGAGGTGGAAGAAG AAGAAGCCGATGATGACGAGGACGATGAGGATGGTGATGAGGTAGAGGAAGAGGCTGAGGAACCCTACGAAGAAGCCACAGAGAGAACCA CCAGCATTGCCACCACCACCACCACCACCACAGAGTCTGTGGAAGAGGTGGTTCGAGAGGTGTGCTCTGAACAAGCCGAGACGGGGCCGT GCCGAGCAATGATCTCCCGCTGGTACTTTGATGTGACTGAAGGGAAGTGTGCCCCATTCTTTTACGGCGGATGTGGCGGCAACCGGAACA ACTTTGACACAGAAGAGTACTGCATGGCCGTGTGTGGCAGCGCCATGTCCCAAAGTTTACTCAAGACTACCCAGGAACCTCTTGCCCGAG ATCCTGTTAAACTTCCTACAACAGCAGCCAGTACCCCTGATGCCGTTGACAAGTATCTCGAGACACCTGGGGATGAGAATGAACATGCCC ATTTCCAGAAAGCCAAAGAGAGGCTTGAGGCCAAGCACCGAGAGAGAATGTCCCAGGTCATGAGAGAATGGGAAGAGGCAGAACGTCAAG CAAAGAACTTGCCTAAAGCTGATAAGAAGGCAGTTATCCAGCATTTCCAGGAGAAAGTGGAATCTTTGGAACAGGAAGCAGCCAACGAGA GACAGCAGCTGGTGGAGACACACATGGCCAGAGTGGAAGCCATGCTCAATGACCGCCGCCGCCTGGCCCTGGAGAACTACATCACCGCTC TGCAGGCTGTTCCTCCTCGGCCTCGTCACGTGTTCAATATGCTAAAGAAGTATGTCCGCGCAGAACAGAAGGACAGACAGCACACCCTAA AGCATTTCGAGCATGTGCGCATGGTGGATCCCAAGAAAGCCGCTCAGATCCGGTCCCAGGTTATGACACACCTCCGTGTGATTTATGAGC GCATGAATCAGTCTCTCTCCCTGCTCTACAACGTGCCTGCAGTGGCCGAGGAGATTCAGGATGAAGTTGATGAGCTGCTTCAGAAAGAGC AAAACTATTCAGATGACGTCTTGGCCAACATGATTAGTGAACCAAGGATCAGTTACGGAAACGATGCTCTCATGCCATCTTTGACCGAAA CGAAAACCACCGTGGAGCTCCTTCCCGTGAATGGAGAGTTCAGCCTGGACGATCTCCAGCCGTGGCATTCTTTTGGGGCTGACTCTGTGC CAGCCAACACAGAAAACGAAGTTGAGCCTGTTGATGCCCGCCCTGCTGCCGACCGAGGACTGACCACTCGACCAGGTTCTGGGTTGACAA ATATCAAGACGGAGGAGATCTCTGAAGTGAAGATGGATGCAGAATTCCGACATGACTCAGGATATGAAGTTCATCATCAAAAATTGGTGT TCTTTGCAGAAGATGTGGGTTCAAACAAAGGTGCAATCATTGGACTCATGGTGGGCGGTGTTGTCATAGCGACAGTGATCGTCATCACCT TGGTGATGCTGAAGAAGAAACAGTACACATCCATTCATCATGGTGTGGTGGAGGTTGACGCCGCTGTCACCCCAGAGGAGCGCCACCTGT CCAAGATGCAGCAGAACGGCTACGAAAATCCAACCTACAAGTTCTTTGAGCAGATGCAGAACTAGACCCCCGCCACAGCAGCCTCTGAAG TTGGACAGCAAAACCATTGCTTCACTACCCATCGGTGTCCATTTATAGAATAATGTGGGAAGAAACAAACCCGTTTTATGATTTACTCAT TATCGCCTTTTGACAGCTGTGCTGTAACACAAGTAGATGCCTGAACTTGAATTAATCCACACATCAGTAATGTATTCTATCTCTCTTTAC ATTTTGGTCTCTATACTACATTATTAATGGGTTTTGTGTACTGTAAAGAATTTAGCTGTATCAAACTAGTGCATGAATAGATTCTCTCCT GATTATTTATCACATAGCCCCTTAGCCAGTTGTATATTATTCTTGTGGTTTGTGACCCAATTAAGTCCTACTTTACATATGCTTTAAGAA TCGATGGGGGATGCTTCATGTGAACGTGGGAGTTCAGCTGCTTCTCTTGCCTAAGTATTCCTTTCCTGATCACTATGCATTTTAAAGTTA AACATTTTTAAGTATTTCAGATGCTTTAGAGAGATTTTTTTTCCATGACTGCATTTTACTGTACAGATTGCTGCTTCTGCTATATTTGTG ATATAGGAATTAAGAGGATACACACGTTTGTTTCTTCGTGCCTGTTTTATGTGCACACATTAGGCATTGAGACTTCAAGCTTTTCTTTTT TTGTCCACGTATCTTTGGGTCTTTGATAAAGAAAAGAATCCCTGTTCATTGTAAGCACTTTTACGGGGCGGGTGGGGAGGGGTGCTCTGC TGGTCTTCAATTACCAAGAATTCTCCAAAACAATTTTCTGCAGGATGATTGTACAGAATCATTGCTTATGACATGATCGCTTTCTACACT GTATTACATAAATAAATTAAATAAAATAACCCCGGGCAAGACTTTTCTTTGAAGGATGACTACAGACATTAAATAATCGAAGTAATTTTG GGTGGGGAGAAGAGGCAGATTCAATTTTCTTTAACCAGTCTGAAGTTTCATTTATGATACAAAAGAAGATGAAAATGGAAGTGGCAATAT AAGGGGATGAGGAAGGCATGCCTGGACAAACCCTTCTTTTAAGATGTGTCTTCAATTTGTATAAAATGGTGTTTTCATGTAAATAAATAC ATTCTTGGAGGAGCA >27334_27334_4_ERG-APP_ERG_chr21_39870286_ENST00000429727_APP_chr21_27484463_ENST00000346798_length(amino acids)=757AA_BP=6 MASTIKVPTDGNAGLLAEPQIAMFCGRLNMHMNVQNGKWDSDPSGTKTCIDTKEGILQYCQEVYPELQITNVVEANQPVTIQNWCKRGRK QCKTHPHFVIPYRCLVGEFVSDALLVPDKCKFLHQERMDVCETHLHWHTVAKETCSEKSTNLHDYGMLLPCGIDKFRGVEFVCCPLAEES DNVDSADAEEDDSDVWWGGADTDYADGSEDKVVEVAEEEEVAEVEEEEADDDEDDEDGDEVEEEAEEPYEEATERTTSIATTTTTTTESV EEVVREVCSEQAETGPCRAMISRWYFDVTEGKCAPFFYGGCGGNRNNFDTEEYCMAVCGSAMSQSLLKTTQEPLARDPVKLPTTAASTPD AVDKYLETPGDENEHAHFQKAKERLEAKHRERMSQVMREWEEAERQAKNLPKADKKAVIQHFQEKVESLEQEAANERQQLVETHMARVEA MLNDRRRLALENYITALQAVPPRPRHVFNMLKKYVRAEQKDRQHTLKHFEHVRMVDPKKAAQIRSQVMTHLRVIYERMNQSLSLLYNVPA VAEEIQDEVDELLQKEQNYSDDVLANMISEPRISYGNDALMPSLTETKTTVELLPVNGEFSLDDLQPWHSFGADSVPANTENEVEPVDAR PAADRGLTTRPGSGLTNIKTEEISEVKMDAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIATVIVITLVMLKKKQYTSIHH GVVEVDAAVTPEERHLSKMQQNGYENPTYKFFEQMQN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ERG-APP |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 732_751 | 19.0 | 771.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 732_751 | 19.0 | 696.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 732_751 | 0 | 640.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 732_751 | 19.0 | 752.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 732_751 | 19.0 | 753.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 732_751 | 19.0 | 715.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 732_751 | 14.0 | 747.0 | G(o)-alpha | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000346798 | 0 | 18 | 695_722 | 19.0 | 771.0 | PSEN1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000348990 | 0 | 16 | 695_722 | 19.0 | 696.0 | PSEN1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000354192 | 0 | 15 | 695_722 | 0 | 640.0 | PSEN1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000357903 | 0 | 17 | 695_722 | 19.0 | 752.0 | PSEN1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000358918 | 0 | 17 | 695_722 | 19.0 | 753.0 | PSEN1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000359726 | 0 | 17 | 695_722 | 19.0 | 715.0 | PSEN1 | |
| Tgene | APP | chr21:39870286 | chr21:27484463 | ENST00000440126 | 0 | 17 | 695_722 | 14.0 | 747.0 | PSEN1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ERG-APP |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ERG-APP |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ERG | C0033578 | Prostatic Neoplasms | 7 | CTD_human |
| Hgene | ERG | C0376358 | Malignant neoplasm of prostate | 7 | CTD_human |
| Hgene | ERG | C0023418 | leukemia | 1 | CTD_human |
| Hgene | ERG | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ERG | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ERG | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | ERG | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | ERG | C0279980 | Extra-osseous Ewing's sarcoma | 1 | ORPHANET |
| Hgene | ERG | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Hgene | ERG | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Tgene | C0002395 | Alzheimer's Disease | 63 | CTD_human;UNIPROT | |
| Tgene | C0011265 | Presenile dementia | 35 | CTD_human | |
| Tgene | C0276496 | Familial Alzheimer Disease (FAD) | 35 | CTD_human | |
| Tgene | C0494463 | Alzheimer Disease, Late Onset | 35 | CTD_human | |
| Tgene | C0546126 | Acute Confusional Senile Dementia | 35 | CTD_human | |
| Tgene | C0750900 | Alzheimer's Disease, Focal Onset | 35 | CTD_human | |
| Tgene | C0750901 | Alzheimer Disease, Early Onset | 35 | CTD_human | |
| Tgene | C0025261 | Memory Disorders | 16 | CTD_human | |
| Tgene | C0233794 | Memory impairment | 16 | CTD_human | |
| Tgene | C0751292 | Age-Related Memory Disorders | 16 | CTD_human | |
| Tgene | C0751293 | Memory Disorder, Semantic | 16 | CTD_human | |
| Tgene | C0751294 | Memory Disorder, Spatial | 16 | CTD_human | |
| Tgene | C0751295 | Memory Loss | 16 | CTD_human | |
| Tgene | C0027746 | Nerve Degeneration | 11 | CTD_human | |
| Tgene | C0023186 | Learning Disorders | 8 | CTD_human | |
| Tgene | C0751262 | Adult Learning Disorders | 8 | CTD_human | |
| Tgene | C0751263 | Learning Disturbance | 8 | CTD_human | |
| Tgene | C0751265 | Learning Disabilities | 8 | CTD_human | |
| Tgene | C1330966 | Developmental Academic Disorder | 8 | CTD_human | |
| Tgene | C2751536 | CEREBRAL AMYLOID ANGIOPATHY, APP-RELATED | 7 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0009241 | Cognition Disorders | 5 | CTD_human | |
| Tgene | C0011570 | Mental Depression | 5 | PSYGENET | |
| Tgene | C0011581 | Depressive disorder | 5 | PSYGENET | |
| Tgene | C0234544 | Todd Paralysis | 5 | CTD_human | |
| Tgene | C0522224 | Paralysed | 5 | CTD_human | |
| Tgene | C0497327 | Dementia | 3 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C2931672 | Cerebral hemorrhage with amyloidosis, hereditary, Dutch type | 3 | CTD_human;ORPHANET | |
| Tgene | C0270715 | Degenerative Diseases, Central Nervous System | 2 | CTD_human | |
| Tgene | C0333463 | Senile Plaques | 2 | CTD_human | |
| Tgene | C0524851 | Neurodegenerative Disorders | 2 | CTD_human | |
| Tgene | C0600467 | Neurogenic Inflammation | 2 | CTD_human | |
| Tgene | C0751733 | Degenerative Diseases, Spinal Cord | 2 | CTD_human | |
| Tgene | C2751494 | CEREBRAL AMYLOID ANGIOPATHY, APP-RELATED, ARCTIC VARIANT | 2 | CTD_human | |
| Tgene | C2936349 | Plaque, Amyloid | 2 | CTD_human | |
| Tgene | C3888307 | CEREBRAL AMYLOID ANGIOPATHY, APP-RELATED, FLEMISH VARIANT | 2 | CTD_human | |
| Tgene | C3888308 | CEREBRAL AMYLOID ANGIOPATHY, APP-RELATED, ITALIAN VARIANT | 2 | CTD_human | |
| Tgene | C3888309 | CEREBRAL AMYLOID ANGIOPATHY, APP-RELATED, IOWA VARIANT | 2 | CTD_human | |
| Tgene | C0002622 | Amnesia | 1 | CTD_human | |
| Tgene | C0002726 | Amyloidosis | 1 | CTD_human | |
| Tgene | C0003469 | Anxiety Disorders | 1 | CTD_human | |
| Tgene | C0005586 | Bipolar Disorder | 1 | PSYGENET | |
| Tgene | C0006111 | Brain Diseases | 1 | CTD_human | |
| Tgene | C0011573 | Endogenous depression | 1 | PSYGENET | |
| Tgene | C0016667 | Fragile X Syndrome | 1 | CTD_human | |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human | |
| Tgene | C0027540 | Necrosis | 1 | CTD_human | |
| Tgene | C0033141 | Cardiomyopathies, Primary | 1 | CTD_human | |
| Tgene | C0036529 | Myocardial Diseases, Secondary | 1 | CTD_human | |
| Tgene | C0036572 | Seizures | 1 | GENOMICS_ENGLAND | |
| Tgene | C0037928 | Spinal Cord Diseases | 1 | CTD_human | |
| Tgene | C0038002 | Splenomegaly | 1 | CTD_human | |
| Tgene | C0038454 | Cerebrovascular accident | 1 | GENOMICS_ENGLAND | |
| Tgene | C0043094 | Weight Gain | 1 | CTD_human | |
| Tgene | C0085220 | Cerebral Amyloid Angiopathy | 1 | CTD_human | |
| Tgene | C0085584 | Encephalopathies | 1 | CTD_human | |
| Tgene | C0231341 | Premature aging syndrome | 1 | CTD_human | |
| Tgene | C0233750 | Hysterical amnesia | 1 | CTD_human | |
| Tgene | C0233796 | Temporary Amnesia | 1 | CTD_human | |
| Tgene | C0234985 | Mental deterioration | 1 | CTD_human | |
| Tgene | C0236795 | Dissociative Amnesia | 1 | CTD_human | |
| Tgene | C0262497 | Global Amnesia | 1 | CTD_human | |
| Tgene | C0270612 | Leukoencephalopathy | 1 | GENOMICS_ENGLAND | |
| Tgene | C0338582 | Sporadic Cerebral Amyloid Angiopathy | 1 | CTD_human | |
| Tgene | C0338630 | Senile Paranoid Dementia | 1 | CTD_human | |
| Tgene | C0338656 | Impaired cognition | 1 | CTD_human | |
| Tgene | C0376280 | Anxiety States, Neurotic | 1 | CTD_human | |
| Tgene | C0553692 | Brain hemorrhage | 1 | GENOMICS_ENGLAND | |
| Tgene | C0750906 | Tactile Amnesia | 1 | CTD_human | |
| Tgene | C0750907 | Amnestic State | 1 | CTD_human | |
| Tgene | C0751071 | Familial Dementia | 1 | CTD_human | |
| Tgene | C0751156 | FRAXA Syndrome | 1 | CTD_human | |
| Tgene | C0751157 | FRAXE Syndrome | 1 | CTD_human | |
| Tgene | C0878544 | Cardiomyopathies | 1 | CTD_human | |
| Tgene | C0948008 | Ischemic stroke | 1 | GENOMICS_ENGLAND | |
| Tgene | C1270972 | Mild cognitive disorder | 1 | CTD_human | |
| Tgene | C1279420 | Anxiety neurosis (finding) | 1 | CTD_human |
