
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ESRRG-ACBD3 (FusionGDB2 ID:HG2104TG64746) |
Fusion Gene Summary for ESRRG-ACBD3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ESRRG-ACBD3 | Fusion gene ID: hg2104tg64746 | Hgene | Tgene | Gene symbol | ESRRG | ACBD3 | Gene ID | 2104 | 64746 |
| Gene name | estrogen related receptor gamma | acyl-CoA binding domain containing 3 | |
| Synonyms | ERR-gamma|ERR3|ERRg|ERRgamma|NR3B3 | GCP60|GOCAP1|GOLPH1|PAP7 | |
| Cytomap | ('ESRRG')('ACBD3') 1q41 | 1q42.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | estrogen-related receptor gammaERR gamma-2estrogen receptor-related protein 3nuclear receptor subfamily 3 group B member 3 | Golgi resident protein GCP60PBR- and PKA-associated protein 7PKA (RIalpha)-associated proteinacyl-Coenzyme A binding domain containing 3golgi complex associated protein 1, 60kDagolgi phosphoprotein 1peripheral benzodiazepine receptor-associated prot | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000359162, ENST00000360012, ENST00000361395, ENST00000361525, ENST00000366937, ENST00000366938, ENST00000366940, ENST00000391890, ENST00000408911, ENST00000463665, ENST00000487276, ENST00000493603, ENST00000493748, ENST00000471371, | ||
| Fusion gene scores | * DoF score | 6 X 6 X 2=72 | 8 X 7 X 5=280 |
| # samples | 6 | 8 | |
| ** MAII score | log2(6/72*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/280*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ESRRG [Title/Abstract] AND ACBD3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ESRRG(216850418)-ACBD3(226353701), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ESRRG-ACBD3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ESRRG-ACBD3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ESRRG-ACBD3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ESRRG-ACBD3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ESRRG-ACBD3 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. ESRRG-ACBD3 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. ESRRG-ACBD3 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ESRRG | GO:0006355 | regulation of transcription, DNA-templated | 23836911 |
 Fusion gene breakpoints across ESRRG (5'-gene) Fusion gene breakpoints across ESRRG (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
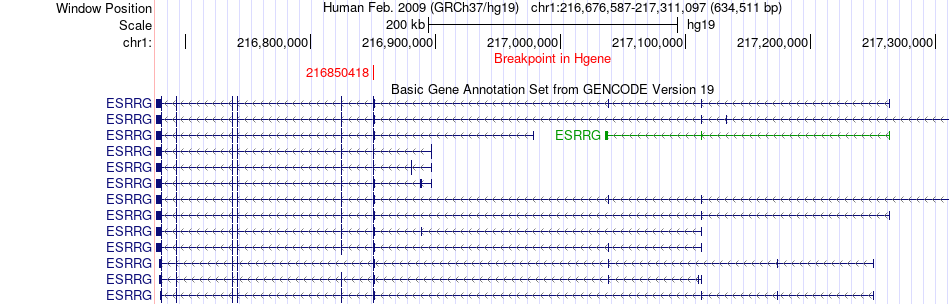 |
 Fusion gene breakpoints across ACBD3 (3'-gene) Fusion gene breakpoints across ACBD3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
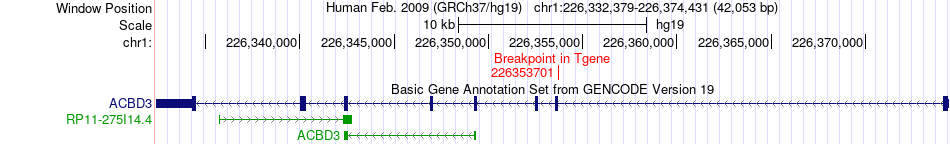 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-B6-A0IQ-01A | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
Top |
Fusion Gene ORF analysis for ESRRG-ACBD3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000359162 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000360012 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000361395 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000361525 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000366937 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000366938 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000366940 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000391890 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000408911 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000463665 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000487276 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000493603 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| 5CDS-intron | ENST00000493748 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000359162 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000360012 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000361525 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000366937 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000366938 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000366940 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000391890 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000463665 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000487276 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| Frame-shift | ENST00000493603 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| In-frame | ENST00000361395 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| In-frame | ENST00000408911 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| In-frame | ENST00000493748 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| intron-3CDS | ENST00000471371 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
| intron-intron | ENST00000471371 | ENST00000464927 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000361395 | ESRRG | chr1 | 216850418 | - | ENST00000366812 | ACBD3 | chr1 | 226353701 | - | 3858 | 626 | 193 | 1926 | 577 |
| ENST00000493748 | ESRRG | chr1 | 216850418 | - | ENST00000366812 | ACBD3 | chr1 | 226353701 | - | 4250 | 1018 | 585 | 2318 | 577 |
| ENST00000408911 | ESRRG | chr1 | 216850418 | - | ENST00000366812 | ACBD3 | chr1 | 226353701 | - | 3858 | 626 | 121 | 1926 | 601 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000361395 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - | 0.000273528 | 0.9997265 |
| ENST00000493748 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - | 0.000282836 | 0.9997172 |
| ENST00000408911 | ENST00000366812 | ESRRG | chr1 | 216850418 | - | ACBD3 | chr1 | 226353701 | - | 0.000219457 | 0.9997806 |
Top |
Fusion Genomic Features for ESRRG-ACBD3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
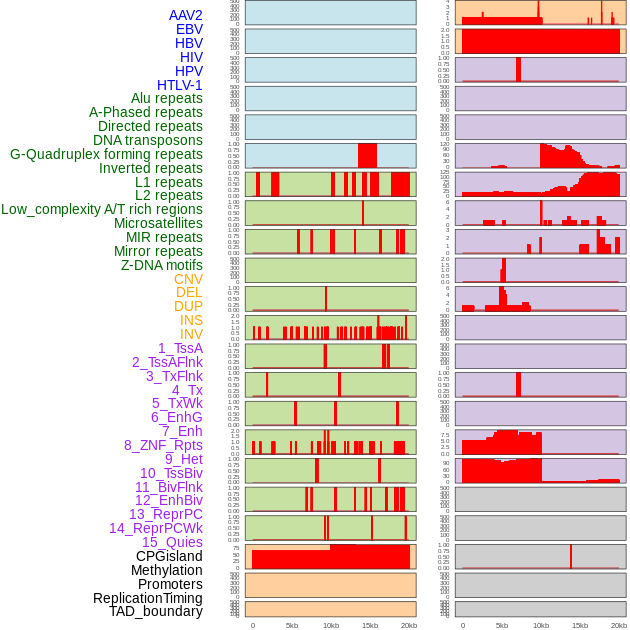 |
Top |
Fusion Protein Features for ESRRG-ACBD3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:216850418/chr1:226353701) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366937 | - | 3 | 8 | 128_148 | 162 | 471.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000408911 | - | 2 | 7 | 128_148 | 157 | 459.0 | Zinc finger | NR C4-type |
| Tgene | ACBD3 | chr1:216850418 | chr1:226353701 | ENST00000366812 | 0 | 8 | 174_257 | 95 | 529.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | ACBD3 | chr1:216850418 | chr1:226353701 | ENST00000366812 | 0 | 8 | 182_240 | 95 | 529.0 | Compositional bias | Note=Glu-rich | |
| Tgene | ACBD3 | chr1:216850418 | chr1:226353701 | ENST00000366812 | 0 | 8 | 196_238 | 95 | 529.0 | Compositional bias | Note=Arg-rich | |
| Tgene | ACBD3 | chr1:216850418 | chr1:226353701 | ENST00000366812 | 0 | 8 | 384_526 | 95 | 529.0 | Domain | GOLD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000359162 | - | 3 | 8 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000360012 | - | 2 | 7 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361395 | - | 3 | 8 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361525 | - | 4 | 9 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366937 | - | 3 | 8 | 125_200 | 162 | 471.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366938 | - | 3 | 8 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366940 | - | 4 | 9 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000391890 | - | 4 | 9 | 125_200 | 134 | 443.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000408911 | - | 2 | 7 | 125_200 | 157 | 459.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000463665 | - | 4 | 8 | 125_200 | 134 | 397.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000487276 | - | 4 | 9 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493603 | - | 4 | 9 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493748 | - | 4 | 9 | 125_200 | 134 | 436.0 | DNA binding | Nuclear receptor |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000359162 | - | 3 | 8 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000360012 | - | 2 | 7 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361395 | - | 3 | 8 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361525 | - | 4 | 9 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366937 | - | 3 | 8 | 233_457 | 162 | 471.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366938 | - | 3 | 8 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366940 | - | 4 | 9 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000391890 | - | 4 | 9 | 233_457 | 134 | 443.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000408911 | - | 2 | 7 | 233_457 | 157 | 459.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000463665 | - | 4 | 8 | 233_457 | 134 | 397.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000487276 | - | 4 | 9 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493603 | - | 4 | 9 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493748 | - | 4 | 9 | 233_457 | 134 | 436.0 | Domain | NR LBD |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000359162 | - | 3 | 8 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000359162 | - | 3 | 8 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000360012 | - | 2 | 7 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000360012 | - | 2 | 7 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361395 | - | 3 | 8 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361395 | - | 3 | 8 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361525 | - | 4 | 9 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000361525 | - | 4 | 9 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366937 | - | 3 | 8 | 164_188 | 162 | 471.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366938 | - | 3 | 8 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366938 | - | 3 | 8 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366940 | - | 4 | 9 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000366940 | - | 4 | 9 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000391890 | - | 4 | 9 | 128_148 | 134 | 443.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000391890 | - | 4 | 9 | 164_188 | 134 | 443.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000408911 | - | 2 | 7 | 164_188 | 157 | 459.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000463665 | - | 4 | 8 | 128_148 | 134 | 397.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000463665 | - | 4 | 8 | 164_188 | 134 | 397.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000487276 | - | 4 | 9 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000487276 | - | 4 | 9 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493603 | - | 4 | 9 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493603 | - | 4 | 9 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493748 | - | 4 | 9 | 128_148 | 134 | 436.0 | Zinc finger | NR C4-type |
| Hgene | ESRRG | chr1:216850418 | chr1:226353701 | ENST00000493748 | - | 4 | 9 | 164_188 | 134 | 436.0 | Zinc finger | NR C4-type |
| Tgene | ACBD3 | chr1:216850418 | chr1:226353701 | ENST00000366812 | 0 | 8 | 21_60 | 95 | 529.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ACBD3 | chr1:216850418 | chr1:226353701 | ENST00000366812 | 0 | 8 | 83_174 | 95 | 529.0 | Domain | ACB |
Top |
Fusion Gene Sequence for ESRRG-ACBD3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27588_27588_1_ESRRG-ACBD3_ESRRG_chr1_216850418_ENST00000361395_ACBD3_chr1_226353701_ENST00000366812_length(transcript)=3858nt_BP=626nt AGCTGACAATTTACTGGTTCATCAATGAAACAATATTAAATTATGAAGATGTAAGGAAAAAATCCTACGCTAACACTGTCGCAGTTTGAA AGCTTTAAAAAGTTGAAGTCTATTGGAAACCTTCTTATAAGAAACCTTCCTAGTCAGAGAATTCAACTTTCTTCTTTTCTTCATTCATGA AAATGCAGATCCACTGAGAAAGGGAATAAGGCTTCTCTGCAGAATGTCAAACAAAGATCGACACATTGATTCCAGCTGTTCGTCCTTCAT CAAGACGGAACCTTCCAGCCCAGCCTCCCTGACGGACAGCGTCAACCACCACAGCCCTGGTGGCTCTTCAGACGCCAGTGGGAGCTACAG TTCAACCATGAATGGCCATCAGAACGGACTTGACTCGCCACCTCTCTACCCTTCTGCTCCTATCCTGGGAGGTAGTGGGCCTGTCAGGAA ACTGTATGATGACTGCTCCAGCACCATTGTTGAAGATCCCCAGACCAAGTGTGAATACATGCTCAACTCGATGCCCAAGAGACTGTGTTT AGTGTGTGGTGACATCGCTTCTGGGTACCACTATGGGGTAGCATCATGTGAAGCCTGCAAGGCATTCTTCAAGAGGACAATTCAAGAAAA AGATGGCAAAGCATTTCATCCAACTTATGAAGAAAAATTGAAGCTTGTGGCACTGCATAAGCAAGTTCTTATGGGCCCATATAATCCAGA CACTTGTCCTGAGGTTGGATTCTTTGATGTGTTGGGGAATGACAGGAGGAGAGAATGGGCAGCCCTGGGAAACATGTCTAAAGAGGATGC CATGGTGGAGTTTGTCAAGCTCTTAAATAGGTGTTGCCATCTCTTTTCAACATATGTTGCGTCCCACAAAATAGAGAAGGAAGAGCAAGA AAAAAAAAGGAAGGAGGAAGAGGAGCGAAGGCGGCGTGAAGAGGAAGAAAGAGAACGTCTGCAAAAGGAGGAAGAGAAACGTAGGAGAGA AGAAGAGGAAAGGCTTCGACGGGAGGAAGAGGAAAGGAGACGGATAGAAGAAGAAAGGCTTCGGTTGGAGCAGCAAAAGCAGCAGATAAT GGCAGCTTTAAACTCCCAGACTGCCGTGCAGTTCCAGCAGTATGCAGCCCAACAGTATCCAGGGAACTACGAACAGCAGCAAATTCTCAT CCGCCAGTTGCAGGAGCAACACTATCAGCAGTACATGCAGCAGTTGTATCAAGTCCAGCTTGCACAGCAACAGGCAGCATTACAGAAACA ACAGGAAGTAGTAGTGGCTGGGTCTTCCTTGCCTACATCATCAAAAGTGAATGCAACTGTACCAAGTAATATGATGTCAGTTAATGGACA GGCCAAAACACACACTGACAGCTCCGAAAAAGAACTGGAACCAGAAGCTGCAGAAGAAGCCCTGGAGAATGGACCAAAAGAATCTCTTCC AGTAATAGCAGCTCCATCCATGTGGACACGACCTCAGATCAAAGACTTCAAAGAGAAGATTCAGCAGGATGCAGATTCCGTGATTACAGT GGGCCGAGGAGAAGTGGTCACTGTTCGAGTACCCACCCATGAAGAAGGATCATATCTCTTTTGGGAATTTGCCACAGACAATTATGACAT TGGGTTTGGGGTGTATTTTGAATGGACAGACTCTCCAAACACTGCTGTCAGCGTGCATGTCAGTGAGTCCAGCGATGACGACGAGGAGGA AGAAGAAAACATCGGTTGTGAAGAGAAAGCCAAAAAGAATGCCAACAAGCCTTTGCTGGATGAGATTGTGCCTGTGTACCGACGGGACTG TCATGAGGAGGTGTATGCTGGCAGCCATCAATATCCAGGGAGAGGAGTCTATCTCCTCAAGTTTGACAACTCCTACTCTTTGTGGCGGTC AAAATCAGTCTACTACAGAGTCTATTATACTAGATAAAAATGTTGTTACAAAGTCTGGAGTCTAGGGTTGGGCAGAAGATGACATTTAAT TTGGAAATTTCTTTTTACTTTTGTGGAGCATTAGAGTCACAGTTTACCTTATTGATATTGGTCTGATGGTTTGTGAACTCTTGCTGGGAA TCAAAATTTCCTTGAGACTCTTTAGCATTCATACTTTGGGGTTAAAGGAGATTCCTCAGACTCATCCAGCCCTTGGGTGCTGACCAGCAG AGTCACTAGTGGATGCTGAAGTTACATGAGCTACATGTTAAATATTTAAAGTCTCCAAAATAAAACACCCCAACGTTGACCTTACCCGGC TGATGGTTAGCCCCTTGCTGCCTGCTCCATGTGTCTTATGAGAGCCCGTAGTTACAGTGTCCTCTAATTTGAAATCCATAAGTTAACAAG TCTATATCAGGTGCAGCTGGCTTTGATTAAAGGCCATTTTTAAAACTTAAAAACTCAACACCTCACAGATTATAATAGAAAAAGAAATGG CCTCAGTTTGATCTCGTTCAGAATGACCCAGATTGTTTCTGCTTTGGGTGCAGCTGTTTAGTTCAGAGTTATATTACAGAGAATTATTTT CTGAGATAATCTTAAACTAGAATGTTCAAAACTAATTGATAATTGAAGTATCAAGATACGTAGAACACCTCAGAGATTTTTCTTCAGGAA CTTCCACAAACTTTGAATCCTTGTATCTTTATTTGGTATTCATACTACTAGTAGCAAAATACAGGTTTTTTGTTTTGTTTTGTTTTGTTT TGGCTTCATAGAGTATCTCAAATTGAAACTTTTCTGCACAAAGAATAAAATTAAGGATTTTATAAACTCAAATTGGCACCTACTGAATTA AAATACATAAAATCATTTAAATATAATTCAGCATATGGGAAGTAACATTGCACTAATATGGAAATCACTGCCAGAGACAGTCTATTTTCT TTTAATTTGTTACTACTTAGTCACAAACCCCACATTATTCCAGTTTGGAATTACTTATTAAGGAGAATTGGAAATACATATGCCCATGCT TAAATTTTATAGCTTTAATTTGTGTTATTTCTTTATTGACGGGAAGAGGTACATCTTTTTTTCCTTACTGAAAACAAATATGGATTAATT GCCTCAAATTTGTATAAGTGATTGGCTAGTGATTCTTGTTTTCAGAAGGGAGAGTGGTATAGATAGAAAATGACAAAGATGGCAATATAC ACTTAATGTTGTTATTGTATGTTGTTACTGAAGTACTTAGATTTTTAAAATTTCAAATCCTAAATCACTTCTTGTAGGAGGGTTTTCATT AACTGCAGTATATACAGTTCACTACATATGGGTTGTTTGAGTTTTTTGTGTGCTGTATTTCTTTCTGTTTTTTAATACCTGGTTTTGTAC ATATCTAACTCTGTTCTCTTTTGGTTGTTCAGAAACTGGATTTTTTTTTTCTTAAGCAGTGCTTAATTTGTGTTTTTTAATTTTGATTCA GAAGTAGTCCCAGCTCATAGGTGTTCATACTGTTACATCCAGAACATTTGTCAGGCTCTCTGTCAGCTTTCATGTACATATGGTATAGAA ACCATGGAGTTAGGCACTTCCTGGATTTTTTTTTTATGAGAAAAATACTGTATTTAAAATGTAAAATAAACTTTTAAAAAGCAGGCACTA ATATATATTTCTTCCAGCCTTTGATTACAAATTTGTCCTTGCACATGTTAAGATGAATTATCTCCTAAAAATATCATTGTTCTTGGGAGC AGTGTATGTTACTTTACATAGCAGCGGTTCCTGTCATGTGTTCATGTCAGAATATTTTTGGTTTTAAACTTTCTTATTGCCTTTGGCTGT TGATTAGTACAGTACAAGTGCGATTTCAAAAAGATCTTGAAAGTAATATATTTAATCAATTAAAATGTTTATCTGTAA >27588_27588_1_ESRRG-ACBD3_ESRRG_chr1_216850418_ENST00000361395_ACBD3_chr1_226353701_ENST00000366812_length(amino acids)=577AA_BP=144 MRKGIRLLCRMSNKDRHIDSSCSSFIKTEPSSPASLTDSVNHHSPGGSSDASGSYSSTMNGHQNGLDSPPLYPSAPILGGSGPVRKLYDD CSSTIVEDPQTKCEYMLNSMPKRLCLVCGDIASGYHYGVASCEACKAFFKRTIQEKDGKAFHPTYEEKLKLVALHKQVLMGPYNPDTCPE VGFFDVLGNDRRREWAALGNMSKEDAMVEFVKLLNRCCHLFSTYVASHKIEKEEQEKKRKEEEERRRREEEERERLQKEEEKRRREEEER LRREEEERRRIEEERLRLEQQKQQIMAALNSQTAVQFQQYAAQQYPGNYEQQQILIRQLQEQHYQQYMQQLYQVQLAQQQAALQKQQEVV VAGSSLPTSSKVNATVPSNMMSVNGQAKTHTDSSEKELEPEAAEEALENGPKESLPVIAAPSMWTRPQIKDFKEKIQQDADSVITVGRGE VVTVRVPTHEEGSYLFWEFATDNYDIGFGVYFEWTDSPNTAVSVHVSESSDDDEEEEENIGCEEKAKKNANKPLLDEIVPVYRRDCHEEV YAGSHQYPGRGVYLLKFDNSYSLWRSKSVYYRVYYTR -------------------------------------------------------------- >27588_27588_2_ESRRG-ACBD3_ESRRG_chr1_216850418_ENST00000408911_ACBD3_chr1_226353701_ENST00000366812_length(transcript)=3858nt_BP=626nt AAGTCCTTGATTAGGAGAGTGTGAGAGCTTTGGTCCCAACTGGCTGTGCCTATAGGCTTGTCACTAGGAGAACATTTGTGTTAATTGCAC TGTGCTCTGTCAAGGAAACTTTGATTTATAGCTGGGGTGCACAAATAATGGTTGCCGGTCGCACATGGATTCGGTAGAACTTTGCCTTCC TGAATCTTTTTCCCTGCACTACGAGGAAGAGCTTCTCTGCAGAATGTCAAACAAAGATCGACACATTGATTCCAGCTGTTCGTCCTTCAT CAAGACGGAACCTTCCAGCCCAGCCTCCCTGACGGACAGCGTCAACCACCACAGCCCTGGTGGCTCTTCAGACGCCAGTGGGAGCTACAG TTCAACCATGAATGGCCATCAGAACGGACTTGACTCGCCACCTCTCTACCCTTCTGCTCCTATCCTGGGAGGTAGTGGGCCTGTCAGGAA ACTGTATGATGACTGCTCCAGCACCATTGTTGAAGATCCCCAGACCAAGTGTGAATACATGCTCAACTCGATGCCCAAGAGACTGTGTTT AGTGTGTGGTGACATCGCTTCTGGGTACCACTATGGGGTAGCATCATGTGAAGCCTGCAAGGCATTCTTCAAGAGGACAATTCAAGAAAA AGATGGCAAAGCATTTCATCCAACTTATGAAGAAAAATTGAAGCTTGTGGCACTGCATAAGCAAGTTCTTATGGGCCCATATAATCCAGA CACTTGTCCTGAGGTTGGATTCTTTGATGTGTTGGGGAATGACAGGAGGAGAGAATGGGCAGCCCTGGGAAACATGTCTAAAGAGGATGC CATGGTGGAGTTTGTCAAGCTCTTAAATAGGTGTTGCCATCTCTTTTCAACATATGTTGCGTCCCACAAAATAGAGAAGGAAGAGCAAGA AAAAAAAAGGAAGGAGGAAGAGGAGCGAAGGCGGCGTGAAGAGGAAGAAAGAGAACGTCTGCAAAAGGAGGAAGAGAAACGTAGGAGAGA AGAAGAGGAAAGGCTTCGACGGGAGGAAGAGGAAAGGAGACGGATAGAAGAAGAAAGGCTTCGGTTGGAGCAGCAAAAGCAGCAGATAAT GGCAGCTTTAAACTCCCAGACTGCCGTGCAGTTCCAGCAGTATGCAGCCCAACAGTATCCAGGGAACTACGAACAGCAGCAAATTCTCAT CCGCCAGTTGCAGGAGCAACACTATCAGCAGTACATGCAGCAGTTGTATCAAGTCCAGCTTGCACAGCAACAGGCAGCATTACAGAAACA ACAGGAAGTAGTAGTGGCTGGGTCTTCCTTGCCTACATCATCAAAAGTGAATGCAACTGTACCAAGTAATATGATGTCAGTTAATGGACA GGCCAAAACACACACTGACAGCTCCGAAAAAGAACTGGAACCAGAAGCTGCAGAAGAAGCCCTGGAGAATGGACCAAAAGAATCTCTTCC AGTAATAGCAGCTCCATCCATGTGGACACGACCTCAGATCAAAGACTTCAAAGAGAAGATTCAGCAGGATGCAGATTCCGTGATTACAGT GGGCCGAGGAGAAGTGGTCACTGTTCGAGTACCCACCCATGAAGAAGGATCATATCTCTTTTGGGAATTTGCCACAGACAATTATGACAT TGGGTTTGGGGTGTATTTTGAATGGACAGACTCTCCAAACACTGCTGTCAGCGTGCATGTCAGTGAGTCCAGCGATGACGACGAGGAGGA AGAAGAAAACATCGGTTGTGAAGAGAAAGCCAAAAAGAATGCCAACAAGCCTTTGCTGGATGAGATTGTGCCTGTGTACCGACGGGACTG TCATGAGGAGGTGTATGCTGGCAGCCATCAATATCCAGGGAGAGGAGTCTATCTCCTCAAGTTTGACAACTCCTACTCTTTGTGGCGGTC AAAATCAGTCTACTACAGAGTCTATTATACTAGATAAAAATGTTGTTACAAAGTCTGGAGTCTAGGGTTGGGCAGAAGATGACATTTAAT TTGGAAATTTCTTTTTACTTTTGTGGAGCATTAGAGTCACAGTTTACCTTATTGATATTGGTCTGATGGTTTGTGAACTCTTGCTGGGAA TCAAAATTTCCTTGAGACTCTTTAGCATTCATACTTTGGGGTTAAAGGAGATTCCTCAGACTCATCCAGCCCTTGGGTGCTGACCAGCAG AGTCACTAGTGGATGCTGAAGTTACATGAGCTACATGTTAAATATTTAAAGTCTCCAAAATAAAACACCCCAACGTTGACCTTACCCGGC TGATGGTTAGCCCCTTGCTGCCTGCTCCATGTGTCTTATGAGAGCCCGTAGTTACAGTGTCCTCTAATTTGAAATCCATAAGTTAACAAG TCTATATCAGGTGCAGCTGGCTTTGATTAAAGGCCATTTTTAAAACTTAAAAACTCAACACCTCACAGATTATAATAGAAAAAGAAATGG CCTCAGTTTGATCTCGTTCAGAATGACCCAGATTGTTTCTGCTTTGGGTGCAGCTGTTTAGTTCAGAGTTATATTACAGAGAATTATTTT CTGAGATAATCTTAAACTAGAATGTTCAAAACTAATTGATAATTGAAGTATCAAGATACGTAGAACACCTCAGAGATTTTTCTTCAGGAA CTTCCACAAACTTTGAATCCTTGTATCTTTATTTGGTATTCATACTACTAGTAGCAAAATACAGGTTTTTTGTTTTGTTTTGTTTTGTTT TGGCTTCATAGAGTATCTCAAATTGAAACTTTTCTGCACAAAGAATAAAATTAAGGATTTTATAAACTCAAATTGGCACCTACTGAATTA AAATACATAAAATCATTTAAATATAATTCAGCATATGGGAAGTAACATTGCACTAATATGGAAATCACTGCCAGAGACAGTCTATTTTCT TTTAATTTGTTACTACTTAGTCACAAACCCCACATTATTCCAGTTTGGAATTACTTATTAAGGAGAATTGGAAATACATATGCCCATGCT TAAATTTTATAGCTTTAATTTGTGTTATTTCTTTATTGACGGGAAGAGGTACATCTTTTTTTCCTTACTGAAAACAAATATGGATTAATT GCCTCAAATTTGTATAAGTGATTGGCTAGTGATTCTTGTTTTCAGAAGGGAGAGTGGTATAGATAGAAAATGACAAAGATGGCAATATAC ACTTAATGTTGTTATTGTATGTTGTTACTGAAGTACTTAGATTTTTAAAATTTCAAATCCTAAATCACTTCTTGTAGGAGGGTTTTCATT AACTGCAGTATATACAGTTCACTACATATGGGTTGTTTGAGTTTTTTGTGTGCTGTATTTCTTTCTGTTTTTTAATACCTGGTTTTGTAC ATATCTAACTCTGTTCTCTTTTGGTTGTTCAGAAACTGGATTTTTTTTTTCTTAAGCAGTGCTTAATTTGTGTTTTTTAATTTTGATTCA GAAGTAGTCCCAGCTCATAGGTGTTCATACTGTTACATCCAGAACATTTGTCAGGCTCTCTGTCAGCTTTCATGTACATATGGTATAGAA ACCATGGAGTTAGGCACTTCCTGGATTTTTTTTTTATGAGAAAAATACTGTATTTAAAATGTAAAATAAACTTTTAAAAAGCAGGCACTA ATATATATTTCTTCCAGCCTTTGATTACAAATTTGTCCTTGCACATGTTAAGATGAATTATCTCCTAAAAATATCATTGTTCTTGGGAGC AGTGTATGTTACTTTACATAGCAGCGGTTCCTGTCATGTGTTCATGTCAGAATATTTTTGGTTTTAAACTTTCTTATTGCCTTTGGCTGT TGATTAGTACAGTACAAGTGCGATTTCAAAAAGATCTTGAAAGTAATATATTTAATCAATTAAAATGTTTATCTGTAA >27588_27588_2_ESRRG-ACBD3_ESRRG_chr1_216850418_ENST00000408911_ACBD3_chr1_226353701_ENST00000366812_length(amino acids)=601AA_BP=168 MGCTNNGCRSHMDSVELCLPESFSLHYEEELLCRMSNKDRHIDSSCSSFIKTEPSSPASLTDSVNHHSPGGSSDASGSYSSTMNGHQNGL DSPPLYPSAPILGGSGPVRKLYDDCSSTIVEDPQTKCEYMLNSMPKRLCLVCGDIASGYHYGVASCEACKAFFKRTIQEKDGKAFHPTYE EKLKLVALHKQVLMGPYNPDTCPEVGFFDVLGNDRRREWAALGNMSKEDAMVEFVKLLNRCCHLFSTYVASHKIEKEEQEKKRKEEEERR RREEEERERLQKEEEKRRREEEERLRREEEERRRIEEERLRLEQQKQQIMAALNSQTAVQFQQYAAQQYPGNYEQQQILIRQLQEQHYQQ YMQQLYQVQLAQQQAALQKQQEVVVAGSSLPTSSKVNATVPSNMMSVNGQAKTHTDSSEKELEPEAAEEALENGPKESLPVIAAPSMWTR PQIKDFKEKIQQDADSVITVGRGEVVTVRVPTHEEGSYLFWEFATDNYDIGFGVYFEWTDSPNTAVSVHVSESSDDDEEEEENIGCEEKA KKNANKPLLDEIVPVYRRDCHEEVYAGSHQYPGRGVYLLKFDNSYSLWRSKSVYYRVYYTR -------------------------------------------------------------- >27588_27588_3_ESRRG-ACBD3_ESRRG_chr1_216850418_ENST00000493748_ACBD3_chr1_226353701_ENST00000366812_length(transcript)=4250nt_BP=1018nt ATTCTGGGCAAAGTGTGGTGACTTGTGGATTTTCACTGTGACGATGTTGCTACACGGTCCTTCACTTGGAGTTAGTGCAACAAGGAGACA TCTGAGCTTAAAATTTATGAAACATCTAAAGAAATCAAGCTTTATATAGGATCACCGTTGTGGGTTGAATTGTGTATCCCCAAAAGATGT TGAAGTCCTGACCACCTCCCTATACCTTACAATATAGCCTTTTACGGAAGTAGGGTCTTTGCAGAAGATCAAGTTAAGATGAAATCATTT GATTGGGCCCTAGTTCAATATATCTTATGTCCTTATAAAAGGAGACAATTTGGACACAGGGACAAGGGGAGAATGCCACATAAAGATTGG AATTATGCTGCCATAAGCCAAGGAACTCCAAAGATCGCTGGCAAGCTGCTAAAAGCTAGCACAGATTCATGGAACAGAGTCTCCGTCATA GCCTACAAAAGGAATCAACCCTGCTGACATCTTGATTTCAGACTTCTAGCCTCCAGAACTAGCTGACAATTTACTGGTTCATCAATGAAA CAATATTAAATTATGAAGATGTAAGGAAAAAATCCTACGCTAACACTGTCGCAGTTTGAAAGGCTTCTCTGCAGAATGTCAAACAAAGAT CGACACATTGATTCCAGCTGTTCGTCCTTCATCAAGACGGAACCTTCCAGCCCAGCCTCCCTGACGGACAGCGTCAACCACCACAGCCCT GGTGGCTCTTCAGACGCCAGTGGGAGCTACAGTTCAACCATGAATGGCCATCAGAACGGACTTGACTCGCCACCTCTCTACCCTTCTGCT CCTATCCTGGGAGGTAGTGGGCCTGTCAGGAAACTGTATGATGACTGCTCCAGCACCATTGTTGAAGATCCCCAGACCAAGTGTGAATAC ATGCTCAACTCGATGCCCAAGAGACTGTGTTTAGTGTGTGGTGACATCGCTTCTGGGTACCACTATGGGGTAGCATCATGTGAAGCCTGC AAGGCATTCTTCAAGAGGACAATTCAAGAAAAAGATGGCAAAGCATTTCATCCAACTTATGAAGAAAAATTGAAGCTTGTGGCACTGCAT AAGCAAGTTCTTATGGGCCCATATAATCCAGACACTTGTCCTGAGGTTGGATTCTTTGATGTGTTGGGGAATGACAGGAGGAGAGAATGG GCAGCCCTGGGAAACATGTCTAAAGAGGATGCCATGGTGGAGTTTGTCAAGCTCTTAAATAGGTGTTGCCATCTCTTTTCAACATATGTT GCGTCCCACAAAATAGAGAAGGAAGAGCAAGAAAAAAAAAGGAAGGAGGAAGAGGAGCGAAGGCGGCGTGAAGAGGAAGAAAGAGAACGT CTGCAAAAGGAGGAAGAGAAACGTAGGAGAGAAGAAGAGGAAAGGCTTCGACGGGAGGAAGAGGAAAGGAGACGGATAGAAGAAGAAAGG CTTCGGTTGGAGCAGCAAAAGCAGCAGATAATGGCAGCTTTAAACTCCCAGACTGCCGTGCAGTTCCAGCAGTATGCAGCCCAACAGTAT CCAGGGAACTACGAACAGCAGCAAATTCTCATCCGCCAGTTGCAGGAGCAACACTATCAGCAGTACATGCAGCAGTTGTATCAAGTCCAG CTTGCACAGCAACAGGCAGCATTACAGAAACAACAGGAAGTAGTAGTGGCTGGGTCTTCCTTGCCTACATCATCAAAAGTGAATGCAACT GTACCAAGTAATATGATGTCAGTTAATGGACAGGCCAAAACACACACTGACAGCTCCGAAAAAGAACTGGAACCAGAAGCTGCAGAAGAA GCCCTGGAGAATGGACCAAAAGAATCTCTTCCAGTAATAGCAGCTCCATCCATGTGGACACGACCTCAGATCAAAGACTTCAAAGAGAAG ATTCAGCAGGATGCAGATTCCGTGATTACAGTGGGCCGAGGAGAAGTGGTCACTGTTCGAGTACCCACCCATGAAGAAGGATCATATCTC TTTTGGGAATTTGCCACAGACAATTATGACATTGGGTTTGGGGTGTATTTTGAATGGACAGACTCTCCAAACACTGCTGTCAGCGTGCAT GTCAGTGAGTCCAGCGATGACGACGAGGAGGAAGAAGAAAACATCGGTTGTGAAGAGAAAGCCAAAAAGAATGCCAACAAGCCTTTGCTG GATGAGATTGTGCCTGTGTACCGACGGGACTGTCATGAGGAGGTGTATGCTGGCAGCCATCAATATCCAGGGAGAGGAGTCTATCTCCTC AAGTTTGACAACTCCTACTCTTTGTGGCGGTCAAAATCAGTCTACTACAGAGTCTATTATACTAGATAAAAATGTTGTTACAAAGTCTGG AGTCTAGGGTTGGGCAGAAGATGACATTTAATTTGGAAATTTCTTTTTACTTTTGTGGAGCATTAGAGTCACAGTTTACCTTATTGATAT TGGTCTGATGGTTTGTGAACTCTTGCTGGGAATCAAAATTTCCTTGAGACTCTTTAGCATTCATACTTTGGGGTTAAAGGAGATTCCTCA GACTCATCCAGCCCTTGGGTGCTGACCAGCAGAGTCACTAGTGGATGCTGAAGTTACATGAGCTACATGTTAAATATTTAAAGTCTCCAA AATAAAACACCCCAACGTTGACCTTACCCGGCTGATGGTTAGCCCCTTGCTGCCTGCTCCATGTGTCTTATGAGAGCCCGTAGTTACAGT GTCCTCTAATTTGAAATCCATAAGTTAACAAGTCTATATCAGGTGCAGCTGGCTTTGATTAAAGGCCATTTTTAAAACTTAAAAACTCAA CACCTCACAGATTATAATAGAAAAAGAAATGGCCTCAGTTTGATCTCGTTCAGAATGACCCAGATTGTTTCTGCTTTGGGTGCAGCTGTT TAGTTCAGAGTTATATTACAGAGAATTATTTTCTGAGATAATCTTAAACTAGAATGTTCAAAACTAATTGATAATTGAAGTATCAAGATA CGTAGAACACCTCAGAGATTTTTCTTCAGGAACTTCCACAAACTTTGAATCCTTGTATCTTTATTTGGTATTCATACTACTAGTAGCAAA ATACAGGTTTTTTGTTTTGTTTTGTTTTGTTTTGGCTTCATAGAGTATCTCAAATTGAAACTTTTCTGCACAAAGAATAAAATTAAGGAT TTTATAAACTCAAATTGGCACCTACTGAATTAAAATACATAAAATCATTTAAATATAATTCAGCATATGGGAAGTAACATTGCACTAATA TGGAAATCACTGCCAGAGACAGTCTATTTTCTTTTAATTTGTTACTACTTAGTCACAAACCCCACATTATTCCAGTTTGGAATTACTTAT TAAGGAGAATTGGAAATACATATGCCCATGCTTAAATTTTATAGCTTTAATTTGTGTTATTTCTTTATTGACGGGAAGAGGTACATCTTT TTTTCCTTACTGAAAACAAATATGGATTAATTGCCTCAAATTTGTATAAGTGATTGGCTAGTGATTCTTGTTTTCAGAAGGGAGAGTGGT ATAGATAGAAAATGACAAAGATGGCAATATACACTTAATGTTGTTATTGTATGTTGTTACTGAAGTACTTAGATTTTTAAAATTTCAAAT CCTAAATCACTTCTTGTAGGAGGGTTTTCATTAACTGCAGTATATACAGTTCACTACATATGGGTTGTTTGAGTTTTTTGTGTGCTGTAT TTCTTTCTGTTTTTTAATACCTGGTTTTGTACATATCTAACTCTGTTCTCTTTTGGTTGTTCAGAAACTGGATTTTTTTTTTCTTAAGCA GTGCTTAATTTGTGTTTTTTAATTTTGATTCAGAAGTAGTCCCAGCTCATAGGTGTTCATACTGTTACATCCAGAACATTTGTCAGGCTC TCTGTCAGCTTTCATGTACATATGGTATAGAAACCATGGAGTTAGGCACTTCCTGGATTTTTTTTTTATGAGAAAAATACTGTATTTAAA ATGTAAAATAAACTTTTAAAAAGCAGGCACTAATATATATTTCTTCCAGCCTTTGATTACAAATTTGTCCTTGCACATGTTAAGATGAAT TATCTCCTAAAAATATCATTGTTCTTGGGAGCAGTGTATGTTACTTTACATAGCAGCGGTTCCTGTCATGTGTTCATGTCAGAATATTTT TGGTTTTAAACTTTCTTATTGCCTTTGGCTGTTGATTAGTACAGTACAAGTGCGATTTCAAAAAGATCTTGAAAGTAATATATTTAATCA ATTAAAATGTTTATCTGTAA >27588_27588_3_ESRRG-ACBD3_ESRRG_chr1_216850418_ENST00000493748_ACBD3_chr1_226353701_ENST00000366812_length(amino acids)=577AA_BP=144 MSQFERLLCRMSNKDRHIDSSCSSFIKTEPSSPASLTDSVNHHSPGGSSDASGSYSSTMNGHQNGLDSPPLYPSAPILGGSGPVRKLYDD CSSTIVEDPQTKCEYMLNSMPKRLCLVCGDIASGYHYGVASCEACKAFFKRTIQEKDGKAFHPTYEEKLKLVALHKQVLMGPYNPDTCPE VGFFDVLGNDRRREWAALGNMSKEDAMVEFVKLLNRCCHLFSTYVASHKIEKEEQEKKRKEEEERRRREEEERERLQKEEEKRRREEEER LRREEEERRRIEEERLRLEQQKQQIMAALNSQTAVQFQQYAAQQYPGNYEQQQILIRQLQEQHYQQYMQQLYQVQLAQQQAALQKQQEVV VAGSSLPTSSKVNATVPSNMMSVNGQAKTHTDSSEKELEPEAAEEALENGPKESLPVIAAPSMWTRPQIKDFKEKIQQDADSVITVGRGE VVTVRVPTHEEGSYLFWEFATDNYDIGFGVYFEWTDSPNTAVSVHVSESSDDDEEEEENIGCEEKAKKNANKPLLDEIVPVYRRDCHEEV YAGSHQYPGRGVYLLKFDNSYSLWRSKSVYYRVYYTR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ESRRG-ACBD3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ESRRG-ACBD3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ESRRG-ACBD3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
