
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ALDOB-C3 (FusionGDB2 ID:HG229TG718) |
Fusion Gene Summary for ALDOB-C3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ALDOB-C3 | Fusion gene ID: hg229tg718 | Hgene | Tgene | Gene symbol | ALDOB | C3 | Gene ID | 229 | 718 |
| Gene name | aldolase, fructose-bisphosphate B | complement C3 | |
| Synonyms | ALDB|ALDO2 | AHUS5|ARMD9|ASP|C3a|C3b|CPAMD1|HEL-S-62p | |
| Cytomap | ('ALDOB')('C3') 9q31.1 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | fructose-bisphosphate aldolase Baldolase 2aldolase B, fructose-bisphosphatasealdolase B, fructose-bisphosphateliver-type aldolase | complement C3C3 and PZP-like alpha-2-macroglobulin domain-containing protein 1C3a anaphylatoxinacylation-stimulating protein cleavage productcomplement component 3complement component C3acomplement component C3bepididymis secretory sperm binding pr | |
| Modification date | 20200329 | 20200327 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000374855, ENST00000468981, | ||
| Fusion gene scores | * DoF score | 5 X 4 X 4=80 | 22 X 20 X 8=3520 |
| # samples | 5 | 22 | |
| ** MAII score | log2(5/80*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/3520*10)=-4 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ALDOB [Title/Abstract] AND C3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ALDOB(104190751)-C3(6713520), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ALDOB-C3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ALDOB-C3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ALDOB | GO:0006096 | glycolytic process | 10625657 |
| Hgene | ALDOB | GO:0006116 | NADH oxidation | 17576770 |
| Hgene | ALDOB | GO:0030388 | fructose 1,6-bisphosphate metabolic process | 9244396|10625657 |
| Tgene | C3 | GO:0001934 | positive regulation of protein phosphorylation | 15833747 |
| Tgene | C3 | GO:0010575 | positive regulation of vascular endothelial growth factor production | 16452172 |
| Tgene | C3 | GO:0010828 | positive regulation of glucose transmembrane transport | 9059512|15833747 |
| Tgene | C3 | GO:0010866 | regulation of triglyceride biosynthetic process | 10432298 |
| Tgene | C3 | GO:0010884 | positive regulation of lipid storage | 9555951 |
| Tgene | C3 | GO:0045745 | positive regulation of G protein-coupled receptor signaling pathway | 15833747 |
 Fusion gene breakpoints across ALDOB (5'-gene) Fusion gene breakpoints across ALDOB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
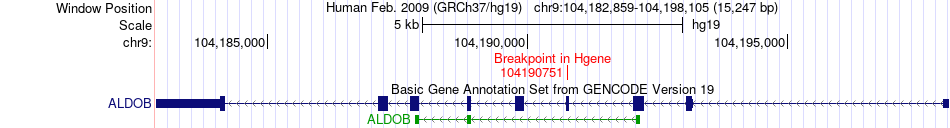 |
 Fusion gene breakpoints across C3 (3'-gene) Fusion gene breakpoints across C3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
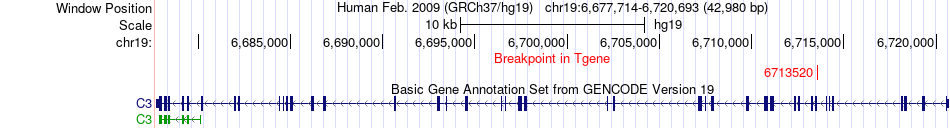 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-DD-A4NV-01A | ALDOB | chr9 | 104190751 | - | C3 | chr19 | 6713520 | - |
Top |
Fusion Gene ORF analysis for ALDOB-C3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000374855 | ENST00000599668 | ALDOB | chr9 | 104190751 | - | C3 | chr19 | 6713520 | - |
| In-frame | ENST00000374855 | ENST00000245907 | ALDOB | chr9 | 104190751 | - | C3 | chr19 | 6713520 | - |
| intron-3CDS | ENST00000468981 | ENST00000245907 | ALDOB | chr9 | 104190751 | - | C3 | chr19 | 6713520 | - |
| intron-intron | ENST00000468981 | ENST00000599668 | ALDOB | chr9 | 104190751 | - | C3 | chr19 | 6713520 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000374855 | ALDOB | chr9 | 104190751 | - | ENST00000245907 | C3 | chr19 | 6713520 | - | 4901 | 504 | 580 | 4722 | 1380 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000374855 | ENST00000245907 | ALDOB | chr9 | 104190751 | - | C3 | chr19 | 6713520 | - | 0.005644127 | 0.99435586 |
Top |
Fusion Genomic Features for ALDOB-C3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
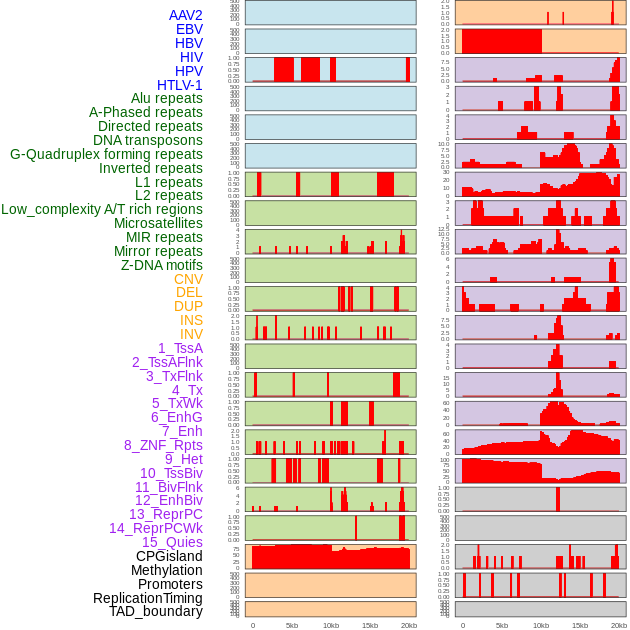 |
Top |
Fusion Protein Features for ALDOB-C3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:104190751/chr19:6713520) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | C3 | chr9:104190751 | chr19:6713520 | ENST00000245907 | 6 | 41 | 1518_1661 | 257 | 1664.0 | Domain | NTR | |
| Tgene | C3 | chr9:104190751 | chr19:6713520 | ENST00000245907 | 6 | 41 | 693_728 | 257 | 1664.0 | Domain | Anaphylatoxin-like |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for ALDOB-C3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >3973_3973_1_ALDOB-C3_ALDOB_chr9_104190751_ENST00000374855_C3_chr19_6713520_ENST00000245907_length(transcript)=4901nt_BP=504nt AAAAACATGATGAGAAGTCTATAAAAATTGTGTGCTACCAAAGATCTGTCTTATTTGGCAGCTGCTGCCTCACCCACAGCTTTTGATATC TAGGAGGACTCTTCTCTCCCAAACTACCTGTCACCATGGCCCACCGATTTCCAGCCCTCACCCAGGAGCAGAAGAAGGAGCTCTCAGAAA TTGCCCAGAGCATTGTTGCCAATGGAAAGGGGATCCTGGCTGCAGATGAATCTGTAGGTACCATGGGGAACCGCCTGCAGAGGATCAAGG TGGAAAACACTGAAGAGAACCGCCGGCAGTTCCGAGAAATCCTCTTCTCTGTGGACAGTTCCATCAACCAGAGCATCGGGGGTGTGATCC TTTTCCACGAGACCCTCTACCAGAAGGACAGCCAGGGAAAGCTGTTCAGAAACATCCTCAAGGAAAAGGGGATCGTGGTGGGAATCAAGT TAGACCAAGGAGGTGCTCCTCTTGCAGGAACAAACAAAGAAACCACCATTCAAGGTTCCTCTACGGGAAGAAAGTGGAGGGAACTGCCTT TGTCATCTTCGGGATCCAGGATGGCGAACAGAGGATTTCCCTGCCTGAATCCCTCAAGCGCATTCCGATTGAGGATGGCTCGGGGGAGGT TGTGCTGAGCCGGAAGGTACTGCTGGACGGGGTGCAGAACCCCCGAGCAGAAGACCTGGTGGGGAAGTCTTTGTACGTGTCTGCCACCGT CATCTTGCACTCAGGCAGTGACATGGTGCAGGCAGAGCGCAGCGGGATCCCCATCGTGACCTCTCCCTACCAGATCCACTTCACCAAGAC ACCCAAGTACTTCAAACCAGGAATGCCCTTTGACCTCATGGTGTTCGTGACGAACCCTGATGGCTCTCCAGCCTACCGAGTCCCCGTGGC AGTCCAGGGCGAGGACACTGTGCAGTCTCTAACCCAGGGAGATGGCGTGGCCAAACTCAGCATCAACACACACCCCAGCCAGAAGCCCTT GAGCATCACGGTGCGCACGAAGAAGCAGGAGCTCTCGGAGGCAGAGCAGGCTACCAGGACCATGCAGGCTCTGCCCTACAGCACCGTGGG CAACTCCAACAATTACCTGCATCTCTCAGTGCTACGTACAGAGCTCAGACCCGGGGAGACCCTCAACGTCAACTTCCTCCTGCGAATGGA CCGCGCCCACGAGGCCAAGATCCGCTACTACACCTACCTGATCATGAACAAGGGCAGGCTGTTGAAGGCGGGACGCCAGGTGCGAGAGCC CGGCCAGGACCTGGTGGTGCTGCCCCTGTCCATCACCACCGACTTCATCCCTTCCTTCCGCCTGGTGGCGTACTACACGCTGATCGGTGC CAGCGGCCAGAGGGAGGTGGTGGCCGACTCCGTGTGGGTGGACGTCAAGGACTCCTGCGTGGGCTCGCTGGTGGTAAAAAGCGGCCAGTC AGAAGACCGGCAGCCTGTACCTGGGCAGCAGATGACCCTGAAGATAGAGGGTGACCACGGGGCCCGGGTGGTACTGGTGGCCGTGGACAA GGGCGTGTTCGTGCTGAATAAGAAGAACAAACTGACGCAGAGTAAGATCTGGGACGTGGTGGAGAAGGCAGACATCGGCTGCACCCCGGG CAGTGGGAAGGATTACGCCGGTGTCTTCTCCGACGCAGGGCTGACCTTCACGAGCAGCAGTGGCCAGCAGACCGCCCAGAGGGCAGAACT TCAGTGCCCGCAGCCAGCCGCCCGCCGACGCCGTTCCGTGCAGCTCACGGAGAAGCGAATGGACAAAGTCGGCAAGTACCCCAAGGAGCT GCGCAAGTGCTGCGAGGACGGCATGCGGGAGAACCCCATGAGGTTCTCGTGCCAGCGCCGGACCCGTTTCATCTCCCTGGGCGAGGCGTG CAAGAAGGTCTTCCTGGACTGCTGCAACTACATCACAGAGCTGCGGCGGCAGCACGCGCGGGCCAGCCACCTGGGCCTGGCCAGGAGTAA CCTGGATGAGGACATCATTGCAGAAGAGAACATCGTTTCCCGAAGTGAGTTCCCAGAGAGCTGGCTGTGGAACGTTGAGGACTTGAAAGA GCCACCGAAAAATGGAATCTCTACGAAGCTCATGAATATATTTTTGAAAGACTCCATCACCACGTGGGAGATTCTGGCTGTGAGCATGTC GGACAAGAAAGGGATCTGTGTGGCAGACCCCTTCGAGGTCACAGTAATGCAGGACTTCTTCATCGACCTGCGGCTACCCTACTCTGTTGT TCGAAACGAGCAGGTGGAAATCCGAGCCGTTCTCTACAATTACCGGCAGAACCAAGAGCTCAAGGTGAGGGTGGAACTACTCCACAATCC AGCCTTCTGCAGCCTGGCCACCACCAAGAGGCGTCACCAGCAGACCGTAACCATCCCCCCCAAGTCCTCGTTGTCCGTTCCATATGTCAT CGTGCCGCTAAAGACCGGCCTGCAGGAAGTGGAAGTCAAGGCTGCTGTCTACCATCATTTCATCAGTGACGGTGTCAGGAAGTCCCTGAA GGTCGTGCCGGAAGGAATCAGAATGAACAAAACTGTGGCTGTTCGCACCCTGGATCCAGAACGCCTGGGCCGTGAAGGAGTGCAGAAAGA GGACATCCCACCTGCAGACCTCAGTGACCAAGTCCCGGACACCGAGTCTGAGACCAGAATTCTCCTGCAAGGGACCCCAGTGGCCCAGAT GACAGAGGATGCCGTCGACGCGGAACGGCTGAAGCACCTCATTGTGACCCCCTCGGGCTGCGGGGAACAGAACATGATCGGCATGACGCC CACGGTCATCGCTGTGCATTACCTGGATGAAACGGAGCAGTGGGAGAAGTTCGGCCTAGAGAAGCGGCAGGGGGCCTTGGAGCTCATCAA GAAGGGGTACACCCAGCAGCTGGCCTTCAGACAACCCAGCTCTGCCTTTGCGGCCTTCGTGAAACGGGCACCCAGCACCTGGCTGACCGC CTACGTGGTCAAGGTCTTCTCTCTGGCTGTCAACCTCATCGCCATCGACTCCCAAGTCCTCTGCGGGGCTGTTAAATGGCTGATCCTGGA GAAGCAGAAGCCCGACGGGGTCTTCCAGGAGGATGCGCCCGTGATACACCAAGAAATGATTGGTGGATTACGGAACAACAACGAGAAAGA CATGGCCCTCACGGCCTTTGTTCTCATCTCGCTGCAGGAGGCTAAAGATATTTGCGAGGAGCAGGTCAACAGCCTGCCAGGCAGCATCAC TAAAGCAGGAGACTTCCTTGAAGCCAACTACATGAACCTACAGAGATCCTACACTGTGGCCATTGCTGGCTATGCTCTGGCCCAGATGGG CAGGCTGAAGGGGCCTCTTCTTAACAAATTTCTGACCACAGCCAAAGATAAGAACCGCTGGGAGGACCCTGGTAAGCAGCTCTACAACGT GGAGGCCACATCCTATGCCCTCTTGGCCCTACTGCAGCTAAAAGACTTTGACTTTGTGCCTCCCGTCGTGCGTTGGCTCAATGAACAGAG ATACTACGGTGGTGGCTATGGCTCTACCCAGGCCACCTTCATGGTGTTCCAAGCCTTGGCTCAATACCAAAAGGACGCCCCTGACCACCA GGAACTGAACCTTGATGTGTCCCTCCAACTGCCCAGCCGCAGCTCCAAGATCACCCACCGTATCCACTGGGAATCTGCCAGCCTCCTGCG ATCAGAAGAGACCAAGGAAAATGAGGGTTTCACAGTCACAGCTGAAGGAAAAGGCCAAGGCACCTTGTCGGTGGTGACAATGTACCATGC TAAGGCCAAAGATCAACTCACCTGTAATAAATTCGACCTCAAGGTCACCATAAAACCAGCACCGGAAACAGAAAAGAGGCCTCAGGATGC CAAGAACACTATGATCCTTGAGATCTGTACCAGGTACCGGGGAGACCAGGATGCCACTATGTCTATATTGGACATATCCATGATGACTGG CTTTGCTCCAGACACAGATGACCTGAAGCAGCTGGCCAATGGTGTTGACAGATACATCTCCAAGTATGAGCTGGACAAAGCCTTCTCCGA TAGGAACACCCTCATCATCTACCTGGACAAGGTCTCACACTCTGAGGATGACTGTCTAGCTTTCAAAGTTCACCAATACTTTAATGTAGA GCTTATCCAGCCTGGAGCAGTCAAGGTCTACGCCTATTACAACCTGGAGGAAAGCTGTACCCGGTTCTACCATCCGGAAAAGGAGGATGG AAAGCTGAACAAGCTCTGCCGTGATGAACTGTGCCGCTGTGCTGAGGAGAATTGCTTCATACAAAAGTCGGATGACAAGGTCACCCTGGA AGAACGGCTGGACAAGGCCTGTGAGCCAGGAGTGGACTATGTGTACAAGACCCGACTGGTCAAGGTTCAGCTGTCCAATGACTTTGACGA GTACATCATGGCCATTGAGCAGACCATCAAGTCAGGCTCGGATGAGGTGCAGGTTGGACAGCAGCGCACGTTCATCAGCCCCATCAAGTG CAGAGAAGCCCTGAAGCTGGAGGAGAAGAAACACTACCTCATGTGGGGTCTCTCCTCCGATTTCTGGGGAGAGAAGCCCAACCTCAGCTA CATCATCGGGAAGGACACTTGGGTGGAGCACTGGCCCGAGGAGGACGAATGCCAAGACGAAGAGAACCAGAAACAATGCCAGGACCTCGG CGCCTTCACCGAGAGCATGGTTGTCTTTGGGTGCCCCAACTGACCACACCCCCATTCCCCCACTCCAGATAAAGCTTCAGTTATATCTCA CGTGTCTGGAGTTCTTTGCCAAGAGGGAGAGGCTGAAATCCCCAGCCGCCTCACCTGCAGCTCAGCTCCATCCTACTTGAAACCTCACCT GTTCCCACCGCATTTTCTCCTGGCGTTCGCCTGCTAGTGTG >3973_3973_1_ALDOB-C3_ALDOB_chr9_104190751_ENST00000374855_C3_chr19_6713520_ENST00000245907_length(amino acids)=1380AA_BP= MPESLKRIPIEDGSGEVVLSRKVLLDGVQNPRAEDLVGKSLYVSATVILHSGSDMVQAERSGIPIVTSPYQIHFTKTPKYFKPGMPFDLM VFVTNPDGSPAYRVPVAVQGEDTVQSLTQGDGVAKLSINTHPSQKPLSITVRTKKQELSEAEQATRTMQALPYSTVGNSNNYLHLSVLRT ELRPGETLNVNFLLRMDRAHEAKIRYYTYLIMNKGRLLKAGRQVREPGQDLVVLPLSITTDFIPSFRLVAYYTLIGASGQREVVADSVWV DVKDSCVGSLVVKSGQSEDRQPVPGQQMTLKIEGDHGARVVLVAVDKGVFVLNKKNKLTQSKIWDVVEKADIGCTPGSGKDYAGVFSDAG LTFTSSSGQQTAQRAELQCPQPAARRRRSVQLTEKRMDKVGKYPKELRKCCEDGMRENPMRFSCQRRTRFISLGEACKKVFLDCCNYITE LRRQHARASHLGLARSNLDEDIIAEENIVSRSEFPESWLWNVEDLKEPPKNGISTKLMNIFLKDSITTWEILAVSMSDKKGICVADPFEV TVMQDFFIDLRLPYSVVRNEQVEIRAVLYNYRQNQELKVRVELLHNPAFCSLATTKRRHQQTVTIPPKSSLSVPYVIVPLKTGLQEVEVK AAVYHHFISDGVRKSLKVVPEGIRMNKTVAVRTLDPERLGREGVQKEDIPPADLSDQVPDTESETRILLQGTPVAQMTEDAVDAERLKHL IVTPSGCGEQNMIGMTPTVIAVHYLDETEQWEKFGLEKRQGALELIKKGYTQQLAFRQPSSAFAAFVKRAPSTWLTAYVVKVFSLAVNLI AIDSQVLCGAVKWLILEKQKPDGVFQEDAPVIHQEMIGGLRNNNEKDMALTAFVLISLQEAKDICEEQVNSLPGSITKAGDFLEANYMNL QRSYTVAIAGYALAQMGRLKGPLLNKFLTTAKDKNRWEDPGKQLYNVEATSYALLALLQLKDFDFVPPVVRWLNEQRYYGGGYGSTQATF MVFQALAQYQKDAPDHQELNLDVSLQLPSRSSKITHRIHWESASLLRSEETKENEGFTVTAEGKGQGTLSVVTMYHAKAKDQLTCNKFDL KVTIKPAPETEKRPQDAKNTMILEICTRYRGDQDATMSILDISMMTGFAPDTDDLKQLANGVDRYISKYELDKAFSDRNTLIIYLDKVSH SEDDCLAFKVHQYFNVELIQPGAVKVYAYYNLEESCTRFYHPEKEDGKLNKLCRDELCRCAEENCFIQKSDDKVTLEERLDKACEPGVDY VYKTRLVKVQLSNDFDEYIMAIEQTIKSGSDEVQVGQQRTFISPIKCREALKLEEKKHYLMWGLSSDFWGEKPNLSYIIGKDTWVEHWPE EDECQDEENQKQCQDLGAFTESMVVFGCPN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ALDOB-C3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | C3 | chr9:104190751 | chr19:6713520 | ENST00000245907 | 6 | 41 | 1634_1659 | 257.6666666666667 | 1664.0 | CFP/properdin |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ALDOB-C3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ALDOB-C3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ALDOB | C0016751 | Hereditary fructose intolerance syndrome | 14 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ALDOB | C0003129 | Anoxemia | 1 | CTD_human |
| Hgene | ALDOB | C0003130 | Anoxia | 1 | CTD_human |
| Hgene | ALDOB | C0019193 | Hepatitis, Toxic | 1 | CTD_human |
| Hgene | ALDOB | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Hgene | ALDOB | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Hgene | ALDOB | C0241910 | Autoimmune Chronic Hepatitis | 1 | CTD_human |
| Hgene | ALDOB | C0242184 | Hypoxia | 1 | CTD_human |
| Hgene | ALDOB | C0700292 | Hypoxemia | 1 | CTD_human |
| Hgene | ALDOB | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human |
| Hgene | ALDOB | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human |
| Hgene | ALDOB | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Hgene | ALDOB | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human |
| Hgene | ALDOB | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human |
| Hgene | ALDOB | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
| Tgene | C3151071 | COMPLEMENT COMPONENT 3 DEFICIENCY, AUTOSOMAL RECESSIVE | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0242383 | Age related macular degeneration | 4 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C2752037 | HEMOLYTIC UREMIC SYNDROME, ATYPICAL, SUSCEPTIBILITY TO, 5 | 4 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C1969651 | Macular Degeneration, Age-Related, 9 | 2 | CTD_human;UNIPROT | |
| Tgene | C0003257 | Antibody Deficiency Syndrome | 1 | CTD_human | |
| Tgene | C0007787 | Transient Ischemic Attack | 1 | CTD_human | |
| Tgene | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human | |
| Tgene | C0013221 | Drug toxicity | 1 | CTD_human | |
| Tgene | C0017665 | Membranous glomerulonephritis | 1 | CTD_human | |
| Tgene | C0019061 | Hemolytic-Uremic Syndrome | 1 | GENOMICS_ENGLAND | |
| Tgene | C0019193 | Hepatitis, Toxic | 1 | CTD_human | |
| Tgene | C0021051 | Immunologic Deficiency Syndromes | 1 | CTD_human | |
| Tgene | C0021655 | Insulin Resistance | 1 | CTD_human | |
| Tgene | C0022660 | Kidney Failure, Acute | 1 | CTD_human | |
| Tgene | C0030524 | Paratuberculosis | 1 | CTD_human | |
| Tgene | C0030807 | Pemphigus | 1 | CTD_human | |
| Tgene | C0030809 | Pemphigus Vulgaris | 1 | CTD_human | |
| Tgene | C0034152 | Henoch-Schoenlein Purpura | 1 | CTD_human | |
| Tgene | C0041755 | Adverse reaction to drug | 1 | CTD_human | |
| Tgene | C0042386 | Vasculitis, Hemorrhagic | 1 | CTD_human | |
| Tgene | C0086445 | Idiopathic Membranous Glomerulonephritis | 1 | CTD_human | |
| Tgene | C0086922 | Rheumatoid Purpura | 1 | CTD_human | |
| Tgene | C0238281 | Middle Cerebral Artery Syndrome | 1 | CTD_human | |
| Tgene | C0242461 | Purpura, Nonthrombocytopenic | 1 | CTD_human | |
| Tgene | C0263313 | Pemphigus Foliaceus | 1 | CTD_human | |
| Tgene | C0272242 | Complement deficiency disease | 1 | GENOMICS_ENGLAND | |
| Tgene | C0376362 | Purpura Hemorrhagica | 1 | CTD_human | |
| Tgene | C0472381 | Posterior Circulation Transient Ischemic Attack | 1 | CTD_human | |
| Tgene | C0740376 | Middle Cerebral Artery Thrombosis | 1 | CTD_human | |
| Tgene | C0740391 | Middle Cerebral Artery Occlusion | 1 | CTD_human | |
| Tgene | C0740392 | Infarction, Middle Cerebral Artery | 1 | CTD_human | |
| Tgene | C0751019 | Carotid Circulation Transient Ischemic Attack | 1 | CTD_human | |
| Tgene | C0751020 | Transient Ischemic Attack, Vertebrobasilar Circulation | 1 | CTD_human | |
| Tgene | C0751021 | Crescendo Transient Ischemic Attacks | 1 | CTD_human | |
| Tgene | C0751022 | Brain Stem Ischemia, Transient | 1 | CTD_human | |
| Tgene | C0751845 | Middle Cerebral Artery Embolus | 1 | CTD_human | |
| Tgene | C0751846 | Left Middle Cerebral Artery Infarction | 1 | CTD_human | |
| Tgene | C0751847 | Embolic Infarction, Middle Cerebral Artery | 1 | CTD_human | |
| Tgene | C0751848 | Thrombotic Infarction, Middle Cerebral Artery | 1 | CTD_human | |
| Tgene | C0751849 | Right Middle Cerebral Artery Infarction | 1 | CTD_human | |
| Tgene | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human | |
| Tgene | C0917805 | Transient Cerebral Ischemia | 1 | CTD_human | |
| Tgene | C0920563 | Insulin Sensitivity | 1 | CTD_human | |
| Tgene | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human | |
| Tgene | C1332655 | Complement component 3 deficiency | 1 | GENOMICS_ENGLAND | |
| Tgene | C1527335 | Transient Ischemic Attack, Anterior Circulation | 1 | CTD_human | |
| Tgene | C1565662 | Acute Kidney Insufficiency | 1 | CTD_human | |
| Tgene | C1704378 | Heymann Nephritis | 1 | CTD_human | |
| Tgene | C2609414 | Acute kidney injury | 1 | CTD_human | |
| Tgene | C2931788 | Atypical Hemolytic Uremic Syndrome | 1 | CTD_human;GENOMICS_ENGLAND | |
| Tgene | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human | |
| Tgene | C4087273 | C3 glomerulopathy | 1 | GENOMICS_ENGLAND | |
| Tgene | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human | |
| Tgene | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
