
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ERC1-TACC3 (FusionGDB2 ID:HG23085TG10460) |
Fusion Gene Summary for ERC1-TACC3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ERC1-TACC3 | Fusion gene ID: hg23085tg10460 | Hgene | Tgene | Gene symbol | ERC1 | TACC3 | Gene ID | 23085 | 10460 |
| Gene name | ELKS/RAB6-interacting/CAST family member 1 | transforming acidic coiled-coil containing protein 3 | |
| Synonyms | Cast2|ELKS|ERC-1|RAB6IP2 | ERIC-1|ERIC1|Tacc4|maskin | |
| Cytomap | ('ERC1')('TACC3') 12p13.33 | 4p16.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ELKS/Rab6-interacting/CAST family member 1RAB6 interacting protein 2 | transforming acidic coiled-coil-containing protein 3 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q8IUD2 | Q9Y6A5 | |
| Ensembl transtripts involved in fusion gene | ENST00000536573, ENST00000355446, ENST00000360905, ENST00000397203, ENST00000543086, ENST00000546231, ENST00000589028, | ||
| Fusion gene scores | * DoF score | 32 X 23 X 16=11776 | 12 X 26 X 14=4368 |
| # samples | 38 | 31 | |
| ** MAII score | log2(38/11776*10)=-4.95370634772607 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(31/4368*10)=-3.81663273564562 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ERC1 [Title/Abstract] AND TACC3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ERC1(1192746)-TACC3(1741429), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | ERC1-TACC3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ERC1-TACC3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ERC1 | GO:0007252 | I-kappaB phosphorylation | 15218148 |
 Fusion gene breakpoints across ERC1 (5'-gene) Fusion gene breakpoints across ERC1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
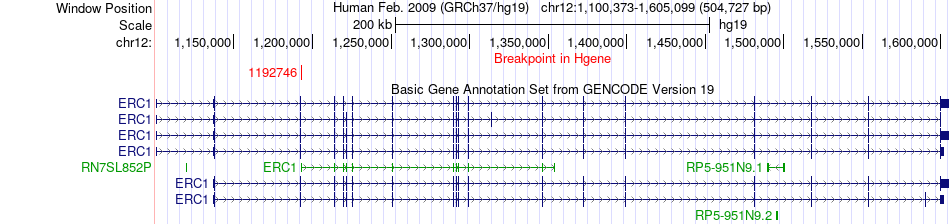 |
 Fusion gene breakpoints across TACC3 (3'-gene) Fusion gene breakpoints across TACC3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
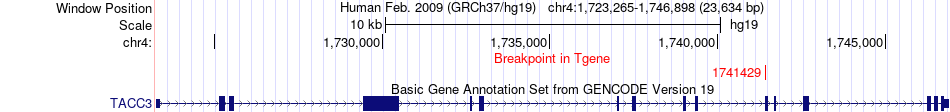 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A9RU-01A | ERC1 | chr12 | 1192746 | - | TACC3 | chr4 | 1741429 | + |
| ChimerDB4 | BRCA | TCGA-E2-A9RU-01A | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
Top |
Fusion Gene ORF analysis for ERC1-TACC3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000536573 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
| In-frame | ENST00000355446 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
| In-frame | ENST00000360905 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
| In-frame | ENST00000397203 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
| In-frame | ENST00000543086 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
| In-frame | ENST00000546231 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
| In-frame | ENST00000589028 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000543086 | ERC1 | chr12 | 1192746 | + | ENST00000313288 | TACC3 | chr4 | 1741429 | + | 2091 | 1357 | 271 | 1932 | 553 |
| ENST00000546231 | ERC1 | chr12 | 1192746 | + | ENST00000313288 | TACC3 | chr4 | 1741429 | + | 2348 | 1614 | 528 | 2189 | 553 |
| ENST00000397203 | ERC1 | chr12 | 1192746 | + | ENST00000313288 | TACC3 | chr4 | 1741429 | + | 2226 | 1492 | 406 | 2067 | 553 |
| ENST00000360905 | ERC1 | chr12 | 1192746 | + | ENST00000313288 | TACC3 | chr4 | 1741429 | + | 2001 | 1267 | 181 | 1842 | 553 |
| ENST00000589028 | ERC1 | chr12 | 1192746 | + | ENST00000313288 | TACC3 | chr4 | 1741429 | + | 1977 | 1243 | 157 | 1818 | 553 |
| ENST00000355446 | ERC1 | chr12 | 1192746 | + | ENST00000313288 | TACC3 | chr4 | 1741429 | + | 1952 | 1218 | 132 | 1793 | 553 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000543086 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + | 0.005787823 | 0.9942122 |
| ENST00000546231 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + | 0.005002321 | 0.99499774 |
| ENST00000397203 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + | 0.005833251 | 0.99416673 |
| ENST00000360905 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + | 0.006062169 | 0.99393785 |
| ENST00000589028 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + | 0.005507231 | 0.99449277 |
| ENST00000355446 | ENST00000313288 | ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741429 | + | 0.005831028 | 0.994169 |
Top |
Fusion Genomic Features for ERC1-TACC3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741428 | + | 0.000714442 | 0.99928564 |
| ERC1 | chr12 | 1192746 | + | TACC3 | chr4 | 1741428 | + | 0.000714442 | 0.99928564 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
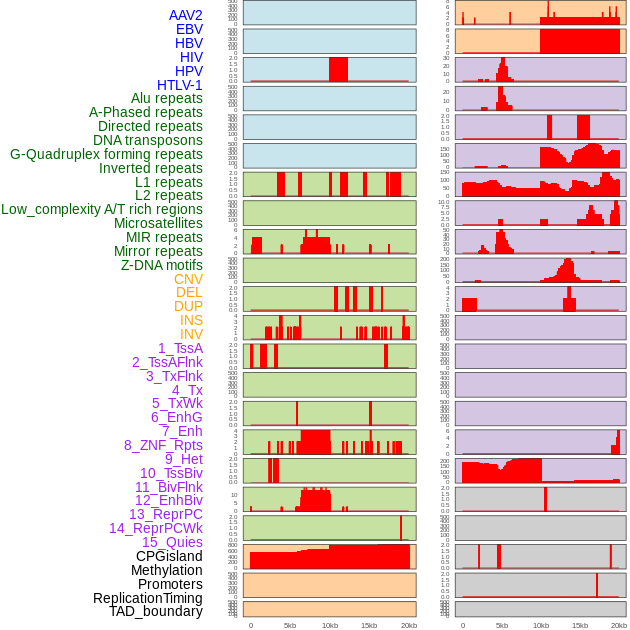 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
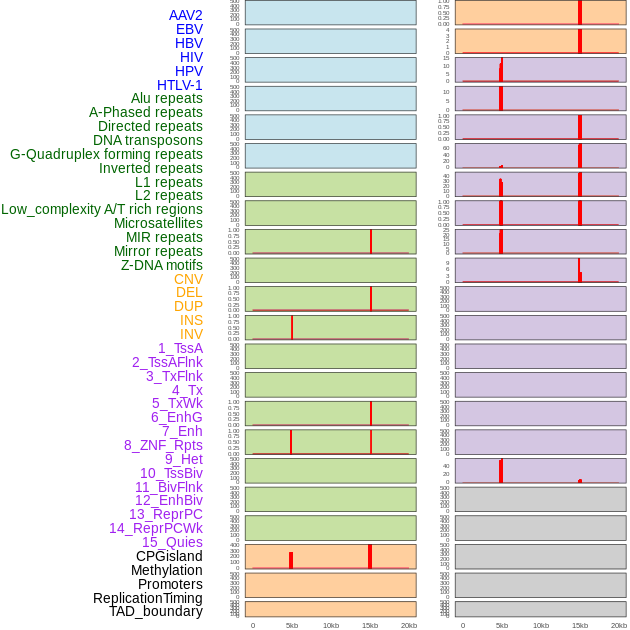 |
Top |
Fusion Protein Features for ERC1-TACC3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:1192746/chr4:1741429) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ERC1 | TACC3 |
| FUNCTION: Regulatory subunit of the IKK complex. Probably recruits IkappaBalpha/NFKBIA to the complex. May be involved in the organization of the cytomatrix at the nerve terminals active zone (CAZ) which regulates neurotransmitter release. May be involved in vesicle trafficking at the CAZ. May be involved in Rab-6 regulated endosomes to Golgi transport. {ECO:0000269|PubMed:15218148}. | FUNCTION: Plays a role in the microtubule-dependent coupling of the nucleus and the centrosome. Involved in the processes that regulate centrosome-mediated interkinetic nuclear migration (INM) of neural progenitors (By similarity). Acts as component of the TACC3/ch-TOG/clathrin complex proposed to contribute to stabilization of kinetochore fibers of the mitotic spindle by acting as inter-microtubule bridge. The TACC3/ch-TOG/clathrin complex is required for the maintenance of kinetochore fiber tension (PubMed:21297582, PubMed:23532825). May be involved in the control of cell growth and differentiation. May contribute to cancer (PubMed:14767476). {ECO:0000250|UniProtKB:Q9JJ11, ECO:0000269|PubMed:14767476, ECO:0000269|PubMed:21297582, ECO:0000269|PubMed:23532825}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000360905 | + | 3 | 19 | 1060_1100 | 362 | 1117.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000360905 | + | 3 | 19 | 144_988 | 362 | 1117.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000397203 | + | 3 | 19 | 1060_1100 | 362 | 1117.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000397203 | + | 3 | 19 | 144_988 | 362 | 1117.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000543086 | + | 3 | 18 | 1060_1100 | 362 | 1089.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000543086 | + | 3 | 18 | 144_988 | 362 | 1089.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000589028 | + | 2 | 18 | 1060_1100 | 362 | 1117.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000589028 | + | 2 | 18 | 144_988 | 362 | 1117.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000360905 | + | 3 | 19 | 1046_1108 | 362 | 1117.0 | Domain | FIP-RBD |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000397203 | + | 3 | 19 | 1046_1108 | 362 | 1117.0 | Domain | FIP-RBD |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000543086 | + | 3 | 18 | 1046_1108 | 362 | 1089.0 | Domain | FIP-RBD |
| Hgene | ERC1 | chr12:1192746 | chr4:1741429 | ENST00000589028 | + | 2 | 18 | 1046_1108 | 362 | 1117.0 | Domain | FIP-RBD |
| Tgene | TACC3 | chr12:1192746 | chr4:1741429 | ENST00000313288 | 9 | 16 | 637_837 | 647 | 839.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | TACC3 | chr12:1192746 | chr4:1741429 | ENST00000313288 | 9 | 16 | 155_160 | 647 | 839.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | TACC3 | chr12:1192746 | chr4:1741429 | ENST00000313288 | 9 | 16 | 522_577 | 647 | 839.0 | Region | Necessary but not sufficient for spindle localization | |
| Tgene | TACC3 | chr12:1192746 | chr4:1741429 | ENST00000313288 | 9 | 16 | 594_838 | 647 | 839.0 | Region | Necessary but not sufficient for spindle localization |
Top |
Fusion Gene Sequence for ERC1-TACC3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >27281_27281_1_ERC1-TACC3_ERC1_chr12_1192746_ENST00000355446_TACC3_chr4_1741429_ENST00000313288_length(transcript)=1952nt_BP=1218nt GATTGGACAGCTGGGGACCTTCTTCTGATTAACCTTAAACCAACTTGTAGCCATAGAGACACCTCACAAGGTTCCCATTTTTGTTGTTGT TGTTGTTGATTTTCTGCTCACACCTTTCCTGACCTTGCAACCATGTATGGAAGTGCCCGCTCTGTTGGGAAGGTGGAGCCGAGCAGCCAG AGCCCTGGGCGTTCACCCAGGCTTCCACGTTCCCCTCGCTTGGGTCACCGTCGAACCAACAGTACGGGAGGGAGTTCGGGAAGCAGTGTT GGAGGTGGCAGTGGGAAAACCCTTTCAATGGAAAATATACAATCTTTAAATGCTGCCTATGCCACCTCTGGCCCTATGTATCTAAGTGAC CATGAAAATGTGGGTTCAGAAACACCTAAAAGCACCATGACACTTGGCCGTTCTGGGGGACGTCTGCCTTACGGTGTTCGGATGACTGCT ATGGGTAGTAGCCCCAATATAGCTAGCAGTGGGGTTGCTAGTGACACCATAGCATTTGGAGAGCATCACCTCCCTCCTGTGAGTATGGCA TCCACTGTACCTCACTCCCTTCGTCAGGCGAGAGATAACACAATCATGGATCTGCAGACACAGCTGAAGGAAGTATTAAGAGAAAATGAT CTCTTGCGGAAGGATGTGGAAGTAAAGGAGAGCAAATTGAGTTCTTCAATGAATAGCATCAAGACCTTCTGGAGCCCAGAGCTGAAGAAG GAACGAGCCCTGAGAAAAGATGAAGCTTCCAAAATCACCATTTGGAAGGAACAGTACAGAGTTGTACAGGAGGAAAACCAGCACATGCAG ATGACAATCCAGGCTCTCCAGGATGAATTGCGGATCCAGAGGGACCTGAATCAGCTGTTTCAGCAGGATAGTAGCAGCAGGACTGGCGAA CCTTGTGTAGCAGAGCTGACAGAGGAGAACTTTCAGAGGCTTCATGCTGAGCATGAGCGGCAGGCCAAAGAGCTGTTTCTTCTTCGAAAG ACATTGGAGGAAATGGAGCTGCGTATTGAGACTCAAAAGCAGACCCTAAATGCTCGGGATGAATCCATTAAGAAGCTTCTGGAAATGTTG CAGAGCAAAGGACTTTCTGCCAAGGCTACCGAGGAAGACCATGAGAGAACAAGACGACTGGCAGAGGCAGAGATGCACGTTCATCACCTA GAAAGCCTTTTGGAGCAGAAGGAAAAAGAGAACAGTATGTTGAGAGAGGTAAAGGCGACACAGGAGGAGAACCGGGAGCTGAGGAGCAGG TGTGAGGAGCTCCACGGGAAGAACCTGGAACTGGGGAAGATCATGGACAGGTTCGAAGAGGTTGTGTACCAGGCCATGGAGGAAGTTCAG AAGCAGAAGGAACTTTCCAAAGCTGAAATCCAGAAAGTTCTAAAAGAAAAAGACCAACTTACCACAGATCTGAACTCCATGGAGAAGTCC TTCTCCGACCTCTTCAAGCGTTTTGAGAAACAGAAAGAGGTGATCGAGGGCTACCGCAAGAACGAAGAGTCACTGAAGAAGTGCGTGGAG GATTACCTGGCAAGGATCACCCAGGAGGGCCAGAGGTACCAAGCCCTGAAGGCCCACGCGGAGGAGAAGCTGCAGCTGGCAAACGAGGAG ATCGCCCAGGTCCGGAGCAAGGCCCAGGCGGAAGCGTTGGCCCTCCAGGCCAGCCTGAGGAAGGAGCAGATGCGCATCCAGTCGCTGGAG AAGACAGTGGAGCAGAAGACTAAAGAGAACGAGGAGCTGACCAGGATCTGCGACGACCTCATCTCCAAGATGGAGAAGATCTGACCTCCA CGGAGCCGCTGTCCCCGCCCCCCTGCTCCCGTCTGTCTGTCCTGTCTGATTCTCTTAGGTGTCATGTTCTTTTTTCTGTCTTGTCTTCAA CTTTTTTAAAAACTAGATTGCTTTGAAAACATGACTCAATAAAAGTTTCCTTTCAATTTAAA >27281_27281_1_ERC1-TACC3_ERC1_chr12_1192746_ENST00000355446_TACC3_chr4_1741429_ENST00000313288_length(amino acids)=553AA_BP=362 MYGSARSVGKVEPSSQSPGRSPRLPRSPRLGHRRTNSTGGSSGSSVGGGSGKTLSMENIQSLNAAYATSGPMYLSDHENVGSETPKSTMT LGRSGGRLPYGVRMTAMGSSPNIASSGVASDTIAFGEHHLPPVSMASTVPHSLRQARDNTIMDLQTQLKEVLRENDLLRKDVEVKESKLS SSMNSIKTFWSPELKKERALRKDEASKITIWKEQYRVVQEENQHMQMTIQALQDELRIQRDLNQLFQQDSSSRTGEPCVAELTEENFQRL HAEHERQAKELFLLRKTLEEMELRIETQKQTLNARDESIKKLLEMLQSKGLSAKATEEDHERTRRLAEAEMHVHHLESLLEQKEKENSML REVKATQEENRELRSRCEELHGKNLELGKIMDRFEEVVYQAMEEVQKQKELSKAEIQKVLKEKDQLTTDLNSMEKSFSDLFKRFEKQKEV IEGYRKNEESLKKCVEDYLARITQEGQRYQALKAHAEEKLQLANEEIAQVRSKAQAEALALQASLRKEQMRIQSLEKTVEQKTKENEELT RICDDLISKMEKI -------------------------------------------------------------- >27281_27281_2_ERC1-TACC3_ERC1_chr12_1192746_ENST00000360905_TACC3_chr4_1741429_ENST00000313288_length(transcript)=2001nt_BP=1267nt AGCAGTAGCGGCAGCCCTGAGGACGATATGGTGTAAGATACTTCTTCAAGATTGGACAGCTGGGGACCTTCTTCTGATTAACCTTAAACC AACTTGTAGCCATAGAGACACCTCACAAGGTTCCCATTTTTGTTGTTGTTGTTGTTGATTTTCTGCTCACACCTTTCCTGACCTTGCAAC CATGTATGGAAGTGCCCGCTCTGTTGGGAAGGTGGAGCCGAGCAGCCAGAGCCCTGGGCGTTCACCCAGGCTTCCACGTTCCCCTCGCTT GGGTCACCGTCGAACCAACAGTACGGGAGGGAGTTCGGGAAGCAGTGTTGGAGGTGGCAGTGGGAAAACCCTTTCAATGGAAAATATACA ATCTTTAAATGCTGCCTATGCCACCTCTGGCCCTATGTATCTAAGTGACCATGAAAATGTGGGTTCAGAAACACCTAAAAGCACCATGAC ACTTGGCCGTTCTGGGGGACGTCTGCCTTACGGTGTTCGGATGACTGCTATGGGTAGTAGCCCCAATATAGCTAGCAGTGGGGTTGCTAG TGACACCATAGCATTTGGAGAGCATCACCTCCCTCCTGTGAGTATGGCATCCACTGTACCTCACTCCCTTCGTCAGGCGAGAGATAACAC AATCATGGATCTGCAGACACAGCTGAAGGAAGTATTAAGAGAAAATGATCTCTTGCGGAAGGATGTGGAAGTAAAGGAGAGCAAATTGAG TTCTTCAATGAATAGCATCAAGACCTTCTGGAGCCCAGAGCTGAAGAAGGAACGAGCCCTGAGAAAAGATGAAGCTTCCAAAATCACCAT TTGGAAGGAACAGTACAGAGTTGTACAGGAGGAAAACCAGCACATGCAGATGACAATCCAGGCTCTCCAGGATGAATTGCGGATCCAGAG GGACCTGAATCAGCTGTTTCAGCAGGATAGTAGCAGCAGGACTGGCGAACCTTGTGTAGCAGAGCTGACAGAGGAGAACTTTCAGAGGCT TCATGCTGAGCATGAGCGGCAGGCCAAAGAGCTGTTTCTTCTTCGAAAGACATTGGAGGAAATGGAGCTGCGTATTGAGACTCAAAAGCA GACCCTAAATGCTCGGGATGAATCCATTAAGAAGCTTCTGGAAATGTTGCAGAGCAAAGGACTTTCTGCCAAGGCTACCGAGGAAGACCA TGAGAGAACAAGACGACTGGCAGAGGCAGAGATGCACGTTCATCACCTAGAAAGCCTTTTGGAGCAGAAGGAAAAAGAGAACAGTATGTT GAGAGAGGTAAAGGCGACACAGGAGGAGAACCGGGAGCTGAGGAGCAGGTGTGAGGAGCTCCACGGGAAGAACCTGGAACTGGGGAAGAT CATGGACAGGTTCGAAGAGGTTGTGTACCAGGCCATGGAGGAAGTTCAGAAGCAGAAGGAACTTTCCAAAGCTGAAATCCAGAAAGTTCT AAAAGAAAAAGACCAACTTACCACAGATCTGAACTCCATGGAGAAGTCCTTCTCCGACCTCTTCAAGCGTTTTGAGAAACAGAAAGAGGT GATCGAGGGCTACCGCAAGAACGAAGAGTCACTGAAGAAGTGCGTGGAGGATTACCTGGCAAGGATCACCCAGGAGGGCCAGAGGTACCA AGCCCTGAAGGCCCACGCGGAGGAGAAGCTGCAGCTGGCAAACGAGGAGATCGCCCAGGTCCGGAGCAAGGCCCAGGCGGAAGCGTTGGC CCTCCAGGCCAGCCTGAGGAAGGAGCAGATGCGCATCCAGTCGCTGGAGAAGACAGTGGAGCAGAAGACTAAAGAGAACGAGGAGCTGAC CAGGATCTGCGACGACCTCATCTCCAAGATGGAGAAGATCTGACCTCCACGGAGCCGCTGTCCCCGCCCCCCTGCTCCCGTCTGTCTGTC CTGTCTGATTCTCTTAGGTGTCATGTTCTTTTTTCTGTCTTGTCTTCAACTTTTTTAAAAACTAGATTGCTTTGAAAACATGACTCAATA AAAGTTTCCTTTCAATTTAAA >27281_27281_2_ERC1-TACC3_ERC1_chr12_1192746_ENST00000360905_TACC3_chr4_1741429_ENST00000313288_length(amino acids)=553AA_BP=362 MYGSARSVGKVEPSSQSPGRSPRLPRSPRLGHRRTNSTGGSSGSSVGGGSGKTLSMENIQSLNAAYATSGPMYLSDHENVGSETPKSTMT LGRSGGRLPYGVRMTAMGSSPNIASSGVASDTIAFGEHHLPPVSMASTVPHSLRQARDNTIMDLQTQLKEVLRENDLLRKDVEVKESKLS SSMNSIKTFWSPELKKERALRKDEASKITIWKEQYRVVQEENQHMQMTIQALQDELRIQRDLNQLFQQDSSSRTGEPCVAELTEENFQRL HAEHERQAKELFLLRKTLEEMELRIETQKQTLNARDESIKKLLEMLQSKGLSAKATEEDHERTRRLAEAEMHVHHLESLLEQKEKENSML REVKATQEENRELRSRCEELHGKNLELGKIMDRFEEVVYQAMEEVQKQKELSKAEIQKVLKEKDQLTTDLNSMEKSFSDLFKRFEKQKEV IEGYRKNEESLKKCVEDYLARITQEGQRYQALKAHAEEKLQLANEEIAQVRSKAQAEALALQASLRKEQMRIQSLEKTVEQKTKENEELT RICDDLISKMEKI -------------------------------------------------------------- >27281_27281_3_ERC1-TACC3_ERC1_chr12_1192746_ENST00000397203_TACC3_chr4_1741429_ENST00000313288_length(transcript)=2226nt_BP=1492nt GTGCTGTGGCGGCGGCGGCGGCGGTAGTGGCGGCGGCGGCGGTGCCTGGGCGGCAGCAGCAGCAGTAGCGGCAGCCCTGAGGACGGTGAG ACGGCCGCGGCGGCGGCGACAGCGCGGAAGTCGCGCGGGACCCAGAGCTTCCCCCCTGTCGCTAGTGCTGAGGCTTTCGCCTTTCCTACC CAGCTTCTTGCTCACGGCCCGGGCTGGGCTGGGCAGGGCAACGCGCCCCCGGGGAGGGGCGGAGCTGTAGATATGGTGTAAGATACTTCT TCAAGATTGGACAGCTGGGGACCTTCTTCTGATTAACCTTAAACCAACTTGTAGCCATAGAGACACCTCACAAGGTTCCCATTTTTGTTG TTGTTGTTGTTGATTTTCTGCTCACACCTTTCCTGACCTTGCAACCATGTATGGAAGTGCCCGCTCTGTTGGGAAGGTGGAGCCGAGCAG CCAGAGCCCTGGGCGTTCACCCAGGCTTCCACGTTCCCCTCGCTTGGGTCACCGTCGAACCAACAGTACGGGAGGGAGTTCGGGAAGCAG TGTTGGAGGTGGCAGTGGGAAAACCCTTTCAATGGAAAATATACAATCTTTAAATGCTGCCTATGCCACCTCTGGCCCTATGTATCTAAG TGACCATGAAAATGTGGGTTCAGAAACACCTAAAAGCACCATGACACTTGGCCGTTCTGGGGGACGTCTGCCTTACGGTGTTCGGATGAC TGCTATGGGTAGTAGCCCCAATATAGCTAGCAGTGGGGTTGCTAGTGACACCATAGCATTTGGAGAGCATCACCTCCCTCCTGTGAGTAT GGCATCCACTGTACCTCACTCCCTTCGTCAGGCGAGAGATAACACAATCATGGATCTGCAGACACAGCTGAAGGAAGTATTAAGAGAAAA TGATCTCTTGCGGAAGGATGTGGAAGTAAAGGAGAGCAAATTGAGTTCTTCAATGAATAGCATCAAGACCTTCTGGAGCCCAGAGCTGAA GAAGGAACGAGCCCTGAGAAAAGATGAAGCTTCCAAAATCACCATTTGGAAGGAACAGTACAGAGTTGTACAGGAGGAAAACCAGCACAT GCAGATGACAATCCAGGCTCTCCAGGATGAATTGCGGATCCAGAGGGACCTGAATCAGCTGTTTCAGCAGGATAGTAGCAGCAGGACTGG CGAACCTTGTGTAGCAGAGCTGACAGAGGAGAACTTTCAGAGGCTTCATGCTGAGCATGAGCGGCAGGCCAAAGAGCTGTTTCTTCTTCG AAAGACATTGGAGGAAATGGAGCTGCGTATTGAGACTCAAAAGCAGACCCTAAATGCTCGGGATGAATCCATTAAGAAGCTTCTGGAAAT GTTGCAGAGCAAAGGACTTTCTGCCAAGGCTACCGAGGAAGACCATGAGAGAACAAGACGACTGGCAGAGGCAGAGATGCACGTTCATCA CCTAGAAAGCCTTTTGGAGCAGAAGGAAAAAGAGAACAGTATGTTGAGAGAGGTAAAGGCGACACAGGAGGAGAACCGGGAGCTGAGGAG CAGGTGTGAGGAGCTCCACGGGAAGAACCTGGAACTGGGGAAGATCATGGACAGGTTCGAAGAGGTTGTGTACCAGGCCATGGAGGAAGT TCAGAAGCAGAAGGAACTTTCCAAAGCTGAAATCCAGAAAGTTCTAAAAGAAAAAGACCAACTTACCACAGATCTGAACTCCATGGAGAA GTCCTTCTCCGACCTCTTCAAGCGTTTTGAGAAACAGAAAGAGGTGATCGAGGGCTACCGCAAGAACGAAGAGTCACTGAAGAAGTGCGT GGAGGATTACCTGGCAAGGATCACCCAGGAGGGCCAGAGGTACCAAGCCCTGAAGGCCCACGCGGAGGAGAAGCTGCAGCTGGCAAACGA GGAGATCGCCCAGGTCCGGAGCAAGGCCCAGGCGGAAGCGTTGGCCCTCCAGGCCAGCCTGAGGAAGGAGCAGATGCGCATCCAGTCGCT GGAGAAGACAGTGGAGCAGAAGACTAAAGAGAACGAGGAGCTGACCAGGATCTGCGACGACCTCATCTCCAAGATGGAGAAGATCTGACC TCCACGGAGCCGCTGTCCCCGCCCCCCTGCTCCCGTCTGTCTGTCCTGTCTGATTCTCTTAGGTGTCATGTTCTTTTTTCTGTCTTGTCT TCAACTTTTTTAAAAACTAGATTGCTTTGAAAACATGACTCAATAAAAGTTTCCTTTCAATTTAAA >27281_27281_3_ERC1-TACC3_ERC1_chr12_1192746_ENST00000397203_TACC3_chr4_1741429_ENST00000313288_length(amino acids)=553AA_BP=362 MYGSARSVGKVEPSSQSPGRSPRLPRSPRLGHRRTNSTGGSSGSSVGGGSGKTLSMENIQSLNAAYATSGPMYLSDHENVGSETPKSTMT LGRSGGRLPYGVRMTAMGSSPNIASSGVASDTIAFGEHHLPPVSMASTVPHSLRQARDNTIMDLQTQLKEVLRENDLLRKDVEVKESKLS SSMNSIKTFWSPELKKERALRKDEASKITIWKEQYRVVQEENQHMQMTIQALQDELRIQRDLNQLFQQDSSSRTGEPCVAELTEENFQRL HAEHERQAKELFLLRKTLEEMELRIETQKQTLNARDESIKKLLEMLQSKGLSAKATEEDHERTRRLAEAEMHVHHLESLLEQKEKENSML REVKATQEENRELRSRCEELHGKNLELGKIMDRFEEVVYQAMEEVQKQKELSKAEIQKVLKEKDQLTTDLNSMEKSFSDLFKRFEKQKEV IEGYRKNEESLKKCVEDYLARITQEGQRYQALKAHAEEKLQLANEEIAQVRSKAQAEALALQASLRKEQMRIQSLEKTVEQKTKENEELT RICDDLISKMEKI -------------------------------------------------------------- >27281_27281_4_ERC1-TACC3_ERC1_chr12_1192746_ENST00000543086_TACC3_chr4_1741429_ENST00000313288_length(transcript)=2091nt_BP=1357nt GGGCGCGGTGACGTCGCGGGCGCCTGGGCCGTGCTGTGGCGGCGGCGGCGGCGGTAGTGGCGGCGGCGGCGGTGCCTGGGCGGCAGCAGC AGCAGTAGCGGCAGCCCTGAGGACGATATGGTGTAAGATACTTCTTCAAGATTGGACAGCTGGGGACCTTCTTCTGATTAACCTTAAACC AACTTGTAGCCATAGAGACACCTCACAAGGTTCCCATTTTTGTTGTTGTTGTTGTTGATTTTCTGCTCACACCTTTCCTGACCTTGCAAC CATGTATGGAAGTGCCCGCTCTGTTGGGAAGGTGGAGCCGAGCAGCCAGAGCCCTGGGCGTTCACCCAGGCTTCCACGTTCCCCTCGCTT GGGTCACCGTCGAACCAACAGTACGGGAGGGAGTTCGGGAAGCAGTGTTGGAGGTGGCAGTGGGAAAACCCTTTCAATGGAAAATATACA ATCTTTAAATGCTGCCTATGCCACCTCTGGCCCTATGTATCTAAGTGACCATGAAAATGTGGGTTCAGAAACACCTAAAAGCACCATGAC ACTTGGCCGTTCTGGGGGACGTCTGCCTTACGGTGTTCGGATGACTGCTATGGGTAGTAGCCCCAATATAGCTAGCAGTGGGGTTGCTAG TGACACCATAGCATTTGGAGAGCATCACCTCCCTCCTGTGAGTATGGCATCCACTGTACCTCACTCCCTTCGTCAGGCGAGAGATAACAC AATCATGGATCTGCAGACACAGCTGAAGGAAGTATTAAGAGAAAATGATCTCTTGCGGAAGGATGTGGAAGTAAAGGAGAGCAAATTGAG TTCTTCAATGAATAGCATCAAGACCTTCTGGAGCCCAGAGCTGAAGAAGGAACGAGCCCTGAGAAAAGATGAAGCTTCCAAAATCACCAT TTGGAAGGAACAGTACAGAGTTGTACAGGAGGAAAACCAGCACATGCAGATGACAATCCAGGCTCTCCAGGATGAATTGCGGATCCAGAG GGACCTGAATCAGCTGTTTCAGCAGGATAGTAGCAGCAGGACTGGCGAACCTTGTGTAGCAGAGCTGACAGAGGAGAACTTTCAGAGGCT TCATGCTGAGCATGAGCGGCAGGCCAAAGAGCTGTTTCTTCTTCGAAAGACATTGGAGGAAATGGAGCTGCGTATTGAGACTCAAAAGCA GACCCTAAATGCTCGGGATGAATCCATTAAGAAGCTTCTGGAAATGTTGCAGAGCAAAGGACTTTCTGCCAAGGCTACCGAGGAAGACCA TGAGAGAACAAGACGACTGGCAGAGGCAGAGATGCACGTTCATCACCTAGAAAGCCTTTTGGAGCAGAAGGAAAAAGAGAACAGTATGTT GAGAGAGGTAAAGGCGACACAGGAGGAGAACCGGGAGCTGAGGAGCAGGTGTGAGGAGCTCCACGGGAAGAACCTGGAACTGGGGAAGAT CATGGACAGGTTCGAAGAGGTTGTGTACCAGGCCATGGAGGAAGTTCAGAAGCAGAAGGAACTTTCCAAAGCTGAAATCCAGAAAGTTCT AAAAGAAAAAGACCAACTTACCACAGATCTGAACTCCATGGAGAAGTCCTTCTCCGACCTCTTCAAGCGTTTTGAGAAACAGAAAGAGGT GATCGAGGGCTACCGCAAGAACGAAGAGTCACTGAAGAAGTGCGTGGAGGATTACCTGGCAAGGATCACCCAGGAGGGCCAGAGGTACCA AGCCCTGAAGGCCCACGCGGAGGAGAAGCTGCAGCTGGCAAACGAGGAGATCGCCCAGGTCCGGAGCAAGGCCCAGGCGGAAGCGTTGGC CCTCCAGGCCAGCCTGAGGAAGGAGCAGATGCGCATCCAGTCGCTGGAGAAGACAGTGGAGCAGAAGACTAAAGAGAACGAGGAGCTGAC CAGGATCTGCGACGACCTCATCTCCAAGATGGAGAAGATCTGACCTCCACGGAGCCGCTGTCCCCGCCCCCCTGCTCCCGTCTGTCTGTC CTGTCTGATTCTCTTAGGTGTCATGTTCTTTTTTCTGTCTTGTCTTCAACTTTTTTAAAAACTAGATTGCTTTGAAAACATGACTCAATA AAAGTTTCCTTTCAATTTAAA >27281_27281_4_ERC1-TACC3_ERC1_chr12_1192746_ENST00000543086_TACC3_chr4_1741429_ENST00000313288_length(amino acids)=553AA_BP=362 MYGSARSVGKVEPSSQSPGRSPRLPRSPRLGHRRTNSTGGSSGSSVGGGSGKTLSMENIQSLNAAYATSGPMYLSDHENVGSETPKSTMT LGRSGGRLPYGVRMTAMGSSPNIASSGVASDTIAFGEHHLPPVSMASTVPHSLRQARDNTIMDLQTQLKEVLRENDLLRKDVEVKESKLS SSMNSIKTFWSPELKKERALRKDEASKITIWKEQYRVVQEENQHMQMTIQALQDELRIQRDLNQLFQQDSSSRTGEPCVAELTEENFQRL HAEHERQAKELFLLRKTLEEMELRIETQKQTLNARDESIKKLLEMLQSKGLSAKATEEDHERTRRLAEAEMHVHHLESLLEQKEKENSML REVKATQEENRELRSRCEELHGKNLELGKIMDRFEEVVYQAMEEVQKQKELSKAEIQKVLKEKDQLTTDLNSMEKSFSDLFKRFEKQKEV IEGYRKNEESLKKCVEDYLARITQEGQRYQALKAHAEEKLQLANEEIAQVRSKAQAEALALQASLRKEQMRIQSLEKTVEQKTKENEELT RICDDLISKMEKI -------------------------------------------------------------- >27281_27281_5_ERC1-TACC3_ERC1_chr12_1192746_ENST00000546231_TACC3_chr4_1741429_ENST00000313288_length(transcript)=2348nt_BP=1614nt GGGCCGTGCTGTGGCGGCGGCGGCGGCGGTAGTGGCGGCGGCGGCGGTGCCTGGGCGGCAGCAGCAGCAGTAGCGGCAGCCCTGAGGACG GTGAGACGGCCGCGGCGGCGGCGACAGCGCGGAAGTCGCGCGGGACCCAGAGCTTCCCCCCTGTCGCTAGTGCTGAGGCTTTCGCCTTTC CTACCCAGCTTCTTGCTCACGGCCCGGGCTGGGCTGGGCAGGGCAACGCGCCCCCGGGGAGGGGCGGAGCTGTAGGTTGGTCTTGAACTC CTGGCCTCAAGCGATCTTCCTACCTTGGCCTCCCAGAGCGCTGGGATTATAGGCATGAGCCATTGCACTTGGCACTTTTTTTGTTTGTTT TCCAGTTGCATGATATGGTGTAAGATACTTCTTCAAGATTGGACAGCTGGGGACCTTCTTCTGATTAACCTTAAACCAACTTGTAGCCAT AGAGACACCTCACAAGGTTCCCATTTTTGTTGTTGTTGTTGTTGATTTTCTGCTCACACCTTTCCTGACCTTGCAACCATGTATGGAAGT GCCCGCTCTGTTGGGAAGGTGGAGCCGAGCAGCCAGAGCCCTGGGCGTTCACCCAGGCTTCCACGTTCCCCTCGCTTGGGTCACCGTCGA ACCAACAGTACGGGAGGGAGTTCGGGAAGCAGTGTTGGAGGTGGCAGTGGGAAAACCCTTTCAATGGAAAATATACAATCTTTAAATGCT GCCTATGCCACCTCTGGCCCTATGTATCTAAGTGACCATGAAAATGTGGGTTCAGAAACACCTAAAAGCACCATGACACTTGGCCGTTCT GGGGGACGTCTGCCTTACGGTGTTCGGATGACTGCTATGGGTAGTAGCCCCAATATAGCTAGCAGTGGGGTTGCTAGTGACACCATAGCA TTTGGAGAGCATCACCTCCCTCCTGTGAGTATGGCATCCACTGTACCTCACTCCCTTCGTCAGGCGAGAGATAACACAATCATGGATCTG CAGACACAGCTGAAGGAAGTATTAAGAGAAAATGATCTCTTGCGGAAGGATGTGGAAGTAAAGGAGAGCAAATTGAGTTCTTCAATGAAT AGCATCAAGACCTTCTGGAGCCCAGAGCTGAAGAAGGAACGAGCCCTGAGAAAAGATGAAGCTTCCAAAATCACCATTTGGAAGGAACAG TACAGAGTTGTACAGGAGGAAAACCAGCACATGCAGATGACAATCCAGGCTCTCCAGGATGAATTGCGGATCCAGAGGGACCTGAATCAG CTGTTTCAGCAGGATAGTAGCAGCAGGACTGGCGAACCTTGTGTAGCAGAGCTGACAGAGGAGAACTTTCAGAGGCTTCATGCTGAGCAT GAGCGGCAGGCCAAAGAGCTGTTTCTTCTTCGAAAGACATTGGAGGAAATGGAGCTGCGTATTGAGACTCAAAAGCAGACCCTAAATGCT CGGGATGAATCCATTAAGAAGCTTCTGGAAATGTTGCAGAGCAAAGGACTTTCTGCCAAGGCTACCGAGGAAGACCATGAGAGAACAAGA CGACTGGCAGAGGCAGAGATGCACGTTCATCACCTAGAAAGCCTTTTGGAGCAGAAGGAAAAAGAGAACAGTATGTTGAGAGAGGTAAAG GCGACACAGGAGGAGAACCGGGAGCTGAGGAGCAGGTGTGAGGAGCTCCACGGGAAGAACCTGGAACTGGGGAAGATCATGGACAGGTTC GAAGAGGTTGTGTACCAGGCCATGGAGGAAGTTCAGAAGCAGAAGGAACTTTCCAAAGCTGAAATCCAGAAAGTTCTAAAAGAAAAAGAC CAACTTACCACAGATCTGAACTCCATGGAGAAGTCCTTCTCCGACCTCTTCAAGCGTTTTGAGAAACAGAAAGAGGTGATCGAGGGCTAC CGCAAGAACGAAGAGTCACTGAAGAAGTGCGTGGAGGATTACCTGGCAAGGATCACCCAGGAGGGCCAGAGGTACCAAGCCCTGAAGGCC CACGCGGAGGAGAAGCTGCAGCTGGCAAACGAGGAGATCGCCCAGGTCCGGAGCAAGGCCCAGGCGGAAGCGTTGGCCCTCCAGGCCAGC CTGAGGAAGGAGCAGATGCGCATCCAGTCGCTGGAGAAGACAGTGGAGCAGAAGACTAAAGAGAACGAGGAGCTGACCAGGATCTGCGAC GACCTCATCTCCAAGATGGAGAAGATCTGACCTCCACGGAGCCGCTGTCCCCGCCCCCCTGCTCCCGTCTGTCTGTCCTGTCTGATTCTC TTAGGTGTCATGTTCTTTTTTCTGTCTTGTCTTCAACTTTTTTAAAAACTAGATTGCTTTGAAAACATGACTCAATAAAAGTTTCCTTTC AATTTAAA >27281_27281_5_ERC1-TACC3_ERC1_chr12_1192746_ENST00000546231_TACC3_chr4_1741429_ENST00000313288_length(amino acids)=553AA_BP=362 MYGSARSVGKVEPSSQSPGRSPRLPRSPRLGHRRTNSTGGSSGSSVGGGSGKTLSMENIQSLNAAYATSGPMYLSDHENVGSETPKSTMT LGRSGGRLPYGVRMTAMGSSPNIASSGVASDTIAFGEHHLPPVSMASTVPHSLRQARDNTIMDLQTQLKEVLRENDLLRKDVEVKESKLS SSMNSIKTFWSPELKKERALRKDEASKITIWKEQYRVVQEENQHMQMTIQALQDELRIQRDLNQLFQQDSSSRTGEPCVAELTEENFQRL HAEHERQAKELFLLRKTLEEMELRIETQKQTLNARDESIKKLLEMLQSKGLSAKATEEDHERTRRLAEAEMHVHHLESLLEQKEKENSML REVKATQEENRELRSRCEELHGKNLELGKIMDRFEEVVYQAMEEVQKQKELSKAEIQKVLKEKDQLTTDLNSMEKSFSDLFKRFEKQKEV IEGYRKNEESLKKCVEDYLARITQEGQRYQALKAHAEEKLQLANEEIAQVRSKAQAEALALQASLRKEQMRIQSLEKTVEQKTKENEELT RICDDLISKMEKI -------------------------------------------------------------- >27281_27281_6_ERC1-TACC3_ERC1_chr12_1192746_ENST00000589028_TACC3_chr4_1741429_ENST00000313288_length(transcript)=1977nt_BP=1243nt GATATGGTGTAAGATACTTCTTCAAGATTGGACAGCTGGGGACCTTCTTCTGATTAACCTTAAACCAACTTGTAGCCATAGAGACACCTC ACAAGGTTCCCATTTTTGTTGTTGTTGTTGTTGATTTTCTGCTCACACCTTTCCTGACCTTGCAACCATGTATGGAAGTGCCCGCTCTGT TGGGAAGGTGGAGCCGAGCAGCCAGAGCCCTGGGCGTTCACCCAGGCTTCCACGTTCCCCTCGCTTGGGTCACCGTCGAACCAACAGTAC GGGAGGGAGTTCGGGAAGCAGTGTTGGAGGTGGCAGTGGGAAAACCCTTTCAATGGAAAATATACAATCTTTAAATGCTGCCTATGCCAC CTCTGGCCCTATGTATCTAAGTGACCATGAAAATGTGGGTTCAGAAACACCTAAAAGCACCATGACACTTGGCCGTTCTGGGGGACGTCT GCCTTACGGTGTTCGGATGACTGCTATGGGTAGTAGCCCCAATATAGCTAGCAGTGGGGTTGCTAGTGACACCATAGCATTTGGAGAGCA TCACCTCCCTCCTGTGAGTATGGCATCCACTGTACCTCACTCCCTTCGTCAGGCGAGAGATAACACAATCATGGATCTGCAGACACAGCT GAAGGAAGTATTAAGAGAAAATGATCTCTTGCGGAAGGATGTGGAAGTAAAGGAGAGCAAATTGAGTTCTTCAATGAATAGCATCAAGAC CTTCTGGAGCCCAGAGCTGAAGAAGGAACGAGCCCTGAGAAAAGATGAAGCTTCCAAAATCACCATTTGGAAGGAACAGTACAGAGTTGT ACAGGAGGAAAACCAGCACATGCAGATGACAATCCAGGCTCTCCAGGATGAATTGCGGATCCAGAGGGACCTGAATCAGCTGTTTCAGCA GGATAGTAGCAGCAGGACTGGCGAACCTTGTGTAGCAGAGCTGACAGAGGAGAACTTTCAGAGGCTTCATGCTGAGCATGAGCGGCAGGC CAAAGAGCTGTTTCTTCTTCGAAAGACATTGGAGGAAATGGAGCTGCGTATTGAGACTCAAAAGCAGACCCTAAATGCTCGGGATGAATC CATTAAGAAGCTTCTGGAAATGTTGCAGAGCAAAGGACTTTCTGCCAAGGCTACCGAGGAAGACCATGAGAGAACAAGACGACTGGCAGA GGCAGAGATGCACGTTCATCACCTAGAAAGCCTTTTGGAGCAGAAGGAAAAAGAGAACAGTATGTTGAGAGAGGTAAAGGCGACACAGGA GGAGAACCGGGAGCTGAGGAGCAGGTGTGAGGAGCTCCACGGGAAGAACCTGGAACTGGGGAAGATCATGGACAGGTTCGAAGAGGTTGT GTACCAGGCCATGGAGGAAGTTCAGAAGCAGAAGGAACTTTCCAAAGCTGAAATCCAGAAAGTTCTAAAAGAAAAAGACCAACTTACCAC AGATCTGAACTCCATGGAGAAGTCCTTCTCCGACCTCTTCAAGCGTTTTGAGAAACAGAAAGAGGTGATCGAGGGCTACCGCAAGAACGA AGAGTCACTGAAGAAGTGCGTGGAGGATTACCTGGCAAGGATCACCCAGGAGGGCCAGAGGTACCAAGCCCTGAAGGCCCACGCGGAGGA GAAGCTGCAGCTGGCAAACGAGGAGATCGCCCAGGTCCGGAGCAAGGCCCAGGCGGAAGCGTTGGCCCTCCAGGCCAGCCTGAGGAAGGA GCAGATGCGCATCCAGTCGCTGGAGAAGACAGTGGAGCAGAAGACTAAAGAGAACGAGGAGCTGACCAGGATCTGCGACGACCTCATCTC CAAGATGGAGAAGATCTGACCTCCACGGAGCCGCTGTCCCCGCCCCCCTGCTCCCGTCTGTCTGTCCTGTCTGATTCTCTTAGGTGTCAT GTTCTTTTTTCTGTCTTGTCTTCAACTTTTTTAAAAACTAGATTGCTTTGAAAACATGACTCAATAAAAGTTTCCTTTCAATTTAAA >27281_27281_6_ERC1-TACC3_ERC1_chr12_1192746_ENST00000589028_TACC3_chr4_1741429_ENST00000313288_length(amino acids)=553AA_BP=362 MYGSARSVGKVEPSSQSPGRSPRLPRSPRLGHRRTNSTGGSSGSSVGGGSGKTLSMENIQSLNAAYATSGPMYLSDHENVGSETPKSTMT LGRSGGRLPYGVRMTAMGSSPNIASSGVASDTIAFGEHHLPPVSMASTVPHSLRQARDNTIMDLQTQLKEVLRENDLLRKDVEVKESKLS SSMNSIKTFWSPELKKERALRKDEASKITIWKEQYRVVQEENQHMQMTIQALQDELRIQRDLNQLFQQDSSSRTGEPCVAELTEENFQRL HAEHERQAKELFLLRKTLEEMELRIETQKQTLNARDESIKKLLEMLQSKGLSAKATEEDHERTRRLAEAEMHVHHLESLLEQKEKENSML REVKATQEENRELRSRCEELHGKNLELGKIMDRFEEVVYQAMEEVQKQKELSKAEIQKVLKEKDQLTTDLNSMEKSFSDLFKRFEKQKEV IEGYRKNEESLKKCVEDYLARITQEGQRYQALKAHAEEKLQLANEEIAQVRSKAQAEALALQASLRKEQMRIQSLEKTVEQKTKENEELT RICDDLISKMEKI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ERC1-TACC3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ERC1-TACC3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ERC1-TACC3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ERC1 | C0238463 | Papillary thyroid carcinoma | 1 | ORPHANET |
| Hgene | ERC1 | C4749581 | Distal monosomy 12p | 1 | ORPHANET |
| Tgene | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human | |
| Tgene | C0005695 | Bladder Neoplasm | 1 | CTD_human | |
| Tgene | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human | |
| Tgene | C0206726 | gliosarcoma | 1 | ORPHANET | |
| Tgene | C0334588 | Giant Cell Glioblastoma | 1 | ORPHANET | |
| Tgene | C2239176 | Liver carcinoma | 1 | CTD_human |
