
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:NEDD4L-MBD2 (FusionGDB2 ID:HG23327TG8932) |
Fusion Gene Summary for NEDD4L-MBD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: NEDD4L-MBD2 | Fusion gene ID: hg23327tg8932 | Hgene | Tgene | Gene symbol | NEDD4L | MBD2 | Gene ID | 23327 | 8932 |
| Gene name | NEDD4 like E3 ubiquitin protein ligase | methyl-CpG binding domain protein 2 | |
| Synonyms | NEDD4-2|NEDD4.2|PVNH7|RSP5|hNEDD4-2 | DMTase|NY-CO-41 | |
| Cytomap | ('NEDD4L')('MBD2') 18q21.31 | 18q21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase NEDD4-likeHECT-type E3 ubiquitin transferase NED4Lneural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligaseubiquitin-protein ligase Rsp5 | methyl-CpG-binding domain protein 2demethylase | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | Q9UBB5 | |
| Ensembl transtripts involved in fusion gene | ENST00000256830, ENST00000356462, ENST00000382850, ENST00000400345, ENST00000589054, ENST00000256832, ENST00000357895, ENST00000431212, ENST00000435432, ENST00000456173, ENST00000456986, ENST00000586263, ENST00000588516, | ||
| Fusion gene scores | * DoF score | 28 X 18 X 11=5544 | 13 X 7 X 7=637 |
| # samples | 35 | 15 | |
| ** MAII score | log2(35/5544*10)=-3.98550043030488 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/637*10)=-2.08633087176042 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: NEDD4L [Title/Abstract] AND MBD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | NEDD4L(55711940)-MBD2(51715381), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | NEDD4L-MBD2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NEDD4L-MBD2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NEDD4L-MBD2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NEDD4L-MBD2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NEDD4L | GO:0003254 | regulation of membrane depolarization | 15217910 |
| Hgene | NEDD4L | GO:0006511 | ubiquitin-dependent protein catabolic process | 21463633 |
| Hgene | NEDD4L | GO:0010038 | response to metal ion | 11244092 |
| Hgene | NEDD4L | GO:0016567 | protein ubiquitination | 15217910|25631046 |
| Hgene | NEDD4L | GO:0034765 | regulation of ion transmembrane transport | 17289006 |
| Hgene | NEDD4L | GO:0042391 | regulation of membrane potential | 17289006 |
| Hgene | NEDD4L | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 21463633 |
| Hgene | NEDD4L | GO:0060306 | regulation of membrane repolarization | 21463633 |
| Hgene | NEDD4L | GO:0070936 | protein K48-linked ubiquitination | 21463633 |
| Hgene | NEDD4L | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 17289006 |
| Hgene | NEDD4L | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity | 21463633 |
| Hgene | NEDD4L | GO:1901380 | negative regulation of potassium ion transmembrane transport | 21463633 |
| Hgene | NEDD4L | GO:1902306 | negative regulation of sodium ion transmembrane transport | 15217910 |
| Hgene | NEDD4L | GO:1903861 | positive regulation of dendrite extension | 23999003 |
| Hgene | NEDD4L | GO:2000009 | negative regulation of protein localization to cell surface | 21463633 |
| Hgene | NEDD4L | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity | 15217910 |
 Fusion gene breakpoints across NEDD4L (5'-gene) Fusion gene breakpoints across NEDD4L (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
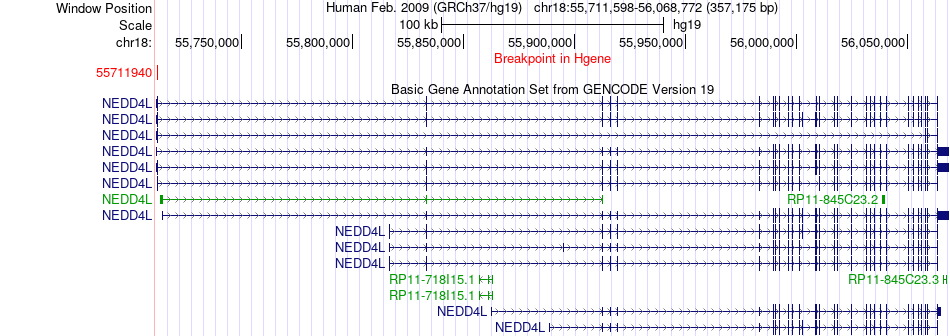 |
 Fusion gene breakpoints across MBD2 (3'-gene) Fusion gene breakpoints across MBD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
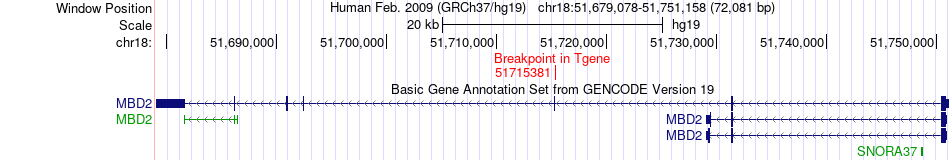 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-ET-A40P | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
Top |
Fusion Gene ORF analysis for NEDD4L-MBD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000256830 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000256830 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000256830 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000356462 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000356462 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000356462 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000382850 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000382850 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000382850 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000400345 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000400345 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000400345 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000589054 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000589054 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5CDS-intron | ENST00000589054 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5UTR-3CDS | ENST00000256832 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5UTR-intron | ENST00000256832 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5UTR-intron | ENST00000256832 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| 5UTR-intron | ENST00000256832 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| In-frame | ENST00000256830 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| In-frame | ENST00000356462 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| In-frame | ENST00000382850 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| In-frame | ENST00000400345 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| In-frame | ENST00000589054 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000357895 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000431212 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000435432 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000456173 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000456986 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000586263 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-3CDS | ENST00000588516 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000357895 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000357895 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000357895 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000431212 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000431212 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000431212 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000435432 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000435432 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000435432 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000456173 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000456173 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000456173 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000456986 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000456986 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000456986 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000586263 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000586263 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000586263 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000588516 | ENST00000398398 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000588516 | ENST00000579025 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
| intron-intron | ENST00000588516 | ENST00000583046 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000356462 | NEDD4L | chr18 | 55711940 | + | ENST00000256429 | MBD2 | chr18 | 51715381 | - | 3491 | 342 | 210 | 875 | 221 |
| ENST00000400345 | NEDD4L | chr18 | 55711940 | + | ENST00000256429 | MBD2 | chr18 | 51715381 | - | 3480 | 331 | 199 | 864 | 221 |
| ENST00000589054 | NEDD4L | chr18 | 55711940 | + | ENST00000256429 | MBD2 | chr18 | 51715381 | - | 3474 | 325 | 193 | 858 | 221 |
| ENST00000382850 | NEDD4L | chr18 | 55711940 | + | ENST00000256429 | MBD2 | chr18 | 51715381 | - | 3310 | 161 | 29 | 694 | 221 |
| ENST00000256830 | NEDD4L | chr18 | 55711940 | + | ENST00000256429 | MBD2 | chr18 | 51715381 | - | 3197 | 48 | 0 | 581 | 193 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000356462 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - | 0.001273654 | 0.9987263 |
| ENST00000400345 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - | 0.001276249 | 0.9987237 |
| ENST00000589054 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - | 0.001255378 | 0.9987446 |
| ENST00000382850 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - | 0.001116108 | 0.9988839 |
| ENST00000256830 | ENST00000256429 | NEDD4L | chr18 | 55711940 | + | MBD2 | chr18 | 51715381 | - | 0.001012064 | 0.998988 |
Top |
Fusion Genomic Features for NEDD4L-MBD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
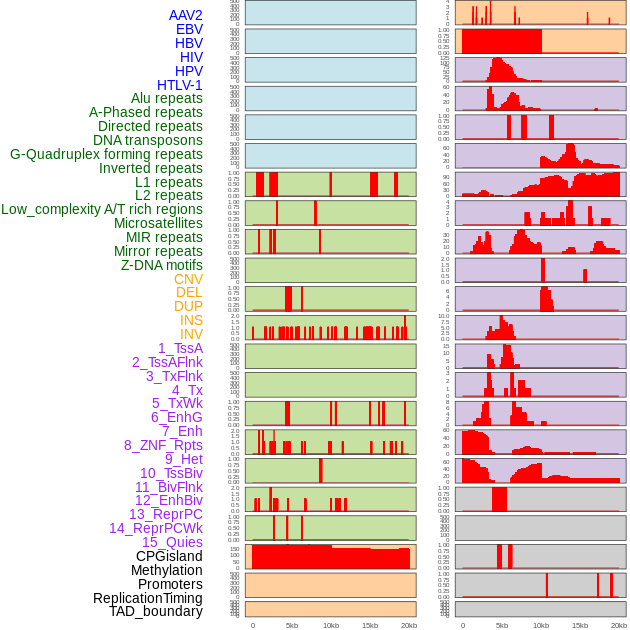 |
Top |
Fusion Protein Features for NEDD4L-MBD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:55711940/chr18:51715381) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MBD2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Binds CpG islands in promoters where the DNA is methylated at position 5 of cytosine within CpG dinucleotides. Binds hemimethylated DNA as well. Recruits histone deacetylases and DNA methyltransferases. Acts as transcriptional repressor and plays a role in gene silencing. Functions as a scaffold protein, targeting GATAD2A and GATAD2B to chromatin to promote repression. May enhance the activation of some unmethylated cAMP-responsive promoters. {ECO:0000269|PubMed:10471499, ECO:0000269|PubMed:10947852, ECO:0000269|PubMed:12665568, ECO:0000269|PubMed:16415179, ECO:0000269|PubMed:24307175, ECO:0000269|PubMed:9774669}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MBD2 | chr18:55711940 | chr18:51715381 | ENST00000583046 | 0 | 3 | 50_96 | 0 | 303.0 | Compositional bias | Note=Arg-rich | |
| Tgene | MBD2 | chr18:55711940 | chr18:51715381 | ENST00000583046 | 0 | 3 | 6_140 | 0 | 303.0 | Compositional bias | Note=Gly-rich | |
| Tgene | MBD2 | chr18:55711940 | chr18:51715381 | ENST00000583046 | 0 | 3 | 145_213 | 0 | 303.0 | Domain | MBD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000256830 | + | 1 | 28 | 193_226 | 16 | 872.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000256830 | + | 1 | 28 | 385_418 | 16 | 872.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000256830 | + | 1 | 28 | 497_530 | 16 | 872.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000256830 | + | 1 | 28 | 4_126 | 16 | 872.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000256830 | + | 1 | 28 | 548_581 | 16 | 872.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000256830 | + | 1 | 28 | 640_974 | 16 | 872.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000356462 | + | 1 | 29 | 193_226 | 16 | 912.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000356462 | + | 1 | 29 | 385_418 | 16 | 912.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000356462 | + | 1 | 29 | 497_530 | 16 | 912.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000356462 | + | 1 | 29 | 4_126 | 16 | 912.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000356462 | + | 1 | 29 | 548_581 | 16 | 912.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000356462 | + | 1 | 29 | 640_974 | 16 | 912.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000357895 | + | 1 | 31 | 193_226 | 0 | 968.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000357895 | + | 1 | 31 | 385_418 | 0 | 968.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000357895 | + | 1 | 31 | 497_530 | 0 | 968.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000357895 | + | 1 | 31 | 4_126 | 0 | 968.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000357895 | + | 1 | 31 | 548_581 | 0 | 968.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000357895 | + | 1 | 31 | 640_974 | 0 | 968.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000382850 | + | 1 | 30 | 193_226 | 16 | 956.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000382850 | + | 1 | 30 | 385_418 | 16 | 956.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000382850 | + | 1 | 30 | 497_530 | 16 | 956.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000382850 | + | 1 | 30 | 4_126 | 16 | 956.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000382850 | + | 1 | 30 | 548_581 | 16 | 956.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000382850 | + | 1 | 30 | 640_974 | 16 | 956.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000400345 | + | 1 | 31 | 193_226 | 16 | 976.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000400345 | + | 1 | 31 | 385_418 | 16 | 976.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000400345 | + | 1 | 31 | 497_530 | 16 | 976.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000400345 | + | 1 | 31 | 4_126 | 16 | 976.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000400345 | + | 1 | 31 | 548_581 | 16 | 976.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000400345 | + | 1 | 31 | 640_974 | 16 | 976.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000431212 | + | 1 | 30 | 193_226 | 0 | 855.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000431212 | + | 1 | 30 | 385_418 | 0 | 855.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000431212 | + | 1 | 30 | 497_530 | 0 | 855.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000431212 | + | 1 | 30 | 4_126 | 0 | 855.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000431212 | + | 1 | 30 | 548_581 | 0 | 855.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000431212 | + | 1 | 30 | 640_974 | 0 | 855.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000435432 | + | 1 | 31 | 193_226 | 0 | 835.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000435432 | + | 1 | 31 | 385_418 | 0 | 835.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000435432 | + | 1 | 31 | 497_530 | 0 | 835.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000435432 | + | 1 | 31 | 4_126 | 0 | 835.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000435432 | + | 1 | 31 | 548_581 | 0 | 835.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000435432 | + | 1 | 31 | 640_974 | 0 | 835.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456173 | + | 1 | 29 | 193_226 | 0 | 835.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456173 | + | 1 | 29 | 385_418 | 0 | 835.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456173 | + | 1 | 29 | 497_530 | 0 | 835.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456173 | + | 1 | 29 | 4_126 | 0 | 835.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456173 | + | 1 | 29 | 548_581 | 0 | 835.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456173 | + | 1 | 29 | 640_974 | 0 | 835.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456986 | + | 1 | 31 | 193_226 | 0 | 855.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456986 | + | 1 | 31 | 385_418 | 0 | 855.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456986 | + | 1 | 31 | 497_530 | 0 | 855.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456986 | + | 1 | 31 | 4_126 | 0 | 855.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456986 | + | 1 | 31 | 548_581 | 0 | 855.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000456986 | + | 1 | 31 | 640_974 | 0 | 855.0 | Domain | HECT |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000586263 | + | 1 | 30 | 193_226 | 0 | 948.0 | Domain | WW 1 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000586263 | + | 1 | 30 | 385_418 | 0 | 948.0 | Domain | WW 2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000586263 | + | 1 | 30 | 497_530 | 0 | 948.0 | Domain | WW 3 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000586263 | + | 1 | 30 | 4_126 | 0 | 948.0 | Domain | C2 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000586263 | + | 1 | 30 | 548_581 | 0 | 948.0 | Domain | WW 4 |
| Hgene | NEDD4L | chr18:55711940 | chr18:51715381 | ENST00000586263 | + | 1 | 30 | 640_974 | 0 | 948.0 | Domain | HECT |
| Tgene | MBD2 | chr18:55711940 | chr18:51715381 | ENST00000256429 | 1 | 7 | 50_96 | 234 | 1015.6666666666666 | Compositional bias | Note=Arg-rich | |
| Tgene | MBD2 | chr18:55711940 | chr18:51715381 | ENST00000256429 | 1 | 7 | 6_140 | 234 | 1015.6666666666666 | Compositional bias | Note=Gly-rich | |
| Tgene | MBD2 | chr18:55711940 | chr18:51715381 | ENST00000256429 | 1 | 7 | 145_213 | 234 | 1015.6666666666666 | Domain | MBD |
Top |
Fusion Gene Sequence for NEDD4L-MBD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >58422_58422_1_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000256830_MBD2_chr18_51715381_ENST00000256429_length(transcript)=3197nt_BP=48nt ATGGCGACCGGGCTCGGGGAGCCGGTCTATGGACTTTCCGAAGACGAGGGTAAACCAGACTTGAATACAACATTGCCAATTAGACAAACA GCATCAATTTTCAAACAACCGGTAACCAAAGTCACAAATCATCCTAGTAATAAAGTGAAATCAGACCCACAACGAATGAATGAACAGCCA CGTCAGCTTTTCTGGGAGAAGAGGCTACAAGGACTTAGTGCATCAGATGTAACAGAACAAATTATAAAAACCATGGAACTACCCAAAGGT CTTCAAGGAGTTGGTCCAGGTAGCAATGATGAGACCCTTTTATCTGCTGTTGCCAGTGCTTTGCACACAAGCTCTGCGCCAATCACAGGG CAAGTCTCCGCTGCTGTGGAAAAGAACCCTGCTGTTTGGCTTAACACATCTCAACCCCTCTGCAAAGCTTTTATTGTCACAGATGAAGAC ATCAGGAAACAGGAAGAGCGAGTACAGCAAGTACGCAAGAAATTGGAAGAAGCACTGATGGCAGACATCTTGTCGCGAGCTGCTGATACA GAAGAGATGGATATTGAAATGGACAGTGGAGATGAAGCCTAAGAATATGATCAGGTAACTTTCGACCGACTTTCCCCAAGAGAAAATTCC TAGAAATTGAACAAAAATGTTTCCACTGGCTTTTGCCTGTAAGAAAAAAAATGTACCCGAGCACATAGAGCTTTTTAATAGCACTAACCA ATGCCTTTTTAGATGTATTTTTGATGTATATATCTATTATTCAAAAAATCATGTTTATTTTGAGTCCTAGGACTTAAAATTAGTCTTTTG TAATATCAAGCAGGACCCTAAGATGAAGCTGAGCTTTTGATGCCAGGTGCAATCTACTGGAAATGTAGCACTTACGTAAAACATTTGTTT CCCCCACAGTTTTAATAAGAACAGATCAGGAATTCTAAATAAATTTCCCAGTTAAAGATTATTGTGACTTCACTGTATATAAACATATTT TTATACTTTATTGAAAGGGGACACCTGTACATTCTTCCATCATCACTGTAAAGACAAATAAATGATTATATTCACAGACTGATTGGAATT CTTTCTGTTGAAAAGCACACACAATAAAGAACCCCTCGTTAGCCTTCCTCTGATTTACATTCAACTCTGATCCCTGGGCCTTAGGTTTGA CATGGAGGTGGAGGAAGATAGCGCATATATTTGCAGTATGAACTATTGCCTCTGGACGTTGTGAGAATTGTGCTTTCACCAGAATTTCTA AGAATTTCTGCTAAATATCACCTAGCATGTGTAATTTTTTTTCCTTGCCTGTGACTTGGACTTTTGATAGTTCTATAAGAATAAGGCTTT TTCTTCCCTTGGGCATGAGTCAGATACACAAGGACCCTTCAGGTGTTACTAGAAGGCGTCCATGTTTATTGTTTTTTAAAGAATGTTTGG CACTCTCTAACGTCCACTAGCTTACTGAGTTATCAGGTGCAGGTCAGACTCTTGGCTACAGTGAGAGGCAGCTTCTAGACAGAGTTGCTT AATGAAAGGGTTTGTAATACTTTACAAACCATTACCTGTACCTGGCCTGGCCTCCAAAATATTAACATTCTTTTTCTGTTGAAACTCGCG AGTGTAACTTTCATACCACTTGAATTTATTGATATTTAATTATGAAAACTAGCATTACATTATTAAACGATTTCTAAAATCAAAACATAC TTAATCTGATACCAAGGAAGGGAGGGAGTGGTTATAAGCAAATGAAAACAAATTTTGAGAGACAGAGCAAAAGTAAAATCATTCTATAGA AAAGGTGTGTTTATTTCTTAACTCTTGAGTTCTTTTAAAATTAGAACCTAAATGATGCAGTCAGGGGTATGACCAATTCCACATGAGTGT CACCTGTACATTTTATTACCACTACCTCCAGTTTCCAAGGCAGGGAAGAAGAGGGGAATAGTCAAAGCAATATGACTAGCTAGGTTTGGG CTGCTGTTTGGGCTGCTGTTTTTCCTGGACTTTTATGCCAAACTACATAAATACTTCTTGGAATTCCTGATTTACGCTCAACTTTGATCC CTGGACCCTAGGGCCATTAGTTTCCTTGAAGCAGTCCTGCTGGTGGATGAGCAGGGAAACTTGTAGACCCTGGGAAGCCAATTGGTAGAG GGTGCCTGCCCGCCCCATCAGACTGTCCCAACTGGGGTGGGGGTGAGGATCATAGTGATTTCTTTTTAAAAGCTAGTATGACTAGTGAGA ATGGATGTCCAAGGGCTTTTCTCTTCCTCCCCCGAGTCCACGATTCTTCATTTGTGATCAGGGTTGGGGTAACTTCACCTTGGTGATCAT ATATTCTTTTTACAAACCAATCCAGCCAAGATATATGTGCTTTTCTAAACAGCTTGTAAAAGCAAAAACACAATGTATACACATATAAAA CTGATTTTTTTTTTTTTGCTTATATACTTTGCTCAGGTGGAGAACAAGGTATGAAAGCCATTATATGGTGTCCCTTGGGGACCATCCCAT AAGTCCAGGGTGTTCCCAATATACCCACAAGACAACATGTTCAAAACTTCATCACCTGGTCCCCACAAACCTACCACCCTCTTCCATTCC CTCACTCTGGTAAAAGCAAGCCATCTTCTCCAGTTACGCAAGGCAGAAGCCCAGGAGTGAGCACAATGCTGCCCTTTCCCTTGCTTCTGA CTTCTCATCAATCCTATTAGAACTACTGACTGCACCCCACCTCTCCTCCATCCCCACGATCACTGCCTTCATATGAACTGCCTTTCCTGT CCTCTGGACTTTCAGTGGCTTCCTTTTTTGCTCACCTGTCCACTTTCTTCCCCTTCCCTCTCTCCATCCCTCCACATAGTCATCAGAAAG ATCATCAAAACAGGCAAATGTGACTGTATTATTTCCCTCACTTATCTTTGTATGCAGTTTAGAGTGATTTTTAAGAGCACACACTTTGAA ATTAGGAGAACCTGTGTCTGAATCCTGATTTTTGCCATTTGTGTGACTTGTCAGGTTATTTAATCTCTTTGAGCCTGTCCCTCCTCAGTG AAATAGGAATAATAATACCTACCTCATAGAGTTGTTGTGAGGATTCAGTGAGATAGACTGTTTCTAAAGTGTGTAGCACAGTGCCTGGCA CATAGTGGGTGCTCACATTGGAGCACACTATTGTCGCTATCCCTGCA >58422_58422_1_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000256830_MBD2_chr18_51715381_ENST00000256429_length(amino acids)=193AA_BP=16 MATGLGEPVYGLSEDEGKPDLNTTLPIRQTASIFKQPVTKVTNHPSNKVKSDPQRMNEQPRQLFWEKRLQGLSASDVTEQIIKTMELPKG LQGVGPGSNDETLLSAVASALHTSSAPITGQVSAAVEKNPAVWLNTSQPLCKAFIVTDEDIRKQEERVQQVRKKLEEALMADILSRAADT EEMDIEMDSGDEA -------------------------------------------------------------- >58422_58422_2_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000356462_MBD2_chr18_51715381_ENST00000256429_length(transcript)=3491nt_BP=342nt GCGCCGCCGCCGCGCGCAGTGAGAGGCGCCGGGGCTGCCGCCCGGTGCTCGGCGCGCTCTCGGGAGCCGCCCGCCCGCTGGTCCCGCAGC CTTCCGGGAGGAAGCGGTGCCGGCAGCGTCCAGGGCGCGCTCTCGGGCACCCTCACCTGCCGGCGCCCGGCCGCTTACCCGGCAGGGCGT GCGCAGGGTAGGGTGCGGGACCGGGGGGACCTGGAGGCAGAGGGGAGAACCGGCCGTCCGCGCCGCAGCACAGCCGCTGGGAGCGCCTCA GACCCCGCGCGGGGCGCCGGCTCCATGGCGACCGGGCTCGGGGAGCCGGTCTATGGACTTTCCGAAGACGAGGGTAAACCAGACTTGAAT ACAACATTGCCAATTAGACAAACAGCATCAATTTTCAAACAACCGGTAACCAAAGTCACAAATCATCCTAGTAATAAAGTGAAATCAGAC CCACAACGAATGAATGAACAGCCACGTCAGCTTTTCTGGGAGAAGAGGCTACAAGGACTTAGTGCATCAGATGTAACAGAACAAATTATA AAAACCATGGAACTACCCAAAGGTCTTCAAGGAGTTGGTCCAGGTAGCAATGATGAGACCCTTTTATCTGCTGTTGCCAGTGCTTTGCAC ACAAGCTCTGCGCCAATCACAGGGCAAGTCTCCGCTGCTGTGGAAAAGAACCCTGCTGTTTGGCTTAACACATCTCAACCCCTCTGCAAA GCTTTTATTGTCACAGATGAAGACATCAGGAAACAGGAAGAGCGAGTACAGCAAGTACGCAAGAAATTGGAAGAAGCACTGATGGCAGAC ATCTTGTCGCGAGCTGCTGATACAGAAGAGATGGATATTGAAATGGACAGTGGAGATGAAGCCTAAGAATATGATCAGGTAACTTTCGAC CGACTTTCCCCAAGAGAAAATTCCTAGAAATTGAACAAAAATGTTTCCACTGGCTTTTGCCTGTAAGAAAAAAAATGTACCCGAGCACAT AGAGCTTTTTAATAGCACTAACCAATGCCTTTTTAGATGTATTTTTGATGTATATATCTATTATTCAAAAAATCATGTTTATTTTGAGTC CTAGGACTTAAAATTAGTCTTTTGTAATATCAAGCAGGACCCTAAGATGAAGCTGAGCTTTTGATGCCAGGTGCAATCTACTGGAAATGT AGCACTTACGTAAAACATTTGTTTCCCCCACAGTTTTAATAAGAACAGATCAGGAATTCTAAATAAATTTCCCAGTTAAAGATTATTGTG ACTTCACTGTATATAAACATATTTTTATACTTTATTGAAAGGGGACACCTGTACATTCTTCCATCATCACTGTAAAGACAAATAAATGAT TATATTCACAGACTGATTGGAATTCTTTCTGTTGAAAAGCACACACAATAAAGAACCCCTCGTTAGCCTTCCTCTGATTTACATTCAACT CTGATCCCTGGGCCTTAGGTTTGACATGGAGGTGGAGGAAGATAGCGCATATATTTGCAGTATGAACTATTGCCTCTGGACGTTGTGAGA ATTGTGCTTTCACCAGAATTTCTAAGAATTTCTGCTAAATATCACCTAGCATGTGTAATTTTTTTTCCTTGCCTGTGACTTGGACTTTTG ATAGTTCTATAAGAATAAGGCTTTTTCTTCCCTTGGGCATGAGTCAGATACACAAGGACCCTTCAGGTGTTACTAGAAGGCGTCCATGTT TATTGTTTTTTAAAGAATGTTTGGCACTCTCTAACGTCCACTAGCTTACTGAGTTATCAGGTGCAGGTCAGACTCTTGGCTACAGTGAGA GGCAGCTTCTAGACAGAGTTGCTTAATGAAAGGGTTTGTAATACTTTACAAACCATTACCTGTACCTGGCCTGGCCTCCAAAATATTAAC ATTCTTTTTCTGTTGAAACTCGCGAGTGTAACTTTCATACCACTTGAATTTATTGATATTTAATTATGAAAACTAGCATTACATTATTAA ACGATTTCTAAAATCAAAACATACTTAATCTGATACCAAGGAAGGGAGGGAGTGGTTATAAGCAAATGAAAACAAATTTTGAGAGACAGA GCAAAAGTAAAATCATTCTATAGAAAAGGTGTGTTTATTTCTTAACTCTTGAGTTCTTTTAAAATTAGAACCTAAATGATGCAGTCAGGG GTATGACCAATTCCACATGAGTGTCACCTGTACATTTTATTACCACTACCTCCAGTTTCCAAGGCAGGGAAGAAGAGGGGAATAGTCAAA GCAATATGACTAGCTAGGTTTGGGCTGCTGTTTGGGCTGCTGTTTTTCCTGGACTTTTATGCCAAACTACATAAATACTTCTTGGAATTC CTGATTTACGCTCAACTTTGATCCCTGGACCCTAGGGCCATTAGTTTCCTTGAAGCAGTCCTGCTGGTGGATGAGCAGGGAAACTTGTAG ACCCTGGGAAGCCAATTGGTAGAGGGTGCCTGCCCGCCCCATCAGACTGTCCCAACTGGGGTGGGGGTGAGGATCATAGTGATTTCTTTT TAAAAGCTAGTATGACTAGTGAGAATGGATGTCCAAGGGCTTTTCTCTTCCTCCCCCGAGTCCACGATTCTTCATTTGTGATCAGGGTTG GGGTAACTTCACCTTGGTGATCATATATTCTTTTTACAAACCAATCCAGCCAAGATATATGTGCTTTTCTAAACAGCTTGTAAAAGCAAA AACACAATGTATACACATATAAAACTGATTTTTTTTTTTTTGCTTATATACTTTGCTCAGGTGGAGAACAAGGTATGAAAGCCATTATAT GGTGTCCCTTGGGGACCATCCCATAAGTCCAGGGTGTTCCCAATATACCCACAAGACAACATGTTCAAAACTTCATCACCTGGTCCCCAC AAACCTACCACCCTCTTCCATTCCCTCACTCTGGTAAAAGCAAGCCATCTTCTCCAGTTACGCAAGGCAGAAGCCCAGGAGTGAGCACAA TGCTGCCCTTTCCCTTGCTTCTGACTTCTCATCAATCCTATTAGAACTACTGACTGCACCCCACCTCTCCTCCATCCCCACGATCACTGC CTTCATATGAACTGCCTTTCCTGTCCTCTGGACTTTCAGTGGCTTCCTTTTTTGCTCACCTGTCCACTTTCTTCCCCTTCCCTCTCTCCA TCCCTCCACATAGTCATCAGAAAGATCATCAAAACAGGCAAATGTGACTGTATTATTTCCCTCACTTATCTTTGTATGCAGTTTAGAGTG ATTTTTAAGAGCACACACTTTGAAATTAGGAGAACCTGTGTCTGAATCCTGATTTTTGCCATTTGTGTGACTTGTCAGGTTATTTAATCT CTTTGAGCCTGTCCCTCCTCAGTGAAATAGGAATAATAATACCTACCTCATAGAGTTGTTGTGAGGATTCAGTGAGATAGACTGTTTCTA AAGTGTGTAGCACAGTGCCTGGCACATAGTGGGTGCTCACATTGGAGCACACTATTGTCGCTATCCCTGCA >58422_58422_2_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000356462_MBD2_chr18_51715381_ENST00000256429_length(amino acids)=221AA_BP=44 MEAEGRTGRPRRSTAAGSASDPARGAGSMATGLGEPVYGLSEDEGKPDLNTTLPIRQTASIFKQPVTKVTNHPSNKVKSDPQRMNEQPRQ LFWEKRLQGLSASDVTEQIIKTMELPKGLQGVGPGSNDETLLSAVASALHTSSAPITGQVSAAVEKNPAVWLNTSQPLCKAFIVTDEDIR KQEERVQQVRKKLEEALMADILSRAADTEEMDIEMDSGDEA -------------------------------------------------------------- >58422_58422_3_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000382850_MBD2_chr18_51715381_ENST00000256429_length(transcript)=3310nt_BP=161nt CGCAGGGTAGGGTGCGGGACCGGGGGGACCTGGAGGCAGAGGGGAGAACCGGCCGTCCGCGCCGCAGCACAGCCGCTGGGAGCGCCTCAG ACCCCGCGCGGGGCGCCGGCTCCATGGCGACCGGGCTCGGGGAGCCGGTCTATGGACTTTCCGAAGACGAGGGTAAACCAGACTTGAATA CAACATTGCCAATTAGACAAACAGCATCAATTTTCAAACAACCGGTAACCAAAGTCACAAATCATCCTAGTAATAAAGTGAAATCAGACC CACAACGAATGAATGAACAGCCACGTCAGCTTTTCTGGGAGAAGAGGCTACAAGGACTTAGTGCATCAGATGTAACAGAACAAATTATAA AAACCATGGAACTACCCAAAGGTCTTCAAGGAGTTGGTCCAGGTAGCAATGATGAGACCCTTTTATCTGCTGTTGCCAGTGCTTTGCACA CAAGCTCTGCGCCAATCACAGGGCAAGTCTCCGCTGCTGTGGAAAAGAACCCTGCTGTTTGGCTTAACACATCTCAACCCCTCTGCAAAG CTTTTATTGTCACAGATGAAGACATCAGGAAACAGGAAGAGCGAGTACAGCAAGTACGCAAGAAATTGGAAGAAGCACTGATGGCAGACA TCTTGTCGCGAGCTGCTGATACAGAAGAGATGGATATTGAAATGGACAGTGGAGATGAAGCCTAAGAATATGATCAGGTAACTTTCGACC GACTTTCCCCAAGAGAAAATTCCTAGAAATTGAACAAAAATGTTTCCACTGGCTTTTGCCTGTAAGAAAAAAAATGTACCCGAGCACATA GAGCTTTTTAATAGCACTAACCAATGCCTTTTTAGATGTATTTTTGATGTATATATCTATTATTCAAAAAATCATGTTTATTTTGAGTCC TAGGACTTAAAATTAGTCTTTTGTAATATCAAGCAGGACCCTAAGATGAAGCTGAGCTTTTGATGCCAGGTGCAATCTACTGGAAATGTA GCACTTACGTAAAACATTTGTTTCCCCCACAGTTTTAATAAGAACAGATCAGGAATTCTAAATAAATTTCCCAGTTAAAGATTATTGTGA CTTCACTGTATATAAACATATTTTTATACTTTATTGAAAGGGGACACCTGTACATTCTTCCATCATCACTGTAAAGACAAATAAATGATT ATATTCACAGACTGATTGGAATTCTTTCTGTTGAAAAGCACACACAATAAAGAACCCCTCGTTAGCCTTCCTCTGATTTACATTCAACTC TGATCCCTGGGCCTTAGGTTTGACATGGAGGTGGAGGAAGATAGCGCATATATTTGCAGTATGAACTATTGCCTCTGGACGTTGTGAGAA TTGTGCTTTCACCAGAATTTCTAAGAATTTCTGCTAAATATCACCTAGCATGTGTAATTTTTTTTCCTTGCCTGTGACTTGGACTTTTGA TAGTTCTATAAGAATAAGGCTTTTTCTTCCCTTGGGCATGAGTCAGATACACAAGGACCCTTCAGGTGTTACTAGAAGGCGTCCATGTTT ATTGTTTTTTAAAGAATGTTTGGCACTCTCTAACGTCCACTAGCTTACTGAGTTATCAGGTGCAGGTCAGACTCTTGGCTACAGTGAGAG GCAGCTTCTAGACAGAGTTGCTTAATGAAAGGGTTTGTAATACTTTACAAACCATTACCTGTACCTGGCCTGGCCTCCAAAATATTAACA TTCTTTTTCTGTTGAAACTCGCGAGTGTAACTTTCATACCACTTGAATTTATTGATATTTAATTATGAAAACTAGCATTACATTATTAAA CGATTTCTAAAATCAAAACATACTTAATCTGATACCAAGGAAGGGAGGGAGTGGTTATAAGCAAATGAAAACAAATTTTGAGAGACAGAG CAAAAGTAAAATCATTCTATAGAAAAGGTGTGTTTATTTCTTAACTCTTGAGTTCTTTTAAAATTAGAACCTAAATGATGCAGTCAGGGG TATGACCAATTCCACATGAGTGTCACCTGTACATTTTATTACCACTACCTCCAGTTTCCAAGGCAGGGAAGAAGAGGGGAATAGTCAAAG CAATATGACTAGCTAGGTTTGGGCTGCTGTTTGGGCTGCTGTTTTTCCTGGACTTTTATGCCAAACTACATAAATACTTCTTGGAATTCC TGATTTACGCTCAACTTTGATCCCTGGACCCTAGGGCCATTAGTTTCCTTGAAGCAGTCCTGCTGGTGGATGAGCAGGGAAACTTGTAGA CCCTGGGAAGCCAATTGGTAGAGGGTGCCTGCCCGCCCCATCAGACTGTCCCAACTGGGGTGGGGGTGAGGATCATAGTGATTTCTTTTT AAAAGCTAGTATGACTAGTGAGAATGGATGTCCAAGGGCTTTTCTCTTCCTCCCCCGAGTCCACGATTCTTCATTTGTGATCAGGGTTGG GGTAACTTCACCTTGGTGATCATATATTCTTTTTACAAACCAATCCAGCCAAGATATATGTGCTTTTCTAAACAGCTTGTAAAAGCAAAA ACACAATGTATACACATATAAAACTGATTTTTTTTTTTTTGCTTATATACTTTGCTCAGGTGGAGAACAAGGTATGAAAGCCATTATATG GTGTCCCTTGGGGACCATCCCATAAGTCCAGGGTGTTCCCAATATACCCACAAGACAACATGTTCAAAACTTCATCACCTGGTCCCCACA AACCTACCACCCTCTTCCATTCCCTCACTCTGGTAAAAGCAAGCCATCTTCTCCAGTTACGCAAGGCAGAAGCCCAGGAGTGAGCACAAT GCTGCCCTTTCCCTTGCTTCTGACTTCTCATCAATCCTATTAGAACTACTGACTGCACCCCACCTCTCCTCCATCCCCACGATCACTGCC TTCATATGAACTGCCTTTCCTGTCCTCTGGACTTTCAGTGGCTTCCTTTTTTGCTCACCTGTCCACTTTCTTCCCCTTCCCTCTCTCCAT CCCTCCACATAGTCATCAGAAAGATCATCAAAACAGGCAAATGTGACTGTATTATTTCCCTCACTTATCTTTGTATGCAGTTTAGAGTGA TTTTTAAGAGCACACACTTTGAAATTAGGAGAACCTGTGTCTGAATCCTGATTTTTGCCATTTGTGTGACTTGTCAGGTTATTTAATCTC TTTGAGCCTGTCCCTCCTCAGTGAAATAGGAATAATAATACCTACCTCATAGAGTTGTTGTGAGGATTCAGTGAGATAGACTGTTTCTAA AGTGTGTAGCACAGTGCCTGGCACATAGTGGGTGCTCACATTGGAGCACACTATTGTCGCTATCCCTGCA >58422_58422_3_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000382850_MBD2_chr18_51715381_ENST00000256429_length(amino acids)=221AA_BP=44 MEAEGRTGRPRRSTAAGSASDPARGAGSMATGLGEPVYGLSEDEGKPDLNTTLPIRQTASIFKQPVTKVTNHPSNKVKSDPQRMNEQPRQ LFWEKRLQGLSASDVTEQIIKTMELPKGLQGVGPGSNDETLLSAVASALHTSSAPITGQVSAAVEKNPAVWLNTSQPLCKAFIVTDEDIR KQEERVQQVRKKLEEALMADILSRAADTEEMDIEMDSGDEA -------------------------------------------------------------- >58422_58422_4_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000400345_MBD2_chr18_51715381_ENST00000256429_length(transcript)=3480nt_BP=331nt GCGCGCAGTGAGAGGCGCCGGGGCTGCCGCCCGGTGCTCGGCGCGCTCTCGGGAGCCGCCCGCCCGCTGGTCCCGCAGCCTTCCGGGAGG AAGCGGTGCCGGCAGCGTCCAGGGCGCGCTCTCGGGCACCCTCACCTGCCGGCGCCCGGCCGCTTACCCGGCAGGGCGTGCGCAGGGTAG GGTGCGGGACCGGGGGGACCTGGAGGCAGAGGGGAGAACCGGCCGTCCGCGCCGCAGCACAGCCGCTGGGAGCGCCTCAGACCCCGCGCG GGGCGCCGGCTCCATGGCGACCGGGCTCGGGGAGCCGGTCTATGGACTTTCCGAAGACGAGGGTAAACCAGACTTGAATACAACATTGCC AATTAGACAAACAGCATCAATTTTCAAACAACCGGTAACCAAAGTCACAAATCATCCTAGTAATAAAGTGAAATCAGACCCACAACGAAT GAATGAACAGCCACGTCAGCTTTTCTGGGAGAAGAGGCTACAAGGACTTAGTGCATCAGATGTAACAGAACAAATTATAAAAACCATGGA ACTACCCAAAGGTCTTCAAGGAGTTGGTCCAGGTAGCAATGATGAGACCCTTTTATCTGCTGTTGCCAGTGCTTTGCACACAAGCTCTGC GCCAATCACAGGGCAAGTCTCCGCTGCTGTGGAAAAGAACCCTGCTGTTTGGCTTAACACATCTCAACCCCTCTGCAAAGCTTTTATTGT CACAGATGAAGACATCAGGAAACAGGAAGAGCGAGTACAGCAAGTACGCAAGAAATTGGAAGAAGCACTGATGGCAGACATCTTGTCGCG AGCTGCTGATACAGAAGAGATGGATATTGAAATGGACAGTGGAGATGAAGCCTAAGAATATGATCAGGTAACTTTCGACCGACTTTCCCC AAGAGAAAATTCCTAGAAATTGAACAAAAATGTTTCCACTGGCTTTTGCCTGTAAGAAAAAAAATGTACCCGAGCACATAGAGCTTTTTA ATAGCACTAACCAATGCCTTTTTAGATGTATTTTTGATGTATATATCTATTATTCAAAAAATCATGTTTATTTTGAGTCCTAGGACTTAA AATTAGTCTTTTGTAATATCAAGCAGGACCCTAAGATGAAGCTGAGCTTTTGATGCCAGGTGCAATCTACTGGAAATGTAGCACTTACGT AAAACATTTGTTTCCCCCACAGTTTTAATAAGAACAGATCAGGAATTCTAAATAAATTTCCCAGTTAAAGATTATTGTGACTTCACTGTA TATAAACATATTTTTATACTTTATTGAAAGGGGACACCTGTACATTCTTCCATCATCACTGTAAAGACAAATAAATGATTATATTCACAG ACTGATTGGAATTCTTTCTGTTGAAAAGCACACACAATAAAGAACCCCTCGTTAGCCTTCCTCTGATTTACATTCAACTCTGATCCCTGG GCCTTAGGTTTGACATGGAGGTGGAGGAAGATAGCGCATATATTTGCAGTATGAACTATTGCCTCTGGACGTTGTGAGAATTGTGCTTTC ACCAGAATTTCTAAGAATTTCTGCTAAATATCACCTAGCATGTGTAATTTTTTTTCCTTGCCTGTGACTTGGACTTTTGATAGTTCTATA AGAATAAGGCTTTTTCTTCCCTTGGGCATGAGTCAGATACACAAGGACCCTTCAGGTGTTACTAGAAGGCGTCCATGTTTATTGTTTTTT AAAGAATGTTTGGCACTCTCTAACGTCCACTAGCTTACTGAGTTATCAGGTGCAGGTCAGACTCTTGGCTACAGTGAGAGGCAGCTTCTA GACAGAGTTGCTTAATGAAAGGGTTTGTAATACTTTACAAACCATTACCTGTACCTGGCCTGGCCTCCAAAATATTAACATTCTTTTTCT GTTGAAACTCGCGAGTGTAACTTTCATACCACTTGAATTTATTGATATTTAATTATGAAAACTAGCATTACATTATTAAACGATTTCTAA AATCAAAACATACTTAATCTGATACCAAGGAAGGGAGGGAGTGGTTATAAGCAAATGAAAACAAATTTTGAGAGACAGAGCAAAAGTAAA ATCATTCTATAGAAAAGGTGTGTTTATTTCTTAACTCTTGAGTTCTTTTAAAATTAGAACCTAAATGATGCAGTCAGGGGTATGACCAAT TCCACATGAGTGTCACCTGTACATTTTATTACCACTACCTCCAGTTTCCAAGGCAGGGAAGAAGAGGGGAATAGTCAAAGCAATATGACT AGCTAGGTTTGGGCTGCTGTTTGGGCTGCTGTTTTTCCTGGACTTTTATGCCAAACTACATAAATACTTCTTGGAATTCCTGATTTACGC TCAACTTTGATCCCTGGACCCTAGGGCCATTAGTTTCCTTGAAGCAGTCCTGCTGGTGGATGAGCAGGGAAACTTGTAGACCCTGGGAAG CCAATTGGTAGAGGGTGCCTGCCCGCCCCATCAGACTGTCCCAACTGGGGTGGGGGTGAGGATCATAGTGATTTCTTTTTAAAAGCTAGT ATGACTAGTGAGAATGGATGTCCAAGGGCTTTTCTCTTCCTCCCCCGAGTCCACGATTCTTCATTTGTGATCAGGGTTGGGGTAACTTCA CCTTGGTGATCATATATTCTTTTTACAAACCAATCCAGCCAAGATATATGTGCTTTTCTAAACAGCTTGTAAAAGCAAAAACACAATGTA TACACATATAAAACTGATTTTTTTTTTTTTGCTTATATACTTTGCTCAGGTGGAGAACAAGGTATGAAAGCCATTATATGGTGTCCCTTG GGGACCATCCCATAAGTCCAGGGTGTTCCCAATATACCCACAAGACAACATGTTCAAAACTTCATCACCTGGTCCCCACAAACCTACCAC CCTCTTCCATTCCCTCACTCTGGTAAAAGCAAGCCATCTTCTCCAGTTACGCAAGGCAGAAGCCCAGGAGTGAGCACAATGCTGCCCTTT CCCTTGCTTCTGACTTCTCATCAATCCTATTAGAACTACTGACTGCACCCCACCTCTCCTCCATCCCCACGATCACTGCCTTCATATGAA CTGCCTTTCCTGTCCTCTGGACTTTCAGTGGCTTCCTTTTTTGCTCACCTGTCCACTTTCTTCCCCTTCCCTCTCTCCATCCCTCCACAT AGTCATCAGAAAGATCATCAAAACAGGCAAATGTGACTGTATTATTTCCCTCACTTATCTTTGTATGCAGTTTAGAGTGATTTTTAAGAG CACACACTTTGAAATTAGGAGAACCTGTGTCTGAATCCTGATTTTTGCCATTTGTGTGACTTGTCAGGTTATTTAATCTCTTTGAGCCTG TCCCTCCTCAGTGAAATAGGAATAATAATACCTACCTCATAGAGTTGTTGTGAGGATTCAGTGAGATAGACTGTTTCTAAAGTGTGTAGC ACAGTGCCTGGCACATAGTGGGTGCTCACATTGGAGCACACTATTGTCGCTATCCCTGCA >58422_58422_4_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000400345_MBD2_chr18_51715381_ENST00000256429_length(amino acids)=221AA_BP=44 MEAEGRTGRPRRSTAAGSASDPARGAGSMATGLGEPVYGLSEDEGKPDLNTTLPIRQTASIFKQPVTKVTNHPSNKVKSDPQRMNEQPRQ LFWEKRLQGLSASDVTEQIIKTMELPKGLQGVGPGSNDETLLSAVASALHTSSAPITGQVSAAVEKNPAVWLNTSQPLCKAFIVTDEDIR KQEERVQQVRKKLEEALMADILSRAADTEEMDIEMDSGDEA -------------------------------------------------------------- >58422_58422_5_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000589054_MBD2_chr18_51715381_ENST00000256429_length(transcript)=3474nt_BP=325nt AGTGAGAGGCGCCGGGGCTGCCGCCCGGTGCTCGGCGCGCTCTCGGGAGCCGCCCGCCCGCTGGTCCCGCAGCCTTCCGGGAGGAAGCGG TGCCGGCAGCGTCCAGGGCGCGCTCTCGGGCACCCTCACCTGCCGGCGCCCGGCCGCTTACCCGGCAGGGCGTGCGCAGGGTAGGGTGCG GGACCGGGGGGACCTGGAGGCAGAGGGGAGAACCGGCCGTCCGCGCCGCAGCACAGCCGCTGGGAGCGCCTCAGACCCCGCGCGGGGCGC CGGCTCCATGGCGACCGGGCTCGGGGAGCCGGTCTATGGACTTTCCGAAGACGAGGGTAAACCAGACTTGAATACAACATTGCCAATTAG ACAAACAGCATCAATTTTCAAACAACCGGTAACCAAAGTCACAAATCATCCTAGTAATAAAGTGAAATCAGACCCACAACGAATGAATGA ACAGCCACGTCAGCTTTTCTGGGAGAAGAGGCTACAAGGACTTAGTGCATCAGATGTAACAGAACAAATTATAAAAACCATGGAACTACC CAAAGGTCTTCAAGGAGTTGGTCCAGGTAGCAATGATGAGACCCTTTTATCTGCTGTTGCCAGTGCTTTGCACACAAGCTCTGCGCCAAT CACAGGGCAAGTCTCCGCTGCTGTGGAAAAGAACCCTGCTGTTTGGCTTAACACATCTCAACCCCTCTGCAAAGCTTTTATTGTCACAGA TGAAGACATCAGGAAACAGGAAGAGCGAGTACAGCAAGTACGCAAGAAATTGGAAGAAGCACTGATGGCAGACATCTTGTCGCGAGCTGC TGATACAGAAGAGATGGATATTGAAATGGACAGTGGAGATGAAGCCTAAGAATATGATCAGGTAACTTTCGACCGACTTTCCCCAAGAGA AAATTCCTAGAAATTGAACAAAAATGTTTCCACTGGCTTTTGCCTGTAAGAAAAAAAATGTACCCGAGCACATAGAGCTTTTTAATAGCA CTAACCAATGCCTTTTTAGATGTATTTTTGATGTATATATCTATTATTCAAAAAATCATGTTTATTTTGAGTCCTAGGACTTAAAATTAG TCTTTTGTAATATCAAGCAGGACCCTAAGATGAAGCTGAGCTTTTGATGCCAGGTGCAATCTACTGGAAATGTAGCACTTACGTAAAACA TTTGTTTCCCCCACAGTTTTAATAAGAACAGATCAGGAATTCTAAATAAATTTCCCAGTTAAAGATTATTGTGACTTCACTGTATATAAA CATATTTTTATACTTTATTGAAAGGGGACACCTGTACATTCTTCCATCATCACTGTAAAGACAAATAAATGATTATATTCACAGACTGAT TGGAATTCTTTCTGTTGAAAAGCACACACAATAAAGAACCCCTCGTTAGCCTTCCTCTGATTTACATTCAACTCTGATCCCTGGGCCTTA GGTTTGACATGGAGGTGGAGGAAGATAGCGCATATATTTGCAGTATGAACTATTGCCTCTGGACGTTGTGAGAATTGTGCTTTCACCAGA ATTTCTAAGAATTTCTGCTAAATATCACCTAGCATGTGTAATTTTTTTTCCTTGCCTGTGACTTGGACTTTTGATAGTTCTATAAGAATA AGGCTTTTTCTTCCCTTGGGCATGAGTCAGATACACAAGGACCCTTCAGGTGTTACTAGAAGGCGTCCATGTTTATTGTTTTTTAAAGAA TGTTTGGCACTCTCTAACGTCCACTAGCTTACTGAGTTATCAGGTGCAGGTCAGACTCTTGGCTACAGTGAGAGGCAGCTTCTAGACAGA GTTGCTTAATGAAAGGGTTTGTAATACTTTACAAACCATTACCTGTACCTGGCCTGGCCTCCAAAATATTAACATTCTTTTTCTGTTGAA ACTCGCGAGTGTAACTTTCATACCACTTGAATTTATTGATATTTAATTATGAAAACTAGCATTACATTATTAAACGATTTCTAAAATCAA AACATACTTAATCTGATACCAAGGAAGGGAGGGAGTGGTTATAAGCAAATGAAAACAAATTTTGAGAGACAGAGCAAAAGTAAAATCATT CTATAGAAAAGGTGTGTTTATTTCTTAACTCTTGAGTTCTTTTAAAATTAGAACCTAAATGATGCAGTCAGGGGTATGACCAATTCCACA TGAGTGTCACCTGTACATTTTATTACCACTACCTCCAGTTTCCAAGGCAGGGAAGAAGAGGGGAATAGTCAAAGCAATATGACTAGCTAG GTTTGGGCTGCTGTTTGGGCTGCTGTTTTTCCTGGACTTTTATGCCAAACTACATAAATACTTCTTGGAATTCCTGATTTACGCTCAACT TTGATCCCTGGACCCTAGGGCCATTAGTTTCCTTGAAGCAGTCCTGCTGGTGGATGAGCAGGGAAACTTGTAGACCCTGGGAAGCCAATT GGTAGAGGGTGCCTGCCCGCCCCATCAGACTGTCCCAACTGGGGTGGGGGTGAGGATCATAGTGATTTCTTTTTAAAAGCTAGTATGACT AGTGAGAATGGATGTCCAAGGGCTTTTCTCTTCCTCCCCCGAGTCCACGATTCTTCATTTGTGATCAGGGTTGGGGTAACTTCACCTTGG TGATCATATATTCTTTTTACAAACCAATCCAGCCAAGATATATGTGCTTTTCTAAACAGCTTGTAAAAGCAAAAACACAATGTATACACA TATAAAACTGATTTTTTTTTTTTTGCTTATATACTTTGCTCAGGTGGAGAACAAGGTATGAAAGCCATTATATGGTGTCCCTTGGGGACC ATCCCATAAGTCCAGGGTGTTCCCAATATACCCACAAGACAACATGTTCAAAACTTCATCACCTGGTCCCCACAAACCTACCACCCTCTT CCATTCCCTCACTCTGGTAAAAGCAAGCCATCTTCTCCAGTTACGCAAGGCAGAAGCCCAGGAGTGAGCACAATGCTGCCCTTTCCCTTG CTTCTGACTTCTCATCAATCCTATTAGAACTACTGACTGCACCCCACCTCTCCTCCATCCCCACGATCACTGCCTTCATATGAACTGCCT TTCCTGTCCTCTGGACTTTCAGTGGCTTCCTTTTTTGCTCACCTGTCCACTTTCTTCCCCTTCCCTCTCTCCATCCCTCCACATAGTCAT CAGAAAGATCATCAAAACAGGCAAATGTGACTGTATTATTTCCCTCACTTATCTTTGTATGCAGTTTAGAGTGATTTTTAAGAGCACACA CTTTGAAATTAGGAGAACCTGTGTCTGAATCCTGATTTTTGCCATTTGTGTGACTTGTCAGGTTATTTAATCTCTTTGAGCCTGTCCCTC CTCAGTGAAATAGGAATAATAATACCTACCTCATAGAGTTGTTGTGAGGATTCAGTGAGATAGACTGTTTCTAAAGTGTGTAGCACAGTG CCTGGCACATAGTGGGTGCTCACATTGGAGCACACTATTGTCGCTATCCCTGCA >58422_58422_5_NEDD4L-MBD2_NEDD4L_chr18_55711940_ENST00000589054_MBD2_chr18_51715381_ENST00000256429_length(amino acids)=221AA_BP=44 MEAEGRTGRPRRSTAAGSASDPARGAGSMATGLGEPVYGLSEDEGKPDLNTTLPIRQTASIFKQPVTKVTNHPSNKVKSDPQRMNEQPRQ LFWEKRLQGLSASDVTEQIIKTMELPKGLQGVGPGSNDETLLSAVASALHTSSAPITGQVSAAVEKNPAVWLNTSQPLCKAFIVTDEDIR KQEERVQQVRKKLEEALMADILSRAADTEEMDIEMDSGDEA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for NEDD4L-MBD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for NEDD4L-MBD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for NEDD4L-MBD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | NEDD4L | C0020538 | Hypertensive disease | 2 | CTD_human |
| Hgene | NEDD4L | C4310669 | PERIVENTRICULAR NODULAR HETEROTOPIA 7 | 2 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | NEDD4L | C0008925 | Cleft Palate | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | NEDD4L | C0014175 | Endometriosis | 1 | CTD_human |
| Hgene | NEDD4L | C0039075 | Syndactyly | 1 | CTD_human |
| Hgene | NEDD4L | C0265553 | Polysyndactyly | 1 | CTD_human |
| Hgene | NEDD4L | C0269102 | Endometrioma | 1 | CTD_human |
| Hgene | NEDD4L | C0543888 | Epileptic encephalopathy | 1 | GENOMICS_ENGLAND |
| Hgene | NEDD4L | C1837218 | Cleft palate, isolated | 1 | CTD_human |
| Hgene | NEDD4L | C1848213 | Periventricular Heterotopia, X-Linked | 1 | CTD_human |
| Hgene | NEDD4L | C1868720 | Periventricular Nodular Heterotopia | 1 | CTD_human;ORPHANET |
| Hgene | NEDD4L | C4551969 | Bilateral Periventricular Nodular Heterotopia | 1 | CTD_human |
| Tgene | C0033578 | Prostatic Neoplasms | 1 | CTD_human | |
| Tgene | C0036341 | Schizophrenia | 1 | PSYGENET | |
| Tgene | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human | |
| Tgene | C2931456 | Prostate cancer, familial | 1 | CTD_human | |
| Tgene | C4722327 | PROSTATE CANCER, HEREDITARY, 1 | 1 | CTD_human |
