
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACAP2-TP63 (FusionGDB2 ID:HG23527TG8626) |
Fusion Gene Summary for ACAP2-TP63 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACAP2-TP63 | Fusion gene ID: hg23527tg8626 | Hgene | Tgene | Gene symbol | ACAP2 | TP63 | Gene ID | 23527 | 8626 |
| Gene name | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 | tumor protein p63 | |
| Synonyms | CENTB2|CNT-B2 | AIS|B(p51A)|B(p51B)|EEC3|KET|LMS|NBP|OFC8|RHS|SHFM4|TP53CP|TP53L|TP73L|p40|p51|p53CP|p63|p73H|p73L | |
| Cytomap | ('ACAP2')('TP63') 3q29 | 3q28 | |
| Type of gene | protein-coding | protein-coding | |
| Description | arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 2Arf GAP with coiled coil, ANK repeat and PH domains 2centaurin-beta-2 | tumor protein 63amplified in squamous cell carcinomachronic ulcerative stomatitis proteinkeratinocyte transcription factor KETtransformation-related protein 63tumor protein p53-competing protein | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000326793, ENST00000472860, | ||
| Fusion gene scores | * DoF score | 16 X 14 X 11=2464 | 13 X 11 X 8=1144 |
| # samples | 22 | 15 | |
| ** MAII score | log2(22/2464*10)=-3.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/1144*10)=-2.93105264628251 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ACAP2 [Title/Abstract] AND TP63 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACAP2(195065988)-TP63(189455529), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ACAP2-TP63 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACAP2-TP63 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACAP2-TP63 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACAP2-TP63 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACAP2 | GO:0030029 | actin filament-based process | 11062263 |
| Tgene | TP63 | GO:0045747 | positive regulation of Notch signaling pathway | 11641404 |
| Tgene | TP63 | GO:0045892 | negative regulation of transcription, DNA-templated | 12446784 |
| Tgene | TP63 | GO:0045893 | positive regulation of transcription, DNA-templated | 12446784|16343436 |
| Tgene | TP63 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 22521434 |
| Tgene | TP63 | GO:2000271 | positive regulation of fibroblast apoptotic process | 9774969 |
 Fusion gene breakpoints across ACAP2 (5'-gene) Fusion gene breakpoints across ACAP2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
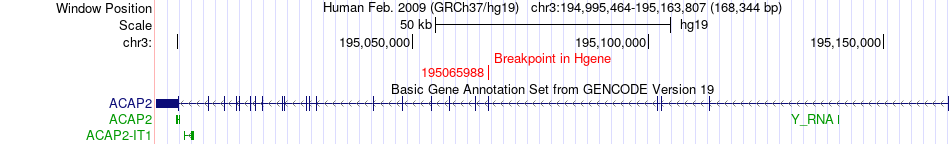 |
 Fusion gene breakpoints across TP63 (3'-gene) Fusion gene breakpoints across TP63 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
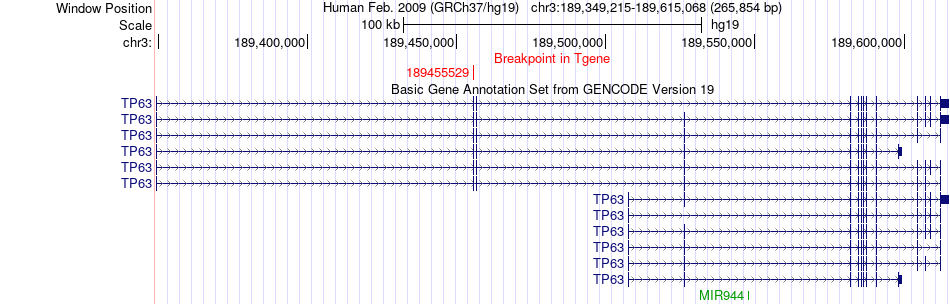 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | TCGA-GD-A2C5-11A | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
Top |
Fusion Gene ORF analysis for ACAP2-TP63 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000326793 | ENST00000354600 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| 5CDS-intron | ENST00000326793 | ENST00000392461 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| 5CDS-intron | ENST00000326793 | ENST00000392463 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| 5CDS-intron | ENST00000326793 | ENST00000437221 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| 5CDS-intron | ENST00000326793 | ENST00000449992 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| 5CDS-intron | ENST00000326793 | ENST00000456148 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| In-frame | ENST00000326793 | ENST00000264731 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| In-frame | ENST00000326793 | ENST00000320472 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| In-frame | ENST00000326793 | ENST00000382063 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| In-frame | ENST00000326793 | ENST00000392460 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| In-frame | ENST00000326793 | ENST00000418709 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| In-frame | ENST00000326793 | ENST00000440651 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-3CDS | ENST00000472860 | ENST00000264731 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-3CDS | ENST00000472860 | ENST00000320472 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-3CDS | ENST00000472860 | ENST00000382063 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-3CDS | ENST00000472860 | ENST00000392460 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-3CDS | ENST00000472860 | ENST00000418709 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-3CDS | ENST00000472860 | ENST00000440651 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-intron | ENST00000472860 | ENST00000354600 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-intron | ENST00000472860 | ENST00000392461 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-intron | ENST00000472860 | ENST00000392463 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-intron | ENST00000472860 | ENST00000437221 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-intron | ENST00000472860 | ENST00000449992 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
| intron-intron | ENST00000472860 | ENST00000456148 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000326793 | ACAP2 | chr3 | 195065988 | - | ENST00000264731 | TP63 | chr3 | 189455529 | + | 5333 | 575 | 213 | 2555 | 780 |
| ENST00000326793 | ACAP2 | chr3 | 195065988 | - | ENST00000382063 | TP63 | chr3 | 189455529 | + | 5078 | 575 | 213 | 2300 | 695 |
| ENST00000326793 | ACAP2 | chr3 | 195065988 | - | ENST00000418709 | TP63 | chr3 | 189455529 | + | 3295 | 575 | 213 | 1976 | 587 |
| ENST00000326793 | ACAP2 | chr3 | 195065988 | - | ENST00000320472 | TP63 | chr3 | 189455529 | + | 2517 | 575 | 213 | 2045 | 610 |
| ENST00000326793 | ACAP2 | chr3 | 195065988 | - | ENST00000392460 | TP63 | chr3 | 189455529 | + | 2181 | 575 | 213 | 2180 | 655 |
| ENST00000326793 | ACAP2 | chr3 | 195065988 | - | ENST00000440651 | TP63 | chr3 | 189455529 | + | 2544 | 575 | 213 | 2543 | 776 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000326793 | ENST00000264731 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + | 0.000292725 | 0.9997073 |
| ENST00000326793 | ENST00000382063 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + | 0.000355945 | 0.99964404 |
| ENST00000326793 | ENST00000418709 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + | 0.000404391 | 0.9995956 |
| ENST00000326793 | ENST00000320472 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + | 0.005500268 | 0.9944997 |
| ENST00000326793 | ENST00000392460 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + | 0.006919185 | 0.9930808 |
| ENST00000326793 | ENST00000440651 | ACAP2 | chr3 | 195065988 | - | TP63 | chr3 | 189455529 | + | 0.006952806 | 0.9930472 |
Top |
Fusion Genomic Features for ACAP2-TP63 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ACAP2 | chr3 | 195065987 | - | TP63 | chr3 | 189455528 | + | 0.001299888 | 0.99870014 |
| ACAP2 | chr3 | 195065987 | - | TP63 | chr3 | 189455528 | + | 0.001299888 | 0.99870014 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
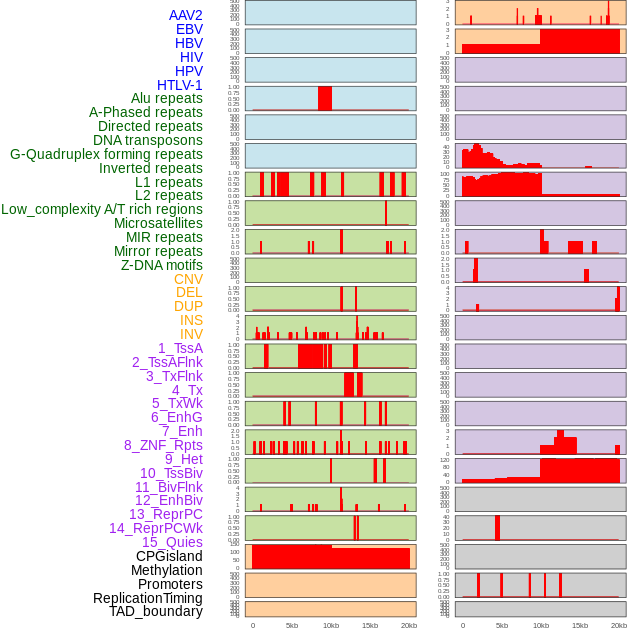 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
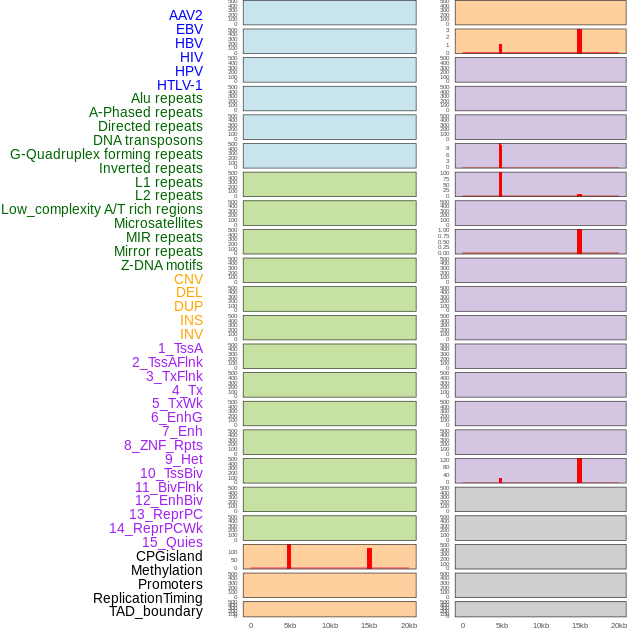 |
Top |
Fusion Protein Features for ACAP2-TP63 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:195065988/chr3:189455529) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 437_444 | 20 | 681.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 437_444 | 20 | 511.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 437_444 | 0 | 587.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 437_444 | 20 | 596.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 437_444 | 20 | 556.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 437_444 | 0 | 417.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 437_444 | 0 | 462.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 437_444 | 20 | 488.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 437_444 | 0 | 394.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 437_444 | 20 | 677.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 437_444 | 0 | 502.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 437_444 | 0 | 583.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 170_362 | 20 | 681.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 170_362 | 20 | 511.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 170_362 | 0 | 587.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 170_362 | 20 | 596.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 170_362 | 20 | 556.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 170_362 | 0 | 417.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 170_362 | 0 | 462.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 170_362 | 20 | 488.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 170_362 | 0 | 394.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 170_362 | 20 | 677.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 170_362 | 0 | 502.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 170_362 | 0 | 583.0 | DNA binding | . | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 541_607 | 20 | 681.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 541_607 | 20 | 511.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 541_607 | 0 | 587.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 541_607 | 20 | 596.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 541_607 | 20 | 556.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 541_607 | 0 | 417.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 541_607 | 0 | 462.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 541_607 | 20 | 488.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 541_607 | 0 | 394.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 541_607 | 20 | 677.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 541_607 | 0 | 502.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 541_607 | 0 | 583.0 | Domain | Note=SAM | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 394_443 | 20 | 681.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 610_680 | 20 | 681.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 394_443 | 20 | 511.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 610_680 | 20 | 511.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 1_107 | 0 | 587.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 394_443 | 0 | 587.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 610_680 | 0 | 587.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 394_443 | 20 | 596.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 610_680 | 20 | 596.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 394_443 | 20 | 556.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 610_680 | 20 | 556.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 1_107 | 0 | 417.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 394_443 | 0 | 417.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 610_680 | 0 | 417.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 1_107 | 0 | 462.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 394_443 | 0 | 462.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 610_680 | 0 | 462.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 394_443 | 20 | 488.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 610_680 | 20 | 488.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 1_107 | 0 | 394.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 394_443 | 0 | 394.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 610_680 | 0 | 394.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 394_443 | 20 | 677.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 610_680 | 20 | 677.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 1_107 | 0 | 502.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 394_443 | 0 | 502.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 610_680 | 0 | 502.0 | Region | Note=Transactivation inhibition | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 1_107 | 0 | 583.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 394_443 | 0 | 583.0 | Region | Note=Oligomerization | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 610_680 | 0 | 583.0 | Region | Note=Transactivation inhibition |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 1_226 | 114 | 779.0 | Domain | Note=BAR |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 266_361 | 114 | 779.0 | Domain | PH |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 399_520 | 114 | 779.0 | Domain | Arf-GAP |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 640_669 | 114 | 779.0 | Repeat | Note=ANK 1 |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 673_702 | 114 | 779.0 | Repeat | Note=ANK 2 |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 706_735 | 114 | 779.0 | Repeat | Note=ANK 3 |
| Hgene | ACAP2 | chr3:195065988 | chr3:189455529 | ENST00000326793 | - | 5 | 23 | 414_437 | 114 | 779.0 | Zinc finger | C4-type |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 1_107 | 20 | 681.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 1_107 | 20 | 511.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 1_107 | 20 | 596.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 1_107 | 20 | 556.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 1_107 | 20 | 488.0 | Region | Note=Transcription activation | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 1_107 | 20 | 677.0 | Region | Note=Transcription activation |
Top |
Fusion Gene Sequence for ACAP2-TP63 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1225_1225_1_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000264731_length(transcript)=5333nt_BP=575nt CGTTTCCCTGCGGCCAGCGCTCTACTTGGGGCGGGAGGAAGAGGGGCGGGACTGGGGCCTGACTAGTCGTCGCCGTCTTCGCGGCTGCTA TGACCAGCGGGCGAGGCGCCCTTCGCAGCTCCGCGCAACGCTAGTCGCGGCCGTGCGCCCGCCGTAGCCGTCTGCGTCCCAGCGCGGCCG CTCCCGCGGCCCCCTCGGCTTTGCCAGCGCCGCCTGCGAGGCGGAGGCAGGATGAAGATGACTGTGGATTTCGAGGAGTGTCTGAAGGAC TCGCCCCGCTTCAGGGCAGCTTTGGAAGAAGTAGAAGGTGATGTGGCAGAATTGGAACTAAAACTTGATAAGCTTGTGAAACTTTGTATT GCAATGATTGATACTGGAAAAGCCTTTTGTGTTGCAAATAAACAGTTCATGAATGGGATTCGAGACCTGGCTCAGTATTCTAGTAATGAT GCTGTCGTTGAGACAAGTTTGACCAAGTTTTCTGACAGTCTTCAAGAAATGATAAATTTTCACACAATCCTGTTTGACCAAACTCAGAGA TCAATTAAGGCACAGCTTCAGAACTTTGTTAAAGATTTCGTAGAAACCCCAGCTCATTTCTCTTGGAAAGAAAGTTATTACCGATCCACC ATGTCCCAGAGCACACAGACAAATGAATTCCTCAGTCCAGAGGTTTTCCAGCATATCTGGGATTTTCTGGAACAGCCTATATGTTCAGTT CAGCCCATTGACTTGAACTTTGTGGATGAACCATCAGAAGATGGTGCGACAAACAAGATTGAGATTAGCATGGACTGTATCCGCATGCAG GACTCGGACCTGAGTGACCCCATGTGGCCACAGTACACGAACCTGGGGCTCCTGAACAGCATGGACCAGCAGATTCAGAACGGCTCCTCG TCCACCAGTCCCTATAACACAGACCACGCGCAGAACAGCGTCACGGCGCCCTCGCCCTACGCACAGCCCAGCTCCACCTTCGATGCTCTC TCTCCATCACCCGCCATCCCCTCCAACACCGACTACCCAGGCCCGCACAGTTTCGACGTGTCCTTCCAGCAGTCGAGCACCGCCAAGTCG GCCACCTGGACGTATTCCACTGAACTGAAGAAACTCTACTGCCAAATTGCAAAGACATGCCCCATCCAGATCAAGGTGATGACCCCACCT CCTCAGGGAGCTGTTATCCGCGCCATGCCTGTCTACAAAAAAGCTGAGCACGTCACGGAGGTGGTGAAGCGGTGCCCCAACCATGAGCTG AGCCGTGAATTCAACGAGGGACAGATTGCCCCTCCTAGTCATTTGATTCGAGTAGAGGGGAACAGCCATGCCCAGTATGTAGAAGATCCC ATCACAGGAAGACAGAGTGTGCTGGTACCTTATGAGCCACCCCAGGTTGGCACTGAATTCACGACAGTCTTGTACAATTTCATGTGTAAC AGCAGTTGTGTTGGAGGGATGAACCGCCGTCCAATTTTAATCATTGTTACTCTGGAAACCAGAGATGGGCAAGTCCTGGGCCGACGCTGC TTTGAGGCCCGGATCTGTGCTTGCCCAGGAAGAGACAGGAAGGCGGATGAAGATAGCATCAGAAAGCAGCAAGTTTCGGACAGTACAAAG AACGGTGATGGTACGAAGCGCCCGTTTCGTCAGAACACACATGGTATCCAGATGACATCCATCAAGAAACGAAGATCCCCAGATGATGAA CTGTTATACTTACCAGTGAGGGGCCGTGAGACTTATGAAATGCTGTTGAAGATCAAAGAGTCCCTGGAACTCATGCAGTACCTTCCTCAG CACACAATTGAAACGTACAGGCAACAGCAACAGCAGCAGCACCAGCACTTACTTCAGAAACAGACCTCAATACAGTCTCCATCTTCATAT GGTAACAGCTCCCCACCTCTGAACAAAATGAACAGCATGAACAAGCTGCCTTCTGTGAGCCAGCTTATCAACCCTCAGCAGCGCAACGCC CTCACTCCTACAACCATTCCTGATGGCATGGGAGCCAACATTCCCATGATGGGCACCCACATGCCAATGGCTGGAGACATGAATGGACTC AGCCCCACCCAGGCACTCCCTCCCCCACTCTCCATGCCATCCACCTCCCACTGCACACCCCCACCTCCGTATCCCACAGATTGCAGCATT GTCAGTTTCTTAGCGAGGTTGGGCTGTTCATCATGTCTGGACTATTTCACGACCCAGGGGCTGACCACCATCTATCAGATTGAGCATTAC TCCATGGATGATCTGGCAAGTCTGAAAATCCCTGAGCAATTTCGACATGCGATCTGGAAGGGCATCCTGGACCACCGGCAGCTCCACGAA TTCTCCTCCCCTTCTCATCTCCTGCGGACCCCAAGCAGTGCCTCTACAGTCAGTGTGGGCTCCAGTGAGACCCGGGGTGAGCGTGTTATT GATGCTGTGCGATTCACCCTCCGCCAGACCATCTCTTTCCCACCCCGAGATGAGTGGAATGACTTCAACTTTGACATGGATGCTCGCCGC AATAAGCAACAGCGCATCAAAGAGGAGGGGGAGTGAGCCTCACCATGTGAGCTCTTCCTATCCCTCTCCTAACTGCCAGCCCCCTAAAAG CACTCCTGCTTAATCTTCAAAGCCTTCTCCCTAGCTCCTCCCCTTCCTCTTGTCTGATTTCTTAGGGGAAGGAGAAGTAAGAGGCTACCT CTTACCTAACATCTGACCTGGCATCTAATTCTGATTCTGGCTTTAAGCCTTCAAAACTATAGCTTGCAGAACTGTAGCTGCCATGGCTAG GTAGAAGTGAGCAAAAAAGAGTTGGGTGTCTCCTTAAGCTGCAGAGATTTCTCATTGACTTTTATAAAGCATGTTCACCCTTATAGTCTA AGACTATATATATAAATGTATAAATATACAGTATAGATTTTTGGGTGGGGGGCATTGAGTATTGTTTAAAATGTAATTTAAATGAAAGAA AATTGAGTTGCACTTATTGACCATTTTTTAATTTACTTGTTTTGGATGGCTTGTCTATACTCCTTCCCTTAAGGGGTATCATGTATGGTG ATAGGTATCTAGAGCTTAATGCTACATGTGAGTGACGATGATGTACAGATTCTTTCAGTTCTTTGGATTCTAAATACATGCCACATCAAA CCTTTGAGTAGATCCATTTCCATTGCTTATTATGTAGGTAAGACTGTAGATATGTATTCTTTTCTCAGTGTTGGTATATTTTATATTACT GACATTTCTTCTAGTGATGATGGTTCACGTTGGGGTGATTTAATCCAGTTATAAGAAGAAGTTCATGTCCAAACGTCCTCTTTAGTTTTT GGTTGGGAATGAGGAAAATTCTTAAAAGGCCCATAGCAGCCAGTTCAAAAACACCCGACGTCATGTATTTGAGCATATCAGTAACCCCCT TAAATTTAATACCAGATACCTTATCTTACAATATTGATTGGGAAAACATTTGCTGCCATTACAGAGGTATTAAAACTAAATTTCACTACT AGATTGACTAACTCAAATACACATTTGCTACTGTTGTAAGAATTCTGATTGATTTGATTGGGATGAATGCCATCTATCTAGTTCTAACAG TGAAGTTTTACTGTCTATTAATATTCAGGGTAAATAGGAATCATTCAGAAATGTTGAGTCTGTACTAAACAGTAAGATATCTCAATGAAC CATAAATTCAACTTTGTAAAAATCTTTTGAAGCATAGATAATATTGTTTGGTAAATGTTTCTTTTGTTTGGTAAATGTTTCTTTTAAAGA CCCTCCTATTCTATAAAACTCTGCATGTAGAGGCTTGTTTACCTTTCTCTCTCTAAGGTTTACAATAGGAGTGGTGATTTGAAAAATATA AAATTATGAGATTGGTTTTCCTGTGGCATAAATTGCATCACTGTATCATTTTCTTTTTTAACCGGTAAGAGTTTCAGTTTGTTGGAAAGT AACTGTGAGAACCCAGTTTCCCGTCCATCTCCCTTAGGGACTACCCATAGACATGAAAGGTCCCCACAGAGCAAGAGATAAGTCTTTCAT GGCTGCTGTTGCTTAAACCACTTAAACGAAGAGTTCCCTTGAAACTTTGGGAAAACATGTTAATGACAATATTCCAGATCTTTCAGAAAT ATAACACATTTTTTTGCATGCATGCAAATGAGCTCTGAAATCTTCCCATGCATTCTGGTCAAGGGCTGTCATTGCACATAAGCTTCCATT TTAATTTTAAAGTGCAAAAGGGCCAGCGTGGCTCTAAAAGGTAATGTGTGGATTGCCTCTGAAAAGTGTGTATATATTTTGTGTGAAATT GCATACTTTGTATTTTGATTATTTTTTTTTTCTTCTTGGGATAGTGGGATTTCCAGAACCACACTTGAAACCTTTTTTTATCGTTTTTGT ATTTTCATGAAAATACCATTTAGTAAGAATACCACATCAAATAAGAAATAATGCTACAATTTTAAGAGGGGAGGGAAGGGAAAGTTTTTT TTTATTATTTTTTTAAAATTTTGTATGTTAAAGAGAATGAGTCCTTGATTTCAAAGTTTTGTTGTACTTAAATGGTAATAAGCACTGTAA ACTTCTGCAACAAGCATGCAGCTTTGCAAACCCATTAAGGGGAAGAATGAAAGCTGTTCCTTGGTCCTAGTAAGAAGACAAACTGCTTCC CTTACTTTGCTGAGGGTTTGAATAAACCTAGGACTTCCGAGCTATGTCAGTACTATTCAGGTAACACTAGGGCCTTGGAAATTCCTGTAC TGTGTCTCATGGATTTGGCACTAGCCAAAGCGAGGCACCCTTACTGGCTTACCTCCTCATGGCAGCCTACTCTCCTTGAGTGTATGAGTA GCCAGGGTAAGGGGTAAAAGGATAGTAAGCATAGAAACCACTAGAAAGTGGGCTTAATGGAGTTCTTGTGGCCTCAGCTCAATGCAGTTA GCTGAAGAATTGAAAAGTTTTTGTTTGGAGACGTTTATAAACAGAAATGGAAAGCAGAGTTTTCATTAAATCCTTTTACCTTTTTTTTTT CTTGGTAATCCCCTAAAATAACAGTATGTGGGATATTGAATGTTAAAGGGATATTTTTTTCTATTATTTTTATAATTGTACAAAATTAAG CAAATGTTAAAAGTTTTATATGCTTTATTAATGTTTTCAAAAGGTATTATACATGTGATACATTTTTTAAGCTTCAGTTGCTTGTCTTCT GGTACTTTCTGTTATGGGCTTTTGGGGAGCCAGAAGCCAATCTACAATCTCTTTTTGTTTGCCAGGACATGCAATAAAATTTAAAAAATA AATAAAAACTAATTAAGAAATTG >1225_1225_1_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000264731_length(amino acids)=780AA_BP=121 MRGGGRMKMTVDFEECLKDSPRFRAALEEVEGDVAELELKLDKLVKLCIAMIDTGKAFCVANKQFMNGIRDLAQYSSNDAVVETSLTKFS DSLQEMINFHTILFDQTQRSIKAQLQNFVKDFVETPAHFSWKESYYRSTMSQSTQTNEFLSPEVFQHIWDFLEQPICSVQPIDLNFVDEP SEDGATNKIEISMDCIRMQDSDLSDPMWPQYTNLGLLNSMDQQIQNGSSSTSPYNTDHAQNSVTAPSPYAQPSSTFDALSPSPAIPSNTD YPGPHSFDVSFQQSSTAKSATWTYSTELKKLYCQIAKTCPIQIKVMTPPPQGAVIRAMPVYKKAEHVTEVVKRCPNHELSREFNEGQIAP PSHLIRVEGNSHAQYVEDPITGRQSVLVPYEPPQVGTEFTTVLYNFMCNSSCVGGMNRRPILIIVTLETRDGQVLGRRCFEARICACPGR DRKADEDSIRKQQVSDSTKNGDGTKRPFRQNTHGIQMTSIKKRRSPDDELLYLPVRGRETYEMLLKIKESLELMQYLPQHTIETYRQQQQ QQHQHLLQKQTSIQSPSSYGNSSPPLNKMNSMNKLPSVSQLINPQQRNALTPTTIPDGMGANIPMMGTHMPMAGDMNGLSPTQALPPPLS MPSTSHCTPPPPYPTDCSIVSFLARLGCSSCLDYFTTQGLTTIYQIEHYSMDDLASLKIPEQFRHAIWKGILDHRQLHEFSSPSHLLRTP SSASTVSVGSSETRGERVIDAVRFTLRQTISFPPRDEWNDFNFDMDARRNKQQRIKEEGE -------------------------------------------------------------- >1225_1225_2_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000320472_length(transcript)=2517nt_BP=575nt CGTTTCCCTGCGGCCAGCGCTCTACTTGGGGCGGGAGGAAGAGGGGCGGGACTGGGGCCTGACTAGTCGTCGCCGTCTTCGCGGCTGCTA TGACCAGCGGGCGAGGCGCCCTTCGCAGCTCCGCGCAACGCTAGTCGCGGCCGTGCGCCCGCCGTAGCCGTCTGCGTCCCAGCGCGGCCG CTCCCGCGGCCCCCTCGGCTTTGCCAGCGCCGCCTGCGAGGCGGAGGCAGGATGAAGATGACTGTGGATTTCGAGGAGTGTCTGAAGGAC TCGCCCCGCTTCAGGGCAGCTTTGGAAGAAGTAGAAGGTGATGTGGCAGAATTGGAACTAAAACTTGATAAGCTTGTGAAACTTTGTATT GCAATGATTGATACTGGAAAAGCCTTTTGTGTTGCAAATAAACAGTTCATGAATGGGATTCGAGACCTGGCTCAGTATTCTAGTAATGAT GCTGTCGTTGAGACAAGTTTGACCAAGTTTTCTGACAGTCTTCAAGAAATGATAAATTTTCACACAATCCTGTTTGACCAAACTCAGAGA TCAATTAAGGCACAGCTTCAGAACTTTGTTAAAGATTTCGTAGAAACCCCAGCTCATTTCTCTTGGAAAGAAAGTTATTACCGATCCACC ATGTCCCAGAGCACACAGACAAATGAATTCCTCAGTCCAGAGGTTTTCCAGCATATCTGGGATTTTCTGGAACAGCCTATATGTTCAGTT CAGCCCATTGACTTGAACTTTGTGGATGAACCATCAGAAGATGGTGCGACAAACAAGATTGAGATTAGCATGGACTGTATCCGCATGCAG GACTCGGACCTGAGTGACCCCATGTGGCCACAGTACACGAACCTGGGGCTCCTGAACAGCATGGACCAGCAGATTCAGAACGGCTCCTCG TCCACCAGTCCCTATAACACAGACCACGCGCAGAACAGCGTCACGGCGCCCTCGCCCTACGCACAGCCCAGCTCCACCTTCGATGCTCTC TCTCCATCACCCGCCATCCCCTCCAACACCGACTACCCAGGCCCGCACAGTTTCGACGTGTCCTTCCAGCAGTCGAGCACCGCCAAGTCG GCCACCTGGACGTATTCCACTGAACTGAAGAAACTCTACTGCCAAATTGCAAAGACATGCCCCATCCAGATCAAGGTGATGACCCCACCT CCTCAGGGAGCTGTTATCCGCGCCATGCCTGTCTACAAAAAAGCTGAGCACGTCACGGAGGTGGTGAAGCGGTGCCCCAACCATGAGCTG AGCCGTGAATTCAACGAGGGACAGATTGCCCCTCCTAGTCATTTGATTCGAGTAGAGGGGAACAGCCATGCCCAGTATGTAGAAGATCCC ATCACAGGAAGACAGAGTGTGCTGGTACCTTATGAGCCACCCCAGGTTGGCACTGAATTCACGACAGTCTTGTACAATTTCATGTGTAAC AGCAGTTGTGTTGGAGGGATGAACCGCCGTCCAATTTTAATCATTGTTACTCTGGAAACCAGAGATGGGCAAGTCCTGGGCCGACGCTGC TTTGAGGCCCGGATCTGTGCTTGCCCAGGAAGAGACAGGAAGGCGGATGAAGATAGCATCAGAAAGCAGCAAGTTTCGGACAGTACAAAG AACGGTGATGGTACGAAGCGCCCGTTTCGTCAGAACACACATGGTATCCAGATGACATCCATCAAGAAACGAAGATCCCCAGATGATGAA CTGTTATACTTACCAGTGAGGGGCCGTGAGACTTATGAAATGCTGTTGAAGATCAAAGAGTCCCTGGAACTCATGCAGTACCTTCCTCAG CACACAATTGAAACGTACAGGCAACAGCAACAGCAGCAGCACCAGCACTTACTTCAGAAACAGACCTCAATACAGTCTCCATCTTCATAT GGTAACAGCTCCCCACCTCTGAACAAAATGAACAGCATGAACAAGCTGCCTTCTGTGAGCCAGCTTATCAACCCTCAGCAGCGCAACGCC CTCACTCCTACAACCATTCCTGATGGCATGGGAGCCAACAGATCTGGCAAGTCTGAAAATCCCTGAGCAATTTCGACATGCGATCTGGAA GGGCATCCTGGACCACCGGCAGCTCCACGAATTCTCCTCCCCTTCTCATCTCCTGCGGACCCCAAGCAGTGCCTCTACAGTCAGTGTGGG CTCCAGTGAGACCCGGGGTGAGCGTGTTATTGATGCTGTGCGATTCACCCTCCGCCAGACCATCTCTTTCCCACCCCGAGATGAGTGGAA TGACTTCAACTTTGACATGGATGCTCGCCGCAATAAGCAACAGCGCATCAAAGAGGAGGGGGAGTGAGCCTCACCATGTGAGCTCTTCCT ATCCCTCTCCTAACTGCCAGCCCCCTAAAAGCACTCCTGCTTAATCTTCAAAGCCTTCTCCCTAGCTCCTCCCCTTCCTCTTGTCTGATT TCTTAGGGGAAGGAGAAGTAAGAGGCTACCTCTTACCTAACATCTGACCTGGCATCTAATTCTGATTCTGGCTTTAAGCCTTCAAAA >1225_1225_2_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000320472_length(amino acids)=610AA_BP=121 MRGGGRMKMTVDFEECLKDSPRFRAALEEVEGDVAELELKLDKLVKLCIAMIDTGKAFCVANKQFMNGIRDLAQYSSNDAVVETSLTKFS DSLQEMINFHTILFDQTQRSIKAQLQNFVKDFVETPAHFSWKESYYRSTMSQSTQTNEFLSPEVFQHIWDFLEQPICSVQPIDLNFVDEP SEDGATNKIEISMDCIRMQDSDLSDPMWPQYTNLGLLNSMDQQIQNGSSSTSPYNTDHAQNSVTAPSPYAQPSSTFDALSPSPAIPSNTD YPGPHSFDVSFQQSSTAKSATWTYSTELKKLYCQIAKTCPIQIKVMTPPPQGAVIRAMPVYKKAEHVTEVVKRCPNHELSREFNEGQIAP PSHLIRVEGNSHAQYVEDPITGRQSVLVPYEPPQVGTEFTTVLYNFMCNSSCVGGMNRRPILIIVTLETRDGQVLGRRCFEARICACPGR DRKADEDSIRKQQVSDSTKNGDGTKRPFRQNTHGIQMTSIKKRRSPDDELLYLPVRGRETYEMLLKIKESLELMQYLPQHTIETYRQQQQ QQHQHLLQKQTSIQSPSSYGNSSPPLNKMNSMNKLPSVSQLINPQQRNALTPTTIPDGMGANRSGKSENP -------------------------------------------------------------- >1225_1225_3_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000382063_length(transcript)=5078nt_BP=575nt CGTTTCCCTGCGGCCAGCGCTCTACTTGGGGCGGGAGGAAGAGGGGCGGGACTGGGGCCTGACTAGTCGTCGCCGTCTTCGCGGCTGCTA TGACCAGCGGGCGAGGCGCCCTTCGCAGCTCCGCGCAACGCTAGTCGCGGCCGTGCGCCCGCCGTAGCCGTCTGCGTCCCAGCGCGGCCG CTCCCGCGGCCCCCTCGGCTTTGCCAGCGCCGCCTGCGAGGCGGAGGCAGGATGAAGATGACTGTGGATTTCGAGGAGTGTCTGAAGGAC TCGCCCCGCTTCAGGGCAGCTTTGGAAGAAGTAGAAGGTGATGTGGCAGAATTGGAACTAAAACTTGATAAGCTTGTGAAACTTTGTATT GCAATGATTGATACTGGAAAAGCCTTTTGTGTTGCAAATAAACAGTTCATGAATGGGATTCGAGACCTGGCTCAGTATTCTAGTAATGAT GCTGTCGTTGAGACAAGTTTGACCAAGTTTTCTGACAGTCTTCAAGAAATGATAAATTTTCACACAATCCTGTTTGACCAAACTCAGAGA TCAATTAAGGCACAGCTTCAGAACTTTGTTAAAGATTTCGTAGAAACCCCAGCTCATTTCTCTTGGAAAGAAAGTTATTACCGATCCACC ATGTCCCAGAGCACACAGACAAATGAATTCCTCAGTCCAGAGGTTTTCCAGCATATCTGGGATTTTCTGGAACAGCCTATATGTTCAGTT CAGCCCATTGACTTGAACTTTGTGGATGAACCATCAGAAGATGGTGCGACAAACAAGATTGAGATTAGCATGGACTGTATCCGCATGCAG GACTCGGACCTGAGTGACCCCATGTGGTATTCCACTGAACTGAAGAAACTCTACTGCCAAATTGCAAAGACATGCCCCATCCAGATCAAG GTGATGACCCCACCTCCTCAGGGAGCTGTTATCCGCGCCATGCCTGTCTACAAAAAAGCTGAGCACGTCACGGAGGTGGTGAAGCGGTGC CCCAACCATGAGCTGAGCCGTGAATTCAACGAGGGACAGATTGCCCCTCCTAGTCATTTGATTCGAGTAGAGGGGAACAGCCATGCCCAG TATGTAGAAGATCCCATCACAGGAAGACAGAGTGTGCTGGTACCTTATGAGCCACCCCAGGTTGGCACTGAATTCACGACAGTCTTGTAC AATTTCATGTGTAACAGCAGTTGTGTTGGAGGGATGAACCGCCGTCCAATTTTAATCATTGTTACTCTGGAAACCAGAGATGGGCAAGTC CTGGGCCGACGCTGCTTTGAGGCCCGGATCTGTGCTTGCCCAGGAAGAGACAGGAAGGCGGATGAAGATAGCATCAGAAAGCAGCAAGTT TCGGACAGTACAAAGAACGGTGATGGTACGAAGCGCCCGTTTCGTCAGAACACACATGGTATCCAGATGACATCCATCAAGAAACGAAGA TCCCCAGATGATGAACTGTTATACTTACCAGTGAGGGGCCGTGAGACTTATGAAATGCTGTTGAAGATCAAAGAGTCCCTGGAACTCATG CAGTACCTTCCTCAGCACACAATTGAAACGTACAGGCAACAGCAACAGCAGCAGCACCAGCACTTACTTCAGAAACAGACCTCAATACAG TCTCCATCTTCATATGGTAACAGCTCCCCACCTCTGAACAAAATGAACAGCATGAACAAGCTGCCTTCTGTGAGCCAGCTTATCAACCCT CAGCAGCGCAACGCCCTCACTCCTACAACCATTCCTGATGGCATGGGAGCCAACATTCCCATGATGGGCACCCACATGCCAATGGCTGGA GACATGAATGGACTCAGCCCCACCCAGGCACTCCCTCCCCCACTCTCCATGCCATCCACCTCCCACTGCACACCCCCACCTCCGTATCCC ACAGATTGCAGCATTGTCAGTTTCTTAGCGAGGTTGGGCTGTTCATCATGTCTGGACTATTTCACGACCCAGGGGCTGACCACCATCTAT CAGATTGAGCATTACTCCATGGATGATCTGGCAAGTCTGAAAATCCCTGAGCAATTTCGACATGCGATCTGGAAGGGCATCCTGGACCAC CGGCAGCTCCACGAATTCTCCTCCCCTTCTCATCTCCTGCGGACCCCAAGCAGTGCCTCTACAGTCAGTGTGGGCTCCAGTGAGACCCGG GGTGAGCGTGTTATTGATGCTGTGCGATTCACCCTCCGCCAGACCATCTCTTTCCCACCCCGAGATGAGTGGAATGACTTCAACTTTGAC ATGGATGCTCGCCGCAATAAGCAACAGCGCATCAAAGAGGAGGGGGAGTGAGCCTCACCATGTGAGCTCTTCCTATCCCTCTCCTAACTG CCAGCCCCCTAAAAGCACTCCTGCTTAATCTTCAAAGCCTTCTCCCTAGCTCCTCCCCTTCCTCTTGTCTGATTTCTTAGGGGAAGGAGA AGTAAGAGGCTACCTCTTACCTAACATCTGACCTGGCATCTAATTCTGATTCTGGCTTTAAGCCTTCAAAACTATAGCTTGCAGAACTGT AGCTGCCATGGCTAGGTAGAAGTGAGCAAAAAAGAGTTGGGTGTCTCCTTAAGCTGCAGAGATTTCTCATTGACTTTTATAAAGCATGTT CACCCTTATAGTCTAAGACTATATATATAAATGTATAAATATACAGTATAGATTTTTGGGTGGGGGGCATTGAGTATTGTTTAAAATGTA ATTTAAATGAAAGAAAATTGAGTTGCACTTATTGACCATTTTTTAATTTACTTGTTTTGGATGGCTTGTCTATACTCCTTCCCTTAAGGG GTATCATGTATGGTGATAGGTATCTAGAGCTTAATGCTACATGTGAGTGACGATGATGTACAGATTCTTTCAGTTCTTTGGATTCTAAAT ACATGCCACATCAAACCTTTGAGTAGATCCATTTCCATTGCTTATTATGTAGGTAAGACTGTAGATATGTATTCTTTTCTCAGTGTTGGT ATATTTTATATTACTGACATTTCTTCTAGTGATGATGGTTCACGTTGGGGTGATTTAATCCAGTTATAAGAAGAAGTTCATGTCCAAACG TCCTCTTTAGTTTTTGGTTGGGAATGAGGAAAATTCTTAAAAGGCCCATAGCAGCCAGTTCAAAAACACCCGACGTCATGTATTTGAGCA TATCAGTAACCCCCTTAAATTTAATACCAGATACCTTATCTTACAATATTGATTGGGAAAACATTTGCTGCCATTACAGAGGTATTAAAA CTAAATTTCACTACTAGATTGACTAACTCAAATACACATTTGCTACTGTTGTAAGAATTCTGATTGATTTGATTGGGATGAATGCCATCT ATCTAGTTCTAACAGTGAAGTTTTACTGTCTATTAATATTCAGGGTAAATAGGAATCATTCAGAAATGTTGAGTCTGTACTAAACAGTAA GATATCTCAATGAACCATAAATTCAACTTTGTAAAAATCTTTTGAAGCATAGATAATATTGTTTGGTAAATGTTTCTTTTGTTTGGTAAA TGTTTCTTTTAAAGACCCTCCTATTCTATAAAACTCTGCATGTAGAGGCTTGTTTACCTTTCTCTCTCTAAGGTTTACAATAGGAGTGGT GATTTGAAAAATATAAAATTATGAGATTGGTTTTCCTGTGGCATAAATTGCATCACTGTATCATTTTCTTTTTTAACCGGTAAGAGTTTC AGTTTGTTGGAAAGTAACTGTGAGAACCCAGTTTCCCGTCCATCTCCCTTAGGGACTACCCATAGACATGAAAGGTCCCCACAGAGCAAG AGATAAGTCTTTCATGGCTGCTGTTGCTTAAACCACTTAAACGAAGAGTTCCCTTGAAACTTTGGGAAAACATGTTAATGACAATATTCC AGATCTTTCAGAAATATAACACATTTTTTTGCATGCATGCAAATGAGCTCTGAAATCTTCCCATGCATTCTGGTCAAGGGCTGTCATTGC ACATAAGCTTCCATTTTAATTTTAAAGTGCAAAAGGGCCAGCGTGGCTCTAAAAGGTAATGTGTGGATTGCCTCTGAAAAGTGTGTATAT ATTTTGTGTGAAATTGCATACTTTGTATTTTGATTATTTTTTTTTTCTTCTTGGGATAGTGGGATTTCCAGAACCACACTTGAAACCTTT TTTTATCGTTTTTGTATTTTCATGAAAATACCATTTAGTAAGAATACCACATCAAATAAGAAATAATGCTACAATTTTAAGAGGGGAGGG AAGGGAAAGTTTTTTTTTATTATTTTTTTAAAATTTTGTATGTTAAAGAGAATGAGTCCTTGATTTCAAAGTTTTGTTGTACTTAAATGG TAATAAGCACTGTAAACTTCTGCAACAAGCATGCAGCTTTGCAAACCCATTAAGGGGAAGAATGAAAGCTGTTCCTTGGTCCTAGTAAGA AGACAAACTGCTTCCCTTACTTTGCTGAGGGTTTGAATAAACCTAGGACTTCCGAGCTATGTCAGTACTATTCAGGTAACACTAGGGCCT TGGAAATTCCTGTACTGTGTCTCATGGATTTGGCACTAGCCAAAGCGAGGCACCCTTACTGGCTTACCTCCTCATGGCAGCCTACTCTCC TTGAGTGTATGAGTAGCCAGGGTAAGGGGTAAAAGGATAGTAAGCATAGAAACCACTAGAAAGTGGGCTTAATGGAGTTCTTGTGGCCTC AGCTCAATGCAGTTAGCTGAAGAATTGAAAAGTTTTTGTTTGGAGACGTTTATAAACAGAAATGGAAAGCAGAGTTTTCATTAAATCCTT TTACCTTTTTTTTTTCTTGGTAATCCCCTAAAATAACAGTATGTGGGATATTGAATGTTAAAGGGATATTTTTTTCTATTATTTTTATAA TTGTACAAAATTAAGCAAATGTTAAAAGTTTTATATGCTTTATTAATGTTTTCAAAAGGTATTATACATGTGATACATTTTTTAAGCTTC AGTTGCTTGTCTTCTGGTACTTTCTGTTATGGGCTTTTGGGGAGCCAGAAGCCAATCTACAATCTCTTTTTGTTTGCCAGGACATGCAAT AAAATTTAAAAAATAAATAAAAACTAATTAAGAAATTG >1225_1225_3_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000382063_length(amino acids)=695AA_BP=121 MRGGGRMKMTVDFEECLKDSPRFRAALEEVEGDVAELELKLDKLVKLCIAMIDTGKAFCVANKQFMNGIRDLAQYSSNDAVVETSLTKFS DSLQEMINFHTILFDQTQRSIKAQLQNFVKDFVETPAHFSWKESYYRSTMSQSTQTNEFLSPEVFQHIWDFLEQPICSVQPIDLNFVDEP SEDGATNKIEISMDCIRMQDSDLSDPMWYSTELKKLYCQIAKTCPIQIKVMTPPPQGAVIRAMPVYKKAEHVTEVVKRCPNHELSREFNE GQIAPPSHLIRVEGNSHAQYVEDPITGRQSVLVPYEPPQVGTEFTTVLYNFMCNSSCVGGMNRRPILIIVTLETRDGQVLGRRCFEARIC ACPGRDRKADEDSIRKQQVSDSTKNGDGTKRPFRQNTHGIQMTSIKKRRSPDDELLYLPVRGRETYEMLLKIKESLELMQYLPQHTIETY RQQQQQQHQHLLQKQTSIQSPSSYGNSSPPLNKMNSMNKLPSVSQLINPQQRNALTPTTIPDGMGANIPMMGTHMPMAGDMNGLSPTQAL PPPLSMPSTSHCTPPPPYPTDCSIVSFLARLGCSSCLDYFTTQGLTTIYQIEHYSMDDLASLKIPEQFRHAIWKGILDHRQLHEFSSPSH LLRTPSSASTVSVGSSETRGERVIDAVRFTLRQTISFPPRDEWNDFNFDMDARRNKQQRIKEEGE -------------------------------------------------------------- >1225_1225_4_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000392460_length(transcript)=2181nt_BP=575nt CGTTTCCCTGCGGCCAGCGCTCTACTTGGGGCGGGAGGAAGAGGGGCGGGACTGGGGCCTGACTAGTCGTCGCCGTCTTCGCGGCTGCTA TGACCAGCGGGCGAGGCGCCCTTCGCAGCTCCGCGCAACGCTAGTCGCGGCCGTGCGCCCGCCGTAGCCGTCTGCGTCCCAGCGCGGCCG CTCCCGCGGCCCCCTCGGCTTTGCCAGCGCCGCCTGCGAGGCGGAGGCAGGATGAAGATGACTGTGGATTTCGAGGAGTGTCTGAAGGAC TCGCCCCGCTTCAGGGCAGCTTTGGAAGAAGTAGAAGGTGATGTGGCAGAATTGGAACTAAAACTTGATAAGCTTGTGAAACTTTGTATT GCAATGATTGATACTGGAAAAGCCTTTTGTGTTGCAAATAAACAGTTCATGAATGGGATTCGAGACCTGGCTCAGTATTCTAGTAATGAT GCTGTCGTTGAGACAAGTTTGACCAAGTTTTCTGACAGTCTTCAAGAAATGATAAATTTTCACACAATCCTGTTTGACCAAACTCAGAGA TCAATTAAGGCACAGCTTCAGAACTTTGTTAAAGATTTCGTAGAAACCCCAGCTCATTTCTCTTGGAAAGAAAGTTATTACCGATCCACC ATGTCCCAGAGCACACAGACAAATGAATTCCTCAGTCCAGAGGTTTTCCAGCATATCTGGGATTTTCTGGAACAGCCTATATGTTCAGTT CAGCCCATTGACTTGAACTTTGTGGATGAACCATCAGAAGATGGTGCGACAAACAAGATTGAGATTAGCATGGACTGTATCCGCATGCAG GACTCGGACCTGAGTGACCCCATGTGGCCACAGTACACGAACCTGGGGCTCCTGAACAGCATGGACCAGCAGATTCAGAACGGCTCCTCG TCCACCAGTCCCTATAACACAGACCACGCGCAGAACAGCGTCACGGCGCCCTCGCCCTACGCACAGCCCAGCTCCACCTTCGATGCTCTC TCTCCATCACCCGCCATCCCCTCCAACACCGACTACCCAGGCCCGCACAGTTTCGACGTGTCCTTCCAGCAGTCGAGCACCGCCAAGTCG GCCACCTGGACGTATTCCACTGAACTGAAGAAACTCTACTGCCAAATTGCAAAGACATGCCCCATCCAGATCAAGGTGATGACCCCACCT CCTCAGGGAGCTGTTATCCGCGCCATGCCTGTCTACAAAAAAGCTGAGCACGTCACGGAGGTGGTGAAGCGGTGCCCCAACCATGAGCTG AGCCGTGAATTCAACGAGGGACAGATTGCCCCTCCTAGTCATTTGATTCGAGTAGAGGGGAACAGCCATGCCCAGTATGTAGAAGATCCC ATCACAGGAAGACAGAGTGTGCTGGTACCTTATGAGCCACCCCAGGTTGGCACTGAATTCACGACAGTCTTGTACAATTTCATGTGTAAC AGCAGTTGTGTTGGAGGGATGAACCGCCGTCCAATTTTAATCATTGTTACTCTGGAAACCAGAGATGGGCAAGTCCTGGGCCGACGCTGC TTTGAGGCCCGGATCTGTGCTTGCCCAGGAAGAGACAGGAAGGCGGATGAAGATAGCATCAGAAAGCAGCAAGTTTCGGACAGTACAAAG AACGGTGATGGTACGAAGCGCCCGTTTCGTCAGAACACACATGGTATCCAGATGACATCCATCAAGAAACGAAGATCCCCAGATGATGAA CTGTTATACTTACCAGTGAGGGGCCGTGAGACTTATGAAATGCTGTTGAAGATCAAAGAGTCCCTGGAACTCATGCAGTACCTTCCTCAG CACACAATTGAAACGTACAGGCAACAGCAACAGCAGCAGCACCAGCACTTACTTCAGAAACAGACCTCAATACAGTCTCCATCTTCATAT GGTAACAGCTCCCCACCTCTGAACAAAATGAACAGCATGAACAAGCTGCCTTCTGTGAGCCAGCTTATCAACCCTCAGCAGCGCAACGCC CTCACTCCTACAACCATTCCTGATGGCATGGGAGCCAACATTCCCATGATGGGCACCCACATGCCAATGGCTGGAGACATGAATGGACTC AGCCCCACCCAGGCACTCCCTCCCCCACTCTCCATGCCATCCACCTCCCACTGCACACCCCCACCTCCGTATCCCACAGATTGCAGCATT GTCAGGATCTGGCAAGTCTGA >1225_1225_4_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000392460_length(amino acids)=655AA_BP=121 MRGGGRMKMTVDFEECLKDSPRFRAALEEVEGDVAELELKLDKLVKLCIAMIDTGKAFCVANKQFMNGIRDLAQYSSNDAVVETSLTKFS DSLQEMINFHTILFDQTQRSIKAQLQNFVKDFVETPAHFSWKESYYRSTMSQSTQTNEFLSPEVFQHIWDFLEQPICSVQPIDLNFVDEP SEDGATNKIEISMDCIRMQDSDLSDPMWPQYTNLGLLNSMDQQIQNGSSSTSPYNTDHAQNSVTAPSPYAQPSSTFDALSPSPAIPSNTD YPGPHSFDVSFQQSSTAKSATWTYSTELKKLYCQIAKTCPIQIKVMTPPPQGAVIRAMPVYKKAEHVTEVVKRCPNHELSREFNEGQIAP PSHLIRVEGNSHAQYVEDPITGRQSVLVPYEPPQVGTEFTTVLYNFMCNSSCVGGMNRRPILIIVTLETRDGQVLGRRCFEARICACPGR DRKADEDSIRKQQVSDSTKNGDGTKRPFRQNTHGIQMTSIKKRRSPDDELLYLPVRGRETYEMLLKIKESLELMQYLPQHTIETYRQQQQ QQHQHLLQKQTSIQSPSSYGNSSPPLNKMNSMNKLPSVSQLINPQQRNALTPTTIPDGMGANIPMMGTHMPMAGDMNGLSPTQALPPPLS MPSTSHCTPPPPYPTDCSIVRIWQV -------------------------------------------------------------- >1225_1225_5_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000418709_length(transcript)=3295nt_BP=575nt CGTTTCCCTGCGGCCAGCGCTCTACTTGGGGCGGGAGGAAGAGGGGCGGGACTGGGGCCTGACTAGTCGTCGCCGTCTTCGCGGCTGCTA TGACCAGCGGGCGAGGCGCCCTTCGCAGCTCCGCGCAACGCTAGTCGCGGCCGTGCGCCCGCCGTAGCCGTCTGCGTCCCAGCGCGGCCG CTCCCGCGGCCCCCTCGGCTTTGCCAGCGCCGCCTGCGAGGCGGAGGCAGGATGAAGATGACTGTGGATTTCGAGGAGTGTCTGAAGGAC TCGCCCCGCTTCAGGGCAGCTTTGGAAGAAGTAGAAGGTGATGTGGCAGAATTGGAACTAAAACTTGATAAGCTTGTGAAACTTTGTATT GCAATGATTGATACTGGAAAAGCCTTTTGTGTTGCAAATAAACAGTTCATGAATGGGATTCGAGACCTGGCTCAGTATTCTAGTAATGAT GCTGTCGTTGAGACAAGTTTGACCAAGTTTTCTGACAGTCTTCAAGAAATGATAAATTTTCACACAATCCTGTTTGACCAAACTCAGAGA TCAATTAAGGCACAGCTTCAGAACTTTGTTAAAGATTTCGTAGAAACCCCAGCTCATTTCTCTTGGAAAGAAAGTTATTACCGATCCACC ATGTCCCAGAGCACACAGACAAATGAATTCCTCAGTCCAGAGGTTTTCCAGCATATCTGGGATTTTCTGGAACAGCCTATATGTTCAGTT CAGCCCATTGACTTGAACTTTGTGGATGAACCATCAGAAGATGGTGCGACAAACAAGATTGAGATTAGCATGGACTGTATCCGCATGCAG GACTCGGACCTGAGTGACCCCATGTGGCCACAGTACACGAACCTGGGGCTCCTGAACAGCATGGACCAGCAGATTCAGAACGGCTCCTCG TCCACCAGTCCCTATAACACAGACCACGCGCAGAACAGCGTCACGGCGCCCTCGCCCTACGCACAGCCCAGCTCCACCTTCGATGCTCTC TCTCCATCACCCGCCATCCCCTCCAACACCGACTACCCAGGCCCGCACAGTTTCGACGTGTCCTTCCAGCAGTCGAGCACCGCCAAGTCG GCCACCTGGACGTATTCCACTGAACTGAAGAAACTCTACTGCCAAATTGCAAAGACATGCCCCATCCAGATCAAGGTGATGACCCCACCT CCTCAGGGAGCTGTTATCCGCGCCATGCCTGTCTACAAAAAAGCTGAGCACGTCACGGAGGTGGTGAAGCGGTGCCCCAACCATGAGCTG AGCCGTGAATTCAACGAGGGACAGATTGCCCCTCCTAGTCATTTGATTCGAGTAGAGGGGAACAGCCATGCCCAGTATGTAGAAGATCCC ATCACAGGAAGACAGAGTGTGCTGGTACCTTATGAGCCACCCCAGGTTGGCACTGAATTCACGACAGTCTTGTACAATTTCATGTGTAAC AGCAGTTGTGTTGGAGGGATGAACCGCCGTCCAATTTTAATCATTGTTACTCTGGAAACCAGAGATGGGCAAGTCCTGGGCCGACGCTGC TTTGAGGCCCGGATCTGTGCTTGCCCAGGAAGAGACAGGAAGGCGGATGAAGATAGCATCAGAAAGCAGCAAGTTTCGGACAGTACAAAG AACGGTGATGGTACGAAGCGCCCGTTTCGTCAGAACACACATGGTATCCAGATGACATCCATCAAGAAACGAAGATCCCCAGATGATGAA CTGTTATACTTACCAGTGAGGGGCCGTGAGACTTATGAAATGCTGTTGAAGATCAAAGAGTCCCTGGAACTCATGCAGTACCTTCCTCAG CACACAATTGAAACGTACAGGCAACAGCAACAGCAGCAGCACCAGCACTTACTTCAGAAACATCTCCTTTCAGCCTGCTTCAGGAATGAG CTTGTGGAGCCCCGGAGAGAAACTCCAAAACAATCTGACGTCTTCTTTAGACATTCCAAGCCCCCAAACCGATCAGTGTACCCATAGAGC CCTATCTCTATATTTTAAGTGTGTGTGTTGTATTTCCATGTGTATATGTGAGTGTGTGTGTGTGTATGTGTGTGCGTGTGTATCTAGCCC TCATAAACAGGACTTGAAGACACTTTGGCTCAGAGACCCAACTGCTCAAAGGCACAAAGCCACTAGTGAGAGAATCTTTTGAAGGGACTC AAACCTTTACAAGAAAGGATGTTTTCTGCAGATTTTGTATCCTTAGACCGGCCATTGGTGGGTGAGGAACCACTGTGTTTGTCTGTGAGC TTTCTGTTGTTTCCTGGGAGGGAGGGGTCAGGTGGGGAAAGGGGCATTAAGATGTTTATTGGAACCCTTTTCTGTCTTCTTCTGTTGTTT TTCTAAAATTCACAGGGAAGCTTTTGAGCAGGTCTCAAACTTAAGATGTCTTTTTAAGAAAAGGAGAAAAAAGTTGTTATTGTCTGTGCA TAAGTAAGTTGTAGGTGACTGAGAGACTCAGTCAGACCCTTTTAATGCTGGTCATGTAATAATATTGCAAGTAGTAAGAAACGAAGGTGT CAAGTGTACTGCTGGGCAGCGAGGTGATCATTACCAAAAGTAATCAACTTTGTGGGTGGAGAGTTCTTTGTGAGAACTTGCATTATTTGT GTCCTCCCCTCATGTGTAGGTAGAACATTTCTTAATGCTGTGTACCTGCCTCTGCCACTGTATGTTGGCATCTGTTATGCTAAAGTTTTT CTTGTACATGAAACCCTGGAAGACCTACTACAAAAAAACTGTTGTTTGGCCCCCATAGCAGGTGAACTCATTTTGTGCTTTTAATAGAAA GACAAATCCACCCCAGTAATATTGCCCTTACGTAGTTGTTTACCATTATTCAAAGCTCAAAATAGAATTTGAAGCCCTCTCACAAAATCT GTGATTAATTTGCTTAATTAGAGCTTCTATCCCTCAAGCCTACCTACCATAAAACCAGCCATATTACTGATACTGTTCAGTGCATTTAGC CAGGAGACTTACGTTTTGAGTAAGTGAGATCCAAGCAGACGTGTTAAAATCAGCACTCCTGGACTGGAAATTAAAGATTGAAAGGGTAGA CTACTTTTCTTTTTTTTACTCAAAAGTTTAGAGAATCTCTGTTTCTTTCCATTTTAAAAACATATTTTAAGATAATAGCATAAAGACTTT AAAAATGTTCCTCCCCTCCATCTTCCCACACCCAGTCACCAGCACTGTATTTTCTGTCACCAAGACAATGATTTCTTGTTATTGAGGCTG TTGCTTTTGTGGATGTGTGATTTTAATTTTCAATAAACTTTTGCATCTTGGTTTA >1225_1225_5_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000418709_length(amino acids)=587AA_BP=121 MRGGGRMKMTVDFEECLKDSPRFRAALEEVEGDVAELELKLDKLVKLCIAMIDTGKAFCVANKQFMNGIRDLAQYSSNDAVVETSLTKFS DSLQEMINFHTILFDQTQRSIKAQLQNFVKDFVETPAHFSWKESYYRSTMSQSTQTNEFLSPEVFQHIWDFLEQPICSVQPIDLNFVDEP SEDGATNKIEISMDCIRMQDSDLSDPMWPQYTNLGLLNSMDQQIQNGSSSTSPYNTDHAQNSVTAPSPYAQPSSTFDALSPSPAIPSNTD YPGPHSFDVSFQQSSTAKSATWTYSTELKKLYCQIAKTCPIQIKVMTPPPQGAVIRAMPVYKKAEHVTEVVKRCPNHELSREFNEGQIAP PSHLIRVEGNSHAQYVEDPITGRQSVLVPYEPPQVGTEFTTVLYNFMCNSSCVGGMNRRPILIIVTLETRDGQVLGRRCFEARICACPGR DRKADEDSIRKQQVSDSTKNGDGTKRPFRQNTHGIQMTSIKKRRSPDDELLYLPVRGRETYEMLLKIKESLELMQYLPQHTIETYRQQQQ QQHQHLLQKHLLSACFRNELVEPRRETPKQSDVFFRHSKPPNRSVYP -------------------------------------------------------------- >1225_1225_6_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000440651_length(transcript)=2544nt_BP=575nt CGTTTCCCTGCGGCCAGCGCTCTACTTGGGGCGGGAGGAAGAGGGGCGGGACTGGGGCCTGACTAGTCGTCGCCGTCTTCGCGGCTGCTA TGACCAGCGGGCGAGGCGCCCTTCGCAGCTCCGCGCAACGCTAGTCGCGGCCGTGCGCCCGCCGTAGCCGTCTGCGTCCCAGCGCGGCCG CTCCCGCGGCCCCCTCGGCTTTGCCAGCGCCGCCTGCGAGGCGGAGGCAGGATGAAGATGACTGTGGATTTCGAGGAGTGTCTGAAGGAC TCGCCCCGCTTCAGGGCAGCTTTGGAAGAAGTAGAAGGTGATGTGGCAGAATTGGAACTAAAACTTGATAAGCTTGTGAAACTTTGTATT GCAATGATTGATACTGGAAAAGCCTTTTGTGTTGCAAATAAACAGTTCATGAATGGGATTCGAGACCTGGCTCAGTATTCTAGTAATGAT GCTGTCGTTGAGACAAGTTTGACCAAGTTTTCTGACAGTCTTCAAGAAATGATAAATTTTCACACAATCCTGTTTGACCAAACTCAGAGA TCAATTAAGGCACAGCTTCAGAACTTTGTTAAAGATTTCGTAGAAACCCCAGCTCATTTCTCTTGGAAAGAAAGTTATTACCGATCCACC ATGTCCCAGAGCACACAGACAAATGAATTCCTCAGTCCAGAGGTTTTCCAGCATATCTGGGATTTTCTGGAACAGCCTATATGTTCAGTT CAGCCCATTGACTTGAACTTTGTGGATGAACCATCAGAAGATGGTGCGACAAACAAGATTGAGATTAGCATGGACTGTATCCGCATGCAG GACTCGGACCTGAGTGACCCCATGTGGCCACAGTACACGAACCTGGGGCTCCTGAACAGCATGGACCAGCAGATTCAGAACGGCTCCTCG TCCACCAGTCCCTATAACACAGACCACGCGCAGAACAGCGTCACGGCGCCCTCGCCCTACGCACAGCCCAGCTCCACCTTCGATGCTCTC TCTCCATCACCCGCCATCCCCTCCAACACCGACTACCCAGGCCCGCACAGTTTCGACGTGTCCTTCCAGCAGTCGAGCACCGCCAAGTCG GCCACCTGGACGTATTCCACTGAACTGAAGAAACTCTACTGCCAAATTGCAAAGACATGCCCCATCCAGATCAAGGTGATGACCCCACCT CCTCAGGGAGCTGTTATCCGCGCCATGCCTGTCTACAAAAAAGCTGAGCACGTCACGGAGGTGGTGAAGCGGTGCCCCAACCATGAGCTG AGCCGTGAATTCAACGAGGGACAGATTGCCCCTCCTAGTCATTTGATTCGAGTAGAGGGGAACAGCCATGCCCAGTATGTAGAAGATCCC ATCACAGGAAGACAGAGTGTGCTGGTACCTTATGAGCCACCCCAGGTTGGCACTGAATTCACGACAGTCTTGTACAATTTCATGTGTAAC AGCAGTTGTGTTGGAGGGATGAACCGCCGTCCAATTTTAATCATTGTTACTCTGGAAACCAGAGATGGGCAAGTCCTGGGCCGACGCTGC TTTGAGGCCCGGATCTGTGCTTGCCCAGGAAGAGACAGGAAGGCGGATGAAGATAGCATCAGAAAGCAGCAAGTTTCGGACAGTACAAAG AACGGTGATGCGTTTCGTCAGAACACACATGGTATCCAGATGACATCCATCAAGAAACGAAGATCCCCAGATGATGAACTGTTATACTTA CCAGTGAGGGGCCGTGAGACTTATGAAATGCTGTTGAAGATCAAAGAGTCCCTGGAACTCATGCAGTACCTTCCTCAGCACACAATTGAA ACGTACAGGCAACAGCAACAGCAGCAGCACCAGCACTTACTTCAGAAACAGACCTCAATACAGTCTCCATCTTCATATGGTAACAGCTCC CCACCTCTGAACAAAATGAACAGCATGAACAAGCTGCCTTCTGTGAGCCAGCTTATCAACCCTCAGCAGCGCAACGCCCTCACTCCTACA ACCATTCCTGATGGCATGGGAGCCAACATTCCCATGATGGGCACCCACATGCCAATGGCTGGAGACATGAATGGACTCAGCCCCACCCAG GCACTCCCTCCCCCACTCTCCATGCCATCCACCTCCCACTGCACACCCCCACCTCCGTATCCCACAGATTGCAGCATTGTCAGTTTCTTA GCGAGGTTGGGCTGTTCATCATGTCTGGACTATTTCACGACCCAGGGGCTGACCACCATCTATCAGATTGAGCATTACTCCATGGATGAT CTGGCAAGTCTGAAAATCCCTGAGCAATTTCGACATGCGATCTGGAAGGGCATCCTGGACCACCGGCAGCTCCACGAATTCTCCTCCCCT TCTCATCTCCTGCGGACCCCAAGCAGTGCCTCTACAGTCAGTGTGGGCTCCAGTGAGACCCGGGGTGAGCGTGTTATTGATGCTGTGCGA TTCACCCTCCGCCAGACCATCTCTTTCCCACCCCGAGATGAGTGGAATGACTTCAACTTTGACATGGATGCTCGCCGCAATAAGCAACAG CGCATCAAAGAGGAGGGGGAGTGA >1225_1225_6_ACAP2-TP63_ACAP2_chr3_195065988_ENST00000326793_TP63_chr3_189455529_ENST00000440651_length(amino acids)=776AA_BP=121 MRGGGRMKMTVDFEECLKDSPRFRAALEEVEGDVAELELKLDKLVKLCIAMIDTGKAFCVANKQFMNGIRDLAQYSSNDAVVETSLTKFS DSLQEMINFHTILFDQTQRSIKAQLQNFVKDFVETPAHFSWKESYYRSTMSQSTQTNEFLSPEVFQHIWDFLEQPICSVQPIDLNFVDEP SEDGATNKIEISMDCIRMQDSDLSDPMWPQYTNLGLLNSMDQQIQNGSSSTSPYNTDHAQNSVTAPSPYAQPSSTFDALSPSPAIPSNTD YPGPHSFDVSFQQSSTAKSATWTYSTELKKLYCQIAKTCPIQIKVMTPPPQGAVIRAMPVYKKAEHVTEVVKRCPNHELSREFNEGQIAP PSHLIRVEGNSHAQYVEDPITGRQSVLVPYEPPQVGTEFTTVLYNFMCNSSCVGGMNRRPILIIVTLETRDGQVLGRRCFEARICACPGR DRKADEDSIRKQQVSDSTKNGDAFRQNTHGIQMTSIKKRRSPDDELLYLPVRGRETYEMLLKIKESLELMQYLPQHTIETYRQQQQQQHQ HLLQKQTSIQSPSSYGNSSPPLNKMNSMNKLPSVSQLINPQQRNALTPTTIPDGMGANIPMMGTHMPMAGDMNGLSPTQALPPPLSMPST SHCTPPPPYPTDCSIVSFLARLGCSSCLDYFTTQGLTTIYQIEHYSMDDLASLKIPEQFRHAIWKGILDHRQLHEFSSPSHLLRTPSSAS TVSVGSSETRGERVIDAVRFTLRQTISFPPRDEWNDFNFDMDARRNKQQRIKEEGE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACAP2-TP63 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000264731 | 0 | 14 | 352_388 | 20.666666666666668 | 681.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000320472 | 0 | 12 | 352_388 | 20.666666666666668 | 511.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000354600 | 0 | 12 | 352_388 | 0 | 587.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000382063 | 0 | 13 | 352_388 | 20.666666666666668 | 596.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392460 | 0 | 13 | 352_388 | 20.666666666666668 | 556.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392461 | 0 | 10 | 352_388 | 0.0 | 417.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000392463 | 0 | 11 | 352_388 | 0.0 | 462.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000418709 | 0 | 11 | 352_388 | 20.666666666666668 | 488.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000437221 | 0 | 9 | 352_388 | 0.0 | 394.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000440651 | 0 | 14 | 352_388 | 20.666666666666668 | 677.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000449992 | 0 | 11 | 352_388 | 0.0 | 502.0 | HIPK2 | |
| Tgene | TP63 | chr3:195065988 | chr3:189455529 | ENST00000456148 | 0 | 12 | 352_388 | 0.0 | 583.0 | HIPK2 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACAP2-TP63 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ACAP2-TP63 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C1785148 | RAPP-HODGKIN SYNDROME | 7 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0406709 | Hay-Wells syndrome | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C1858562 | ECTRODACTYLY, ECTODERMAL DYSPLASIA, AND CLEFT LIP/PALATE SYNDROME 3 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0008924 | Cleft upper lip | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C0406704 | Rudiger syndrome 1 | 3 | CTD_human;ORPHANET | |
| Tgene | C1863204 | ADULT SYNDROME | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0024121 | Lung Neoplasms | 2 | CTD_human | |
| Tgene | C0158646 | Cleft palate with cleft lip | 2 | ORPHANET | |
| Tgene | C0242379 | Malignant neoplasm of lung | 2 | CTD_human | |
| Tgene | C1298692 | Cleft lip and alveolus | 2 | ORPHANET | |
| Tgene | C1851841 | ECTRODACTYLY, ECTODERMAL DYSPLASIA, AND CLEFT LIP/PALATE SYNDROME 1 | 2 | ORPHANET | |
| Tgene | C1854442 | SPLIT-HAND/FOOT MALFORMATION 4 | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0000772 | Multiple congenital anomalies | 1 | CTD_human | |
| Tgene | C0001418 | Adenocarcinoma | 1 | CTD_human | |
| Tgene | C0005689 | Bladder Exstrophy | 1 | ORPHANET | |
| Tgene | C0006145 | Breast Diseases | 1 | CTD_human | |
| Tgene | C0007097 | Carcinoma | 1 | CTD_human | |
| Tgene | C0007137 | Squamous cell carcinoma | 1 | CTD_human | |
| Tgene | C0007621 | Neoplastic Cell Transformation | 1 | CTD_human | |
| Tgene | C0008925 | Cleft Palate | 1 | CTD_human | |
| Tgene | C0009402 | Colorectal Carcinoma | 1 | CTD_human | |
| Tgene | C0009404 | Colorectal Neoplasms | 1 | CTD_human | |
| Tgene | C0016508 | Congenital Foot Deformity | 1 | CTD_human | |
| Tgene | C0018566 | Congenital Hand Deformities | 1 | CTD_human | |
| Tgene | C0027627 | Neoplasm Metastasis | 1 | CTD_human | |
| Tgene | C0030297 | Pancreatic Neoplasm | 1 | CTD_human | |
| Tgene | C0037268 | Skin Abnormalities | 1 | CTD_human | |
| Tgene | C0038987 | Sweat Gland Neoplasms | 1 | CTD_human | |
| Tgene | C0152013 | Adenocarcinoma of lung (disorder) | 1 | CTD_human | |
| Tgene | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human | |
| Tgene | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human | |
| Tgene | C0205643 | Carcinoma, Cribriform | 1 | CTD_human | |
| Tgene | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human | |
| Tgene | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human | |
| Tgene | C0205696 | Anaplastic carcinoma | 1 | CTD_human | |
| Tgene | C0205697 | Carcinoma, Spindle-Cell | 1 | CTD_human | |
| Tgene | C0205698 | Undifferentiated carcinoma | 1 | CTD_human | |
| Tgene | C0205699 | Carcinomatosis | 1 | CTD_human | |
| Tgene | C0206720 | Squamous Cell Neoplasms | 1 | CTD_human | |
| Tgene | C0206762 | Limb Deformities, Congenital | 1 | CTD_human | |
| Tgene | C0265554 | Ectrodactyly | 1 | ORPHANET | |
| Tgene | C0346647 | Malignant neoplasm of pancreas | 1 | CTD_human | |
| Tgene | C0376634 | Craniofacial Abnormalities | 1 | CTD_human | |
| Tgene | C0751688 | Malignant Squamous Cell Neoplasm | 1 | CTD_human | |
| Tgene | C0919267 | ovarian neoplasm | 1 | CTD_human | |
| Tgene | C1140680 | Malignant neoplasm of ovary | 1 | CTD_human;UNIPROT | |
| Tgene | C1449718 | Endocrine Breast Diseases | 1 | CTD_human | |
| Tgene | C1720887 | Female Urogenital Diseases | 1 | CTD_human | |
| Tgene | C1837218 | Cleft palate, isolated | 1 | CTD_human | |
| Tgene | C1851878 | OROFACIAL CLEFT 8 | 1 | UNIPROT | |
| Tgene | C1863753 | LIMB-MAMMARY SYNDROME | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
