
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FPGS-GARNL3 (FusionGDB2 ID:HG2356TG84253) |
Fusion Gene Summary for FPGS-GARNL3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FPGS-GARNL3 | Fusion gene ID: hg2356tg84253 | Hgene | Tgene | Gene symbol | FPGS | GARNL3 | Gene ID | 2356 | 84253 |
| Gene name | folylpolyglutamate synthase | GTPase activating Rap/RanGAP domain like 3 | |
| Synonyms | - | bA356B19.1 | |
| Cytomap | ('FPGS')('GARNL3') 9q34.11 | 9q33.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | folylpolyglutamate synthase, mitochondrialfolylpoly-gamma-glutamate synthetasetetrahydrofolate synthasetetrahydrofolylpolyglutamate synthase | GTPase-activating Rap/Ran-GAP domain-like protein 3GTPase activating RANGAP domain-like 3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q05932 | Q5VVW2 | |
| Ensembl transtripts involved in fusion gene | ENST00000373245, ENST00000460181, ENST00000373225, ENST00000373247, ENST00000393706, | ||
| Fusion gene scores | * DoF score | 6 X 6 X 5=180 | 8 X 10 X 8=640 |
| # samples | 8 | 11 | |
| ** MAII score | log2(8/180*10)=-1.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/640*10)=-2.5405683813627 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: FPGS [Title/Abstract] AND GARNL3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FPGS(130575602)-GARNL3(130145717), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | FPGS-GARNL3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. FPGS-GARNL3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. FPGS-GARNL3 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. FPGS-GARNL3 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FPGS | GO:0006536 | glutamate metabolic process | 3619447 |
| Hgene | FPGS | GO:0006760 | folic acid-containing compound metabolic process | 3619447 |
| Hgene | FPGS | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process | 3619447 |
 Fusion gene breakpoints across FPGS (5'-gene) Fusion gene breakpoints across FPGS (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
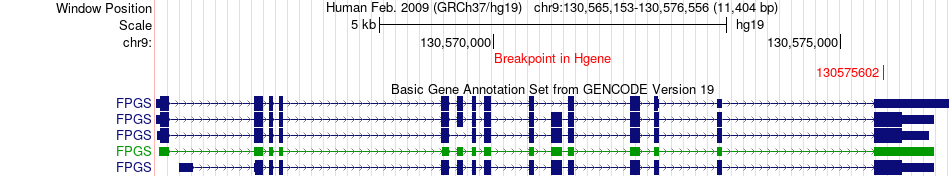 |
 Fusion gene breakpoints across GARNL3 (3'-gene) Fusion gene breakpoints across GARNL3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
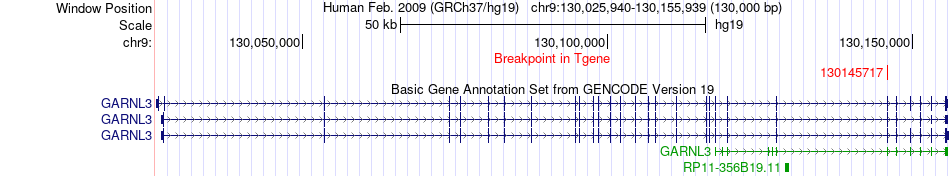 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-EO-A1Y7-01A | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
Top |
Fusion Gene ORF analysis for FPGS-GARNL3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000373245 | ENST00000314904 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3CDS | ENST00000373245 | ENST00000373387 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3CDS | ENST00000373245 | ENST00000435213 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3CDS | ENST00000460181 | ENST00000314904 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3CDS | ENST00000460181 | ENST00000373387 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3CDS | ENST00000460181 | ENST00000435213 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3UTR | ENST00000373245 | ENST00000496711 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 3UTR-3UTR | ENST00000460181 | ENST00000496711 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 5CDS-3UTR | ENST00000373225 | ENST00000496711 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 5CDS-3UTR | ENST00000373247 | ENST00000496711 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| 5CDS-3UTR | ENST00000393706 | ENST00000496711 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| Frame-shift | ENST00000373225 | ENST00000314904 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| Frame-shift | ENST00000373225 | ENST00000373387 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| Frame-shift | ENST00000373225 | ENST00000435213 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| Frame-shift | ENST00000393706 | ENST00000314904 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| Frame-shift | ENST00000393706 | ENST00000373387 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| Frame-shift | ENST00000393706 | ENST00000435213 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| In-frame | ENST00000373247 | ENST00000314904 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| In-frame | ENST00000373247 | ENST00000373387 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
| In-frame | ENST00000373247 | ENST00000435213 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000373247 | FPGS | chr9 | 130575602 | + | ENST00000435213 | GARNL3 | chr9 | 130145717 | + | 3178 | 2145 | 50 | 1684 | 544 |
| ENST00000373247 | FPGS | chr9 | 130575602 | + | ENST00000314904 | GARNL3 | chr9 | 130145717 | + | 3180 | 2145 | 50 | 1684 | 544 |
| ENST00000373247 | FPGS | chr9 | 130575602 | + | ENST00000373387 | GARNL3 | chr9 | 130145717 | + | 3432 | 2145 | 50 | 1684 | 544 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000373247 | ENST00000435213 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + | 0.056083415 | 0.94391656 |
| ENST00000373247 | ENST00000314904 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + | 0.043278966 | 0.95672107 |
| ENST00000373247 | ENST00000373387 | FPGS | chr9 | 130575602 | + | GARNL3 | chr9 | 130145717 | + | 0.033720464 | 0.96627957 |
Top |
Fusion Genomic Features for FPGS-GARNL3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
Top |
Fusion Protein Features for FPGS-GARNL3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:130575602/chr9:130145717) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FPGS | GARNL3 |
| FUNCTION: Catalyzes conversion of folates to polyglutamate derivatives allowing concentration of folate compounds in the cell and the intracellular retention of these cofactors, which are important substrates for most of the folate-dependent enzymes that are involved in one-carbon transfer reactions involved in purine, pyrimidine and amino acid synthesis. Unsubstituted reduced folates are the preferred substrates. Metabolizes methotrexate (MTX) to polyglutamates. {ECO:0000269|PubMed:8408018, ECO:0000269|PubMed:8408019, ECO:0000269|PubMed:8408021, ECO:0000269|PubMed:8662720}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FPGS | chr9:130575602 | chr9:130145717 | ENST00000373225 | + | 1 | 15 | 106_109 | 0 | 538.0 | Nucleotide binding | ATP |
| Hgene | FPGS | chr9:130575602 | chr9:130145717 | ENST00000373247 | + | 1 | 15 | 106_109 | 0 | 588.0 | Nucleotide binding | ATP |
| Hgene | FPGS | chr9:130575602 | chr9:130145717 | ENST00000393706 | + | 1 | 14 | 106_109 | 0 | 562.0 | Nucleotide binding | ATP |
| Tgene | GARNL3 | chr9:130575602 | chr9:130145717 | ENST00000314904 | 21 | 27 | 191_407 | 720 | 811.6666666666666 | Domain | Rap-GAP | |
| Tgene | GARNL3 | chr9:130575602 | chr9:130145717 | ENST00000314904 | 21 | 27 | 489_800 | 720 | 811.6666666666666 | Domain | CNH | |
| Tgene | GARNL3 | chr9:130575602 | chr9:130145717 | ENST00000373387 | 21 | 28 | 191_407 | 720 | 1014.0 | Domain | Rap-GAP | |
| Tgene | GARNL3 | chr9:130575602 | chr9:130145717 | ENST00000373387 | 21 | 28 | 489_800 | 720 | 1014.0 | Domain | CNH |
Top |
Fusion Gene Sequence for FPGS-GARNL3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31261_31261_1_FPGS-GARNL3_FPGS_chr9_130575602_ENST00000373247_GARNL3_chr9_130145717_ENST00000314904_length(transcript)=3180nt_BP=2145nt GCGGGGCGTCTCCCGCCCGGGCCTAGAGCGCTGCCGGGGGCGCCGGGACTATGTCGCGGGCGCGGAGCCACCTGCGCGCCGCTCTATTCC TGGCAGCGGCGTCTGCGCGCGGCATAACGACCCAGGTCGCGGCGCGGCGGGGCTTGAGCGCGTGGCCGGTGCCGCAGGAGCCGAGCATGG AGTACCAGGATGCCGTGCGCATGCTCAATACCCTGCAGACCAATGCCGGCTACCTGGAGCAGGTGAAGCGCCAGCGGGGTGACCCTCAGA CACAGTTGGAAGCCATGGAACTGTACCTGGCACGGAGTGGGCTGCAGGTGGAGGACTTGGACCGGCTGAACATCATCCACGTCACTGGGA CGAAGGGGAAGGGCTCCACCTGTGCCTTCACGGAATGTATCCTCCGAAGCTATGGCCTGAAGACGGGATTCTTTAGCTCTCCCCACCTGG TGCAGGTTCGGGAGCGGATCCGCATCAATGGGCAGCCCATCAGTCCTGAGCTCTTCACCAAGTACTTCTGGCGCCTCTACCACCGGCTGG AGGAGACCAAGGATGGCAGCTGTGTCTCCATGCCCCCCTACTTCCGCTTCCTGACACTCATGGCCTTCCACGTCTTCCTCCAAGAGAAGG TGGACCTGGCAGTGGTGGAGGTGGGCATTGGCGGGGCTTATGACTGCACCAACATCATCAGGAAGCCTGTGGTGTGCGGAGTCTCCTCTC TTGGCATCGACCACACCAGCCTCCTGGGGGATACGGTGGAGAAGATCGCATGGCAGAAAGGGGGCATCTTTAAGCAAGGTGTCCCTGCCT TCACTGTGCTCCAACCTGAAGGTCCCCTGGCAGTGCTGAGGGACCGAGCCCAGCAGATCTCATGTCCTCTATACCTGTGTCCGATGCTGG AGGCCCTCGAGGAAGGGGGGCCGCCGCTGACCCTGGGCCTGGAGGGGGAGCACCAGCGGTCCAACGCCGCCTTGGCCTTGCAGCTGGCCC ACTGCTGGCTGCAGCGGCAGGACCGCCATGGTGCTGGGGAGCCAAAGGCATCCAGGCCAGGGCTCCTGTGGCAGCTGCCCCTGGCACCTG TGTTCCAGCCCACATCCCACATGCGGCTCGGGCTTCGGAACACGGAGTGGCCGGGCCGGACGCAGGTGCTGCGGCGCGGGCCCCTCACCT GGTACCTGGACGGTGCGCACACCGCCAGCAGCGCGCAGGCCTGCGTGCGCTGGTTCCGCCAGGCGCTGCAGGGCCGCGAGAGGCCGAGCG GTGGCCCCGAGGTTCGAGTCTTGCTCTTCAATGCTACCGGGGACCGGGACCCGGCGGCCCTGCTGAAGCTGCTGCAGCCCTGCCAGTTTG ACTATGCCGTCTTCTGCCCTAACCTGACAGAGGTGTCATCCACAGGCAACGCAGGTGGGTCCGCATCCCTGCTTCTGGCGCCCCACCCAC CCCACACCTGCAGTGCCAGCTCCCTCGTCTTCAGCTGCATTTCACATGCCTTGCAATGGATCAGCCAAGGCCGAGACCCCATCTTCCAGC CACCTAGTCCCCCAAAGGGCCTCCTCACCCACCCTGTGGCTCACAGTGGGGCCAGCATACTCCGTGAGGCTGCTGCCATCCATGTGCTAG TCACTGGCAGCCTGCACCTGGTGGGTGGTGTCCTGAAGCTGCTGGAGCCCGCACTGTCCCAGTAGCCAAGGCCCGGGGTTGGAGGTGGGA GCTTCCCACACCTGCCTGCGTTCTCCCCATGAACTTACATACTAGGTGCCTTTTGTTTTTGGCTTTCCTGGTTCTGTCTAGACTGGCCTA GGGGCCAGGGCTTTGGGATGGGAGGCCGGGAGAGGATGTCTTTTTTAAGGCTCTGTGCCTTGGTCTCTCCTTCCTCTTGGCTGAGATAGC AGAGGGGCTCCCCGGGTCTCTCACTGTTGCAGTGGCCTGGCCGTTCAGCCTGTCTCCCCCAACACCCCGCCTGCCTCCTGGCTCAGGCCC AGCTTATTGTGTGCGCTGCCTGGCCAGGCCCTGGGTCTTGCCATGTGCTGGGTGGTAGATTTCCTCCTCCCAGTGCCTTCTGGGAAGGGA GAGGGCCTCTGCCTGGGACACTGCGGGACAGAGGGTGGCTGGAGTGAATTAAAGCCTTTGTTTTTTAAAGAAATGACAGTTGCATCTATA AAAAGGTTTGCCCCTTTAATGGTGGCTCTTTTTTGGTTCAACCTTCTGCGTCAGATTTCCAGTTCTGTTGGAACCAGGCTCCCTATGCAA TTGTCTGTGCTTTCCCGTATCTCCTGGCCTTCACCACCGACTCCATGGAGATCCGCCTGGTGGTGAACGGGAACCTGGTCCACACTGCAG TCGTGCCGCAGCTGCAGCTGGTGGCCTCCAGGGTGAAATTCAATCAAAAAATCTGTACAAGATTCCACTTAGAAACCTCGTGGGCAGAAG CATCGAACGACCTCTGAAGTCACCCTTAGTCTCCAAGGTCATCACCCCACCCACTCCCATCAGTGTGGGCCTTGCTGCCATTCCAGTCAC GCACTCCTTGTCCCTGTCTCGCATGGAGATCAAAGAAATAGCAAGCAGGACCCGCAGGGAACTACTGGGCCTCTCGGATGAAGGTGGACC CAAGTCAGAAGGAGCGCCAAAGGCCAAATCAAAACCCCGGAAGCGGTTAGAAGAAAGCCAAGGAGGCCCCAAGCCAGGGGCAGTGAGGTC ATCTAGCAGTGACAGGATCCCATCAGGCTCCTTGGAAAGTGCTTCTACTTCCGAAGCCAACCCTGAGGGGCACTCAGCCAGCTCTGACCA GGACCCTGTGGCAGACAGAGAGGGCAGCCCGGTCTCCGGCAGCAGCCCCTTCCAGCTCACGGCTTTCTCCGATGAAGACATTATAGACTT GAAGTAACAGAGTTGAATCTCATTTGCCATCTTTAGTTTTCTTATGGAGGTTTATACTCTTTAAACAGTTCTGATGTAATTTCTCAACAA AATGTGGCTTTTAGCCTGTCAGTGATCTATTGGACCAAACCTTCTGCACACTCGGCCAGTTCCCTCTCCAATGTCCGGTGCCATCTTTCC TGACCTTTGTTTCTTTCTGTTCAGGAACCATCAGTCCCCTTGTAATAAAGGTGGTAGATTTCATTGAGGTTTTAGATTGAAACTTTGAAT AAATCAAAAATACTCATTCTTATTTACTGC >31261_31261_1_FPGS-GARNL3_FPGS_chr9_130575602_ENST00000373247_GARNL3_chr9_130145717_ENST00000314904_length(amino acids)=544AA_BP= MSRARSHLRAALFLAAASARGITTQVAARRGLSAWPVPQEPSMEYQDAVRMLNTLQTNAGYLEQVKRQRGDPQTQLEAMELYLARSGLQV EDLDRLNIIHVTGTKGKGSTCAFTECILRSYGLKTGFFSSPHLVQVRERIRINGQPISPELFTKYFWRLYHRLEETKDGSCVSMPPYFRF LTLMAFHVFLQEKVDLAVVEVGIGGAYDCTNIIRKPVVCGVSSLGIDHTSLLGDTVEKIAWQKGGIFKQGVPAFTVLQPEGPLAVLRDRA QQISCPLYLCPMLEALEEGGPPLTLGLEGEHQRSNAALALQLAHCWLQRQDRHGAGEPKASRPGLLWQLPLAPVFQPTSHMRLGLRNTEW PGRTQVLRRGPLTWYLDGAHTASSAQACVRWFRQALQGRERPSGGPEVRVLLFNATGDRDPAALLKLLQPCQFDYAVFCPNLTEVSSTGN AGGSASLLLAPHPPHTCSASSLVFSCISHALQWISQGRDPIFQPPSPPKGLLTHPVAHSGASILREAAAIHVLVTGSLHLVGGVLKLLEP ALSQ -------------------------------------------------------------- >31261_31261_2_FPGS-GARNL3_FPGS_chr9_130575602_ENST00000373247_GARNL3_chr9_130145717_ENST00000373387_length(transcript)=3432nt_BP=2145nt GCGGGGCGTCTCCCGCCCGGGCCTAGAGCGCTGCCGGGGGCGCCGGGACTATGTCGCGGGCGCGGAGCCACCTGCGCGCCGCTCTATTCC TGGCAGCGGCGTCTGCGCGCGGCATAACGACCCAGGTCGCGGCGCGGCGGGGCTTGAGCGCGTGGCCGGTGCCGCAGGAGCCGAGCATGG AGTACCAGGATGCCGTGCGCATGCTCAATACCCTGCAGACCAATGCCGGCTACCTGGAGCAGGTGAAGCGCCAGCGGGGTGACCCTCAGA CACAGTTGGAAGCCATGGAACTGTACCTGGCACGGAGTGGGCTGCAGGTGGAGGACTTGGACCGGCTGAACATCATCCACGTCACTGGGA CGAAGGGGAAGGGCTCCACCTGTGCCTTCACGGAATGTATCCTCCGAAGCTATGGCCTGAAGACGGGATTCTTTAGCTCTCCCCACCTGG TGCAGGTTCGGGAGCGGATCCGCATCAATGGGCAGCCCATCAGTCCTGAGCTCTTCACCAAGTACTTCTGGCGCCTCTACCACCGGCTGG AGGAGACCAAGGATGGCAGCTGTGTCTCCATGCCCCCCTACTTCCGCTTCCTGACACTCATGGCCTTCCACGTCTTCCTCCAAGAGAAGG TGGACCTGGCAGTGGTGGAGGTGGGCATTGGCGGGGCTTATGACTGCACCAACATCATCAGGAAGCCTGTGGTGTGCGGAGTCTCCTCTC TTGGCATCGACCACACCAGCCTCCTGGGGGATACGGTGGAGAAGATCGCATGGCAGAAAGGGGGCATCTTTAAGCAAGGTGTCCCTGCCT TCACTGTGCTCCAACCTGAAGGTCCCCTGGCAGTGCTGAGGGACCGAGCCCAGCAGATCTCATGTCCTCTATACCTGTGTCCGATGCTGG AGGCCCTCGAGGAAGGGGGGCCGCCGCTGACCCTGGGCCTGGAGGGGGAGCACCAGCGGTCCAACGCCGCCTTGGCCTTGCAGCTGGCCC ACTGCTGGCTGCAGCGGCAGGACCGCCATGGTGCTGGGGAGCCAAAGGCATCCAGGCCAGGGCTCCTGTGGCAGCTGCCCCTGGCACCTG TGTTCCAGCCCACATCCCACATGCGGCTCGGGCTTCGGAACACGGAGTGGCCGGGCCGGACGCAGGTGCTGCGGCGCGGGCCCCTCACCT GGTACCTGGACGGTGCGCACACCGCCAGCAGCGCGCAGGCCTGCGTGCGCTGGTTCCGCCAGGCGCTGCAGGGCCGCGAGAGGCCGAGCG GTGGCCCCGAGGTTCGAGTCTTGCTCTTCAATGCTACCGGGGACCGGGACCCGGCGGCCCTGCTGAAGCTGCTGCAGCCCTGCCAGTTTG ACTATGCCGTCTTCTGCCCTAACCTGACAGAGGTGTCATCCACAGGCAACGCAGGTGGGTCCGCATCCCTGCTTCTGGCGCCCCACCCAC CCCACACCTGCAGTGCCAGCTCCCTCGTCTTCAGCTGCATTTCACATGCCTTGCAATGGATCAGCCAAGGCCGAGACCCCATCTTCCAGC CACCTAGTCCCCCAAAGGGCCTCCTCACCCACCCTGTGGCTCACAGTGGGGCCAGCATACTCCGTGAGGCTGCTGCCATCCATGTGCTAG TCACTGGCAGCCTGCACCTGGTGGGTGGTGTCCTGAAGCTGCTGGAGCCCGCACTGTCCCAGTAGCCAAGGCCCGGGGTTGGAGGTGGGA GCTTCCCACACCTGCCTGCGTTCTCCCCATGAACTTACATACTAGGTGCCTTTTGTTTTTGGCTTTCCTGGTTCTGTCTAGACTGGCCTA GGGGCCAGGGCTTTGGGATGGGAGGCCGGGAGAGGATGTCTTTTTTAAGGCTCTGTGCCTTGGTCTCTCCTTCCTCTTGGCTGAGATAGC AGAGGGGCTCCCCGGGTCTCTCACTGTTGCAGTGGCCTGGCCGTTCAGCCTGTCTCCCCCAACACCCCGCCTGCCTCCTGGCTCAGGCCC AGCTTATTGTGTGCGCTGCCTGGCCAGGCCCTGGGTCTTGCCATGTGCTGGGTGGTAGATTTCCTCCTCCCAGTGCCTTCTGGGAAGGGA GAGGGCCTCTGCCTGGGACACTGCGGGACAGAGGGTGGCTGGAGTGAATTAAAGCCTTTGTTTTTTAAAGAAATGACAGTTGCATCTATA AAAAGGTTTGCCCCTTTAATGGTGGCTCTTTTTTGGTTCAACCTTCTGCGTCAGATTTCCAGTTCTGTTGGAACCAGGCTCCCTATGCAA TTGTCTGTGCTTTCCCGTATCTCCTGGCCTTCACCACCGACTCCATGGAGATCCGCCTGGTGGTGAACGGGAACCTGGTCCACACTGCAG TCGTGCCGCAGCTGCAGCTGGTGGCCTCCAGGTCGGATATATACTTCACAGCAACTGCAGCTGTGAATGAGGTCTCATCTGGAGGCAGCT CCAAGGGGGCCAGTGCCCGAAATTCTCCTCAGACACCCCCGGGCCGAGATACTCCAGTATTTCCTTCTTCCCTGGGGGAAGGTGAAATTC AATCAAAAAATCTGTACAAGATTCCACTTAGAAACCTCGTGGGCAGAAGCATCGAACGACCTCTGAAGTCACCCTTAGTCTCCAAGGTCA TCACCCCACCCACTCCCATCAGTGTGGGCCTTGCTGCCATTCCAGTCACGCACTCCTTGTCCCTGTCTCGCATGGAGATCAAAGAAATAG CAAGCAGGACCCGCAGGGAACTACTGGGCCTCTCGGATGAAGGTGGACCCAAGTCAGAAGGAGCGCCAAAGGCCAAATCAAAACCCCGGA AGCGGTTAGAAGAAAGCCAAGGAGGCCCCAAGCCAGGGGCAGTGAGGTCATCTAGCAGTGACAGGATCCCATCAGGCTCCTTGGAAAGTG CTTCTACTTCCGAAGCCAACCCTGAGGGGCACTCAGCCAGCTCTGACCAGGACCCTGTGGCAGACAGAGAGGGCAGCCCGGTCTCCGGCA GCAGCCCCTTCCAGCTCACGGCTTTCTCCGATGAAGACATTATAGACTTGAAGTAACAGAGTTGAATCTCATTTGCCATCTTTAGTTTTC TTATGGAGGTTTATACTCTTTAAACAGTTCTGATGTAATTTCTCAACAAAATGTGGCTTTTAGCCTGTCAGTGATCTATTGGACCAAACC TTCTGCACACTCGGCCAGTTCCCTCTCCAATGTCCGGTGCCATCTTTCCTGACCTTTGTTTCTTTCTGTTCAGGAACCATCAGTCCCCTT GTAATAAAGGTGGTAGATTTCATTGAGGTTTTAGATTGAAACTTTGAATAAATCAAAAATACTCATTCTTATTTACTGCAACCTTCTCAT ATTTTATATACTTAGATGGAAAATTATTTTTTCATTACAGTTTTGATTTATGTCACATAATGTGTTTCTTTTATAAAGAAAAGTCGTTGC TTTTTAACTTTA >31261_31261_2_FPGS-GARNL3_FPGS_chr9_130575602_ENST00000373247_GARNL3_chr9_130145717_ENST00000373387_length(amino acids)=544AA_BP= MSRARSHLRAALFLAAASARGITTQVAARRGLSAWPVPQEPSMEYQDAVRMLNTLQTNAGYLEQVKRQRGDPQTQLEAMELYLARSGLQV EDLDRLNIIHVTGTKGKGSTCAFTECILRSYGLKTGFFSSPHLVQVRERIRINGQPISPELFTKYFWRLYHRLEETKDGSCVSMPPYFRF LTLMAFHVFLQEKVDLAVVEVGIGGAYDCTNIIRKPVVCGVSSLGIDHTSLLGDTVEKIAWQKGGIFKQGVPAFTVLQPEGPLAVLRDRA QQISCPLYLCPMLEALEEGGPPLTLGLEGEHQRSNAALALQLAHCWLQRQDRHGAGEPKASRPGLLWQLPLAPVFQPTSHMRLGLRNTEW PGRTQVLRRGPLTWYLDGAHTASSAQACVRWFRQALQGRERPSGGPEVRVLLFNATGDRDPAALLKLLQPCQFDYAVFCPNLTEVSSTGN AGGSASLLLAPHPPHTCSASSLVFSCISHALQWISQGRDPIFQPPSPPKGLLTHPVAHSGASILREAAAIHVLVTGSLHLVGGVLKLLEP ALSQ -------------------------------------------------------------- >31261_31261_3_FPGS-GARNL3_FPGS_chr9_130575602_ENST00000373247_GARNL3_chr9_130145717_ENST00000435213_length(transcript)=3178nt_BP=2145nt GCGGGGCGTCTCCCGCCCGGGCCTAGAGCGCTGCCGGGGGCGCCGGGACTATGTCGCGGGCGCGGAGCCACCTGCGCGCCGCTCTATTCC TGGCAGCGGCGTCTGCGCGCGGCATAACGACCCAGGTCGCGGCGCGGCGGGGCTTGAGCGCGTGGCCGGTGCCGCAGGAGCCGAGCATGG AGTACCAGGATGCCGTGCGCATGCTCAATACCCTGCAGACCAATGCCGGCTACCTGGAGCAGGTGAAGCGCCAGCGGGGTGACCCTCAGA CACAGTTGGAAGCCATGGAACTGTACCTGGCACGGAGTGGGCTGCAGGTGGAGGACTTGGACCGGCTGAACATCATCCACGTCACTGGGA CGAAGGGGAAGGGCTCCACCTGTGCCTTCACGGAATGTATCCTCCGAAGCTATGGCCTGAAGACGGGATTCTTTAGCTCTCCCCACCTGG TGCAGGTTCGGGAGCGGATCCGCATCAATGGGCAGCCCATCAGTCCTGAGCTCTTCACCAAGTACTTCTGGCGCCTCTACCACCGGCTGG AGGAGACCAAGGATGGCAGCTGTGTCTCCATGCCCCCCTACTTCCGCTTCCTGACACTCATGGCCTTCCACGTCTTCCTCCAAGAGAAGG TGGACCTGGCAGTGGTGGAGGTGGGCATTGGCGGGGCTTATGACTGCACCAACATCATCAGGAAGCCTGTGGTGTGCGGAGTCTCCTCTC TTGGCATCGACCACACCAGCCTCCTGGGGGATACGGTGGAGAAGATCGCATGGCAGAAAGGGGGCATCTTTAAGCAAGGTGTCCCTGCCT TCACTGTGCTCCAACCTGAAGGTCCCCTGGCAGTGCTGAGGGACCGAGCCCAGCAGATCTCATGTCCTCTATACCTGTGTCCGATGCTGG AGGCCCTCGAGGAAGGGGGGCCGCCGCTGACCCTGGGCCTGGAGGGGGAGCACCAGCGGTCCAACGCCGCCTTGGCCTTGCAGCTGGCCC ACTGCTGGCTGCAGCGGCAGGACCGCCATGGTGCTGGGGAGCCAAAGGCATCCAGGCCAGGGCTCCTGTGGCAGCTGCCCCTGGCACCTG TGTTCCAGCCCACATCCCACATGCGGCTCGGGCTTCGGAACACGGAGTGGCCGGGCCGGACGCAGGTGCTGCGGCGCGGGCCCCTCACCT GGTACCTGGACGGTGCGCACACCGCCAGCAGCGCGCAGGCCTGCGTGCGCTGGTTCCGCCAGGCGCTGCAGGGCCGCGAGAGGCCGAGCG GTGGCCCCGAGGTTCGAGTCTTGCTCTTCAATGCTACCGGGGACCGGGACCCGGCGGCCCTGCTGAAGCTGCTGCAGCCCTGCCAGTTTG ACTATGCCGTCTTCTGCCCTAACCTGACAGAGGTGTCATCCACAGGCAACGCAGGTGGGTCCGCATCCCTGCTTCTGGCGCCCCACCCAC CCCACACCTGCAGTGCCAGCTCCCTCGTCTTCAGCTGCATTTCACATGCCTTGCAATGGATCAGCCAAGGCCGAGACCCCATCTTCCAGC CACCTAGTCCCCCAAAGGGCCTCCTCACCCACCCTGTGGCTCACAGTGGGGCCAGCATACTCCGTGAGGCTGCTGCCATCCATGTGCTAG TCACTGGCAGCCTGCACCTGGTGGGTGGTGTCCTGAAGCTGCTGGAGCCCGCACTGTCCCAGTAGCCAAGGCCCGGGGTTGGAGGTGGGA GCTTCCCACACCTGCCTGCGTTCTCCCCATGAACTTACATACTAGGTGCCTTTTGTTTTTGGCTTTCCTGGTTCTGTCTAGACTGGCCTA GGGGCCAGGGCTTTGGGATGGGAGGCCGGGAGAGGATGTCTTTTTTAAGGCTCTGTGCCTTGGTCTCTCCTTCCTCTTGGCTGAGATAGC AGAGGGGCTCCCCGGGTCTCTCACTGTTGCAGTGGCCTGGCCGTTCAGCCTGTCTCCCCCAACACCCCGCCTGCCTCCTGGCTCAGGCCC AGCTTATTGTGTGCGCTGCCTGGCCAGGCCCTGGGTCTTGCCATGTGCTGGGTGGTAGATTTCCTCCTCCCAGTGCCTTCTGGGAAGGGA GAGGGCCTCTGCCTGGGACACTGCGGGACAGAGGGTGGCTGGAGTGAATTAAAGCCTTTGTTTTTTAAAGAAATGACAGTTGCATCTATA AAAAGGTTTGCCCCTTTAATGGTGGCTCTTTTTTGGTTCAACCTTCTGCGTCAGATTTCCAGTTCTGTTGGAACCAGGCTCCCTATGCAA TTGTCTGTGCTTTCCCGTATCTCCTGGCCTTCACCACCGACTCCATGGAGATCCGCCTGGTGGTGAACGGGAACCTGGTCCACACTGCAG TCGTGCCGCAGCTGCAGCTGGTGGCCTCCAGGTCGGATATATACTTCACAGCAACTGCAGCTGTGAATGAGGTCTCATCTGGAGGCAGCT CCAAGGGGGCCAGTGCCCGAAATTCTCCTCAGACACCCCCGGGCCGAGATACTCCAGTATTTCCTTCTTCCCTGGGGGAAGGTGAAATTC AATCAAAAAATCTGTACAAGATTCCACTTAGAAACCTCGTGGGCAGAAGCATCGAACGACCTCTGAAGTCACCCTTAGTCTCCAAGGTCA TCACCCCACCCACTCCCATCAGTGTGGGCCTTGCTGCCATTCCAGTCACGCACTCCTTGTCCCTGTCTCGCATGGAGATCAAAGAAATAG CAAGCAGGACCCGCAGGGAACTACTGGGCCTCTCGGATGAAGGTGGACCCAAGTCAGAAGGAGCGCCAAAGGCCAAATCAAAACCCCGGA AGCGGTTAGAAGAAAGCCAAGGAGGCCCCAAGCCAGGGGCAGTGAGGTCATCTAGCAGTGACAGGATCCCATCAGGCTCCTTGGAAAGTG CTTCTACTTCCGAAGCCAACCCTGAGGGGCACTCAGCCAGCTCTGACCAGGACCCTGTGGCAGACAGAGAGGGCAGCCCGGTCTCCGGCA GCAGCCCCTTCCAGCTCACGGCTTTCTCCGATGAAGACATTATAGACTTGAAGTAACAGAGTTGAATCTCATTTGCCATCTTTAGTTTTC TTATGGAGGTTTATACTCTTTAAACAGTTCTGATGTAATTTCTCAACAAAATGTGGCTTTTAGCCTGTCAGTGATCTATTGGACCAAACC TTCTGCACACTCGGCCAGTTCCCTCTCC >31261_31261_3_FPGS-GARNL3_FPGS_chr9_130575602_ENST00000373247_GARNL3_chr9_130145717_ENST00000435213_length(amino acids)=544AA_BP= MSRARSHLRAALFLAAASARGITTQVAARRGLSAWPVPQEPSMEYQDAVRMLNTLQTNAGYLEQVKRQRGDPQTQLEAMELYLARSGLQV EDLDRLNIIHVTGTKGKGSTCAFTECILRSYGLKTGFFSSPHLVQVRERIRINGQPISPELFTKYFWRLYHRLEETKDGSCVSMPPYFRF LTLMAFHVFLQEKVDLAVVEVGIGGAYDCTNIIRKPVVCGVSSLGIDHTSLLGDTVEKIAWQKGGIFKQGVPAFTVLQPEGPLAVLRDRA QQISCPLYLCPMLEALEEGGPPLTLGLEGEHQRSNAALALQLAHCWLQRQDRHGAGEPKASRPGLLWQLPLAPVFQPTSHMRLGLRNTEW PGRTQVLRRGPLTWYLDGAHTASSAQACVRWFRQALQGRERPSGGPEVRVLLFNATGDRDPAALLKLLQPCQFDYAVFCPNLTEVSSTGN AGGSASLLLAPHPPHTCSASSLVFSCISHALQWISQGRDPIFQPPSPPKGLLTHPVAHSGASILREAAAIHVLVTGSLHLVGGVLKLLEP ALSQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FPGS-GARNL3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FPGS-GARNL3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Hgene | FPGS | Q05932 | DB00293 | Raltitrexed | Antagonist | Small molecule | Approved|Investigational |
| Hgene | FPGS | Q05932 | DB00142 | Glutamic acid | Small molecule | Approved|Nutraceutical |
Top |
Related Diseases for FPGS-GARNL3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | FPGS | C0003873 | Rheumatoid Arthritis | 1 | CTD_human |
| Hgene | FPGS | C0009402 | Colorectal Carcinoma | 1 | CTD_human |
| Hgene | FPGS | C0009404 | Colorectal Neoplasms | 1 | CTD_human |
| Hgene | FPGS | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | FPGS | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | FPGS | C0024302 | Reticulosarcoma | 1 | CTD_human |
| Hgene | FPGS | C0024304 | Lymphoma, Mixed-Cell | 1 | CTD_human |
| Hgene | FPGS | C0024305 | Lymphoma, Non-Hodgkin | 1 | CTD_human |
| Hgene | FPGS | C0024306 | Lymphoma, Undifferentiated | 1 | CTD_human |
| Hgene | FPGS | C0079740 | High Grade Lymphoma (neoplasm) | 1 | CTD_human |
| Hgene | FPGS | C0079741 | Lymphoma, Intermediate-Grade | 1 | CTD_human |
| Hgene | FPGS | C0079747 | Low Grade Lymphoma (neoplasm) | 1 | CTD_human |
| Hgene | FPGS | C0079757 | Diffuse Mixed-Cell Lymphoma | 1 | CTD_human |
| Hgene | FPGS | C0079770 | Lymphoma, Small Noncleaved-Cell | 1 | CTD_human |
| Hgene | FPGS | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Hgene | FPGS | C3714542 | Lymphoma, Diffuse | 1 | CTD_human |
| Hgene | FPGS | C4721532 | Lymphoma, Non-Hodgkin, Familial | 1 | CTD_human |
