
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CADM1-PARD6G (FusionGDB2 ID:HG23705TG84552) |
Fusion Gene Summary for CADM1-PARD6G |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CADM1-PARD6G | Fusion gene ID: hg23705tg84552 | Hgene | Tgene | Gene symbol | CADM1 | PARD6G | Gene ID | 23705 | 84552 |
| Gene name | cell adhesion molecule 1 | par-6 family cell polarity regulator gamma | |
| Synonyms | BL2|IGSF4|IGSF4A|NECL2|Necl-2|RA175|ST17|SYNCAM|TSLC1|sTSLC-1|sgIGSF|synCAM1 | PAR-6G|PAR6gamma | |
| Cytomap | ('CADM1')('PARD6G') 11q23.3 | 18q23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cell adhesion molecule 1TSLC-1TSLC1/Nectin-like 2/IGSF4immunoglobulin superfamily member 4immunoglobulin superfamily, member 4D variant 1immunoglobulin superfamily, member 4D variant 2nectin-like 2nectin-like protein 2spermatogenic immunoglobulin | partitioning defective 6 homolog gammaPAR-6 gamma proteinPAR6Dpar-6 partitioning defective 6 homolog gamma | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000537140, ENST00000331581, ENST00000452722, ENST00000536727, ENST00000537058, ENST00000542447, | ||
| Fusion gene scores | * DoF score | 7 X 5 X 4=140 | 10 X 3 X 8=240 |
| # samples | 8 | 10 | |
| ** MAII score | log2(8/140*10)=-0.807354922057604 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/240*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CADM1 [Title/Abstract] AND PARD6G [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CADM1(115374988)-PARD6G(77960815), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CADM1-PARD6G seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CADM1-PARD6G seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CADM1-PARD6G seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CADM1-PARD6G seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CADM1 | GO:0001913 | T cell mediated cytotoxicity | 15811952 |
| Hgene | CADM1 | GO:0008037 | cell recognition | 15811952 |
| Hgene | CADM1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity | 15811952 |
| Hgene | CADM1 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity | 15811952 |
| Hgene | CADM1 | GO:0050715 | positive regulation of cytokine secretion | 15811952 |
| Hgene | CADM1 | GO:0050798 | activated T cell proliferation | 15811952 |
| Hgene | CADM1 | GO:0051606 | detection of stimulus | 15811952 |
 Fusion gene breakpoints across CADM1 (5'-gene) Fusion gene breakpoints across CADM1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
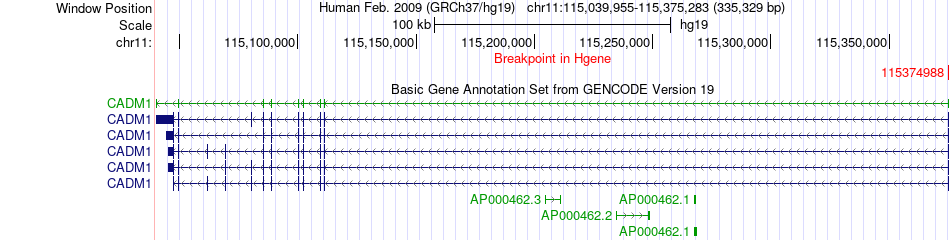 |
 Fusion gene breakpoints across PARD6G (3'-gene) Fusion gene breakpoints across PARD6G (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
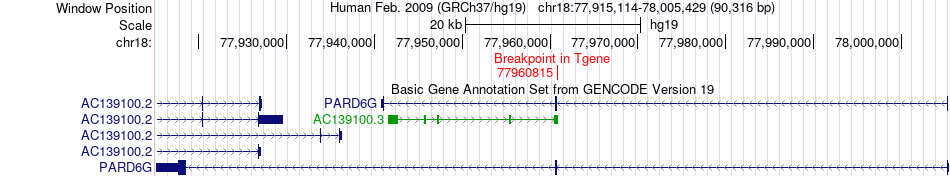 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-61-1910 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
Top |
Fusion Gene ORF analysis for CADM1-PARD6G |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000537140 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| 5UTR-3CDS | ENST00000537140 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000331581 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000331581 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000452722 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000452722 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000536727 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000536727 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000537058 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000537058 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000542447 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
| In-frame | ENST00000542447 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000452722 | CADM1 | chr11 | 115374988 | - | ENST00000353265 | PARD6G | chr18 | 77960815 | - | 3743 | 145 | 172 | 1203 | 343 |
| ENST00000452722 | CADM1 | chr11 | 115374988 | - | ENST00000470488 | PARD6G | chr18 | 77960815 | - | 669 | 145 | 393 | 647 | 84 |
| ENST00000542447 | CADM1 | chr11 | 115374988 | - | ENST00000353265 | PARD6G | chr18 | 77960815 | - | 3851 | 253 | 280 | 1311 | 343 |
| ENST00000542447 | CADM1 | chr11 | 115374988 | - | ENST00000470488 | PARD6G | chr18 | 77960815 | - | 777 | 253 | 2 | 361 | 119 |
| ENST00000537058 | CADM1 | chr11 | 115374988 | - | ENST00000353265 | PARD6G | chr18 | 77960815 | - | 3743 | 145 | 172 | 1203 | 343 |
| ENST00000537058 | CADM1 | chr11 | 115374988 | - | ENST00000470488 | PARD6G | chr18 | 77960815 | - | 669 | 145 | 393 | 647 | 84 |
| ENST00000536727 | CADM1 | chr11 | 115374988 | - | ENST00000353265 | PARD6G | chr18 | 77960815 | - | 3743 | 145 | 172 | 1203 | 343 |
| ENST00000536727 | CADM1 | chr11 | 115374988 | - | ENST00000470488 | PARD6G | chr18 | 77960815 | - | 669 | 145 | 393 | 647 | 84 |
| ENST00000331581 | CADM1 | chr11 | 115374988 | - | ENST00000353265 | PARD6G | chr18 | 77960815 | - | 3893 | 295 | 322 | 1353 | 343 |
| ENST00000331581 | CADM1 | chr11 | 115374988 | - | ENST00000470488 | PARD6G | chr18 | 77960815 | - | 819 | 295 | 44 | 403 | 119 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000452722 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.004610159 | 0.9953898 |
| ENST00000452722 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.16987911 | 0.83012086 |
| ENST00000542447 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.004698597 | 0.99530137 |
| ENST00000542447 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.26564896 | 0.73435104 |
| ENST00000537058 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.004610159 | 0.9953898 |
| ENST00000537058 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.16987911 | 0.83012086 |
| ENST00000536727 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.004610159 | 0.9953898 |
| ENST00000536727 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.16987911 | 0.83012086 |
| ENST00000331581 | ENST00000353265 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.004724399 | 0.9952756 |
| ENST00000331581 | ENST00000470488 | CADM1 | chr11 | 115374988 | - | PARD6G | chr18 | 77960815 | - | 0.26985663 | 0.7301433 |
Top |
Fusion Genomic Features for CADM1-PARD6G |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
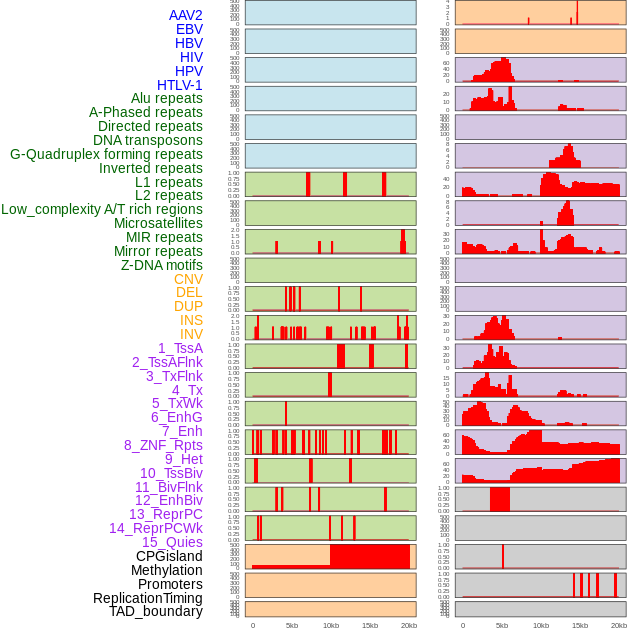 |
Top |
Fusion Protein Features for CADM1-PARD6G |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:115374988/chr18:77960815) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000353265 | 0 | 3 | 134_151 | 24 | 377.0 | Domain | Note=Pseudo-CRIB | |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000353265 | 0 | 3 | 158_251 | 24 | 377.0 | Domain | PDZ | |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000470488 | 0 | 3 | 134_151 | 24 | 108.0 | Domain | Note=Pseudo-CRIB | |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000470488 | 0 | 3 | 158_251 | 24 | 108.0 | Domain | PDZ |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000331581 | - | 1 | 12 | 144_238 | 41 | 472.0 | Domain | Ig-like C2-type 1 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000331581 | - | 1 | 12 | 243_329 | 41 | 472.0 | Domain | Ig-like C2-type 2 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000331581 | - | 1 | 12 | 45_139 | 41 | 472.0 | Domain | Ig-like V-type |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000452722 | - | 1 | 10 | 144_238 | 41 | 443.0 | Domain | Ig-like C2-type 1 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000452722 | - | 1 | 10 | 243_329 | 41 | 443.0 | Domain | Ig-like C2-type 2 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000452722 | - | 1 | 10 | 45_139 | 41 | 443.0 | Domain | Ig-like V-type |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000537058 | - | 1 | 11 | 144_238 | 41 | 454.0 | Domain | Ig-like C2-type 1 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000537058 | - | 1 | 11 | 243_329 | 41 | 454.0 | Domain | Ig-like C2-type 2 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000537058 | - | 1 | 11 | 45_139 | 41 | 454.0 | Domain | Ig-like V-type |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000542447 | - | 1 | 9 | 144_238 | 41 | 415.0 | Domain | Ig-like C2-type 1 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000542447 | - | 1 | 9 | 243_329 | 41 | 415.0 | Domain | Ig-like C2-type 2 |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000542447 | - | 1 | 9 | 45_139 | 41 | 415.0 | Domain | Ig-like V-type |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000331581 | - | 1 | 12 | 396_442 | 41 | 472.0 | Topological domain | Cytoplasmic |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000331581 | - | 1 | 12 | 45_374 | 41 | 472.0 | Topological domain | Extracellular |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000452722 | - | 1 | 10 | 396_442 | 41 | 443.0 | Topological domain | Cytoplasmic |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000452722 | - | 1 | 10 | 45_374 | 41 | 443.0 | Topological domain | Extracellular |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000537058 | - | 1 | 11 | 396_442 | 41 | 454.0 | Topological domain | Cytoplasmic |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000537058 | - | 1 | 11 | 45_374 | 41 | 454.0 | Topological domain | Extracellular |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000542447 | - | 1 | 9 | 396_442 | 41 | 415.0 | Topological domain | Cytoplasmic |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000542447 | - | 1 | 9 | 45_374 | 41 | 415.0 | Topological domain | Extracellular |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000331581 | - | 1 | 12 | 375_395 | 41 | 472.0 | Transmembrane | Helical |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000452722 | - | 1 | 10 | 375_395 | 41 | 443.0 | Transmembrane | Helical |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000537058 | - | 1 | 11 | 375_395 | 41 | 454.0 | Transmembrane | Helical |
| Hgene | CADM1 | chr11:115374988 | chr18:77960815 | ENST00000542447 | - | 1 | 9 | 375_395 | 41 | 415.0 | Transmembrane | Helical |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000353265 | 0 | 3 | 18_98 | 24 | 377.0 | Domain | PB1 | |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000470488 | 0 | 3 | 18_98 | 24 | 108.0 | Domain | PB1 |
Top |
Fusion Gene Sequence for CADM1-PARD6G |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >12445_12445_1_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000331581_PARD6G_chr18_77960815_ENST00000353265_length(transcript)=3893nt_BP=295nt AGGGGGCGGGGTGGGGGAGGGAGCGAGGCCCTCCGAGAGCCGGGTTGGGCTCGCGGCGCTGTGATTGGTCTGCCCGGACTCCGCCTCCAG CGCATGTCATTAGCATCTCATTAGCTGTCCGCTCGGGCTCCGGAGGCAGCCAACGCCGCCAGTCTGAGGCAGGTGCCCGACATGGCGAGT GTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGGCTCCGGCTCCGGCTTCTGCTGTTGCTCTTC TCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCGTCATAAGCCTGGGAAGTTTGAAGATTTCTA CAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGATGTGCACGGAGACCTGCTGCCCATCAACAA TGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCAGAAACGAGAGGAGGCCGAGCGTGGCAGCCT CGGCGCGGGCTCGCTGTGCAGGCGGAGGCGGGCGCTGGGCGCGCTGCGTGATGAAGGACCCCGGCGGCGTGCACACCTGGACATCGGCCT CCCGCGCGACTTCCGCCCCGTATCATCCATCATCGATGTGGACCTGGTCCCCGAGACGCACCGGCGAGTGCGGCTGCACCGGCACGGCTG CGAGAAGCCGCTGGGCTTCTACATCCGCGATGGCGCCAGCGTGCGCGTGACCCCGCACGGGCTGGAGAAGGTGCCCGGCATCTTCATCTC GCGCATGGTACCCGGGGGCCTGGCGGAGAGCACCGGGCTGCTGGCTGTGAATGACGAGGTCCTGGAGGTGAACGGCATTGAGGTGGCCGG GAAGACGCTGGACCAGGTCACGGACATGATGATCGCCAACAGCCACAACCTCATCGTCACCGTCAAGCCCGCCAACCAGCGCAACAACGT GGTGCGCGGCGGCCGCGCGTTGGGCAGCTCGGGACCGCCCTCGGACGGCACCGCGGGCTTCGTGGGTCCCCCCGCCCCGCGCGTCCTGCA GAACTTCCACCCCGACGAGGCGGAGAGCGATGAGGACAACGACGTCGTCATCGAGGGCACACTGGAGCCTGCACGTCCCCCCCAGACCCC GGGCGCGCCCGCAGGCAGCCTCTCCCGGGTCAATGGCGCGGGCCTGGCGCAGCGGCTGCAGCGGGACCTGGCCCTGGACGGCGGCCTCCA GCGGCTGCTCAGCTCCCTGCGGGCCGACCCCCGTCACAGCCTGGCGCTGCCGCCAGGCGGCGTGGAGGAGCACGGGCCCGCGGTCACGCT CTAGACTCCCGAGAGGCCCCCAAATCCTAGCTCCAGTTCCCCGGTAAGGACAGGGACAGGACCTGCAGACTGCATCCGCTCATTTTTTGT TGTTTTTGTGACCACAAAAACAACGCTGCTCTTTGTTTCAACTTCCGGATATAAAAACACAAGTATTGCCTATTTTTATAGAATTTCGCC ACAGAACTTAAAATGAACGCCTGTAGAGACACTCTATACCAGGCCTGAGTGCAGGTCTTGGCGCCTTTGCAAAGTGATAGAAAAGATCTG ACCAAGTGCAAAAAAATGCTTTTTAAAATTTTTACAAATACTTTGTTTTTAAAGTGAAGCTTTAAAATTACTTTTTTAAAATAGTCAAAT TCATGTTTTTAAGTAAGTTGGCCCATGCAACATATCTCAGCATTCTTGCTGTGCTCTCTGCTGACTTGCAGGCGAGGCCGACTGTGCTGA TGTACGTGACAAGAGGCTGGTTTTTAATAACGAATGCAGCCAGGTGCAGTGGCTCACGTCTGTAATCCCAGCACTTTGGGAGGCCTACGT GGGCTGATCACGAGGTCAGGAGGTCGAGACCAGCATGGCCAACATGGTGAAACCCTGTCTCTACTAAAAATATAAAAATTAGCCAGGTGT GGTGGCAGGCGCCTGTAATCCCAGCTACCTTGGGAGGCTGAGGCAGGAGAATCGCTTGAACCCGGGAGGCAGAGGTTGCAGTGAGCTGAG ATCGCACCACTGCACTCCAGCCTGGGTGACAGAGCAAGACTCTGTCTCGGGAAAAAAAAAAAATACACCCAAACCTGTCCATGTACAACC ATGGAGATGAATTCCAGATTTTCACACTGTTCATAGTCATAATTCGGTTGGGACAGGAGAGCTCCCACCAGCACACTTATGGCACCCGCA GCTGTGGAATTGTTCCGGGGCTTTTTTTTGTTTGTTTTGAGACATAGTGTCTTGTTCTGTCATCCAAGCTGGAGGGATCTCAGCTCACTG CAACCTCCGCCTCCTGGGTTCAAGCAATTCTCCTGCCTCAGCCTCCCAAGGAGCTGGGACAGGCACATGCCACCATACCCAGCTAATTTT TGTATTTTTAGTAGAGATGGGATTTCACCACGTTGGTCAGGCTGGTCTTGAACTCCTGACCTCAGGTGATACGGTCCCCCCTCAGCCTCC AAAAATGCTAGGATTATAGACGTGAGCCACAGCACCAGACCAGGGGCTATTTTTGTGACCCACATAGGTGGCCCATTCAGCCTCAAGGGG AAATCCAGGGTAGGAAGCAGCAGCTCTGCTGGACTGGGCTCAGCAGTCCCATTTGGGATGTAAGCCTCTATTGCCTCTAAGATGACCCTT TTCCTGTGGGTTGACTGACTCTGAGGGCGATTTCGGAAATGTTTTCAGCAGAAATGAAGGATAAGCATCGCACATGTCCAGCAATGTCTT TGAGAATGCAGTCATTTGCACGTGAGTTCTCATCTTTGTTTTCTGAAACAGACGTTTCTTTTAGTAAGTCACAATACATGCTAATTTTCA TTTCTAGAATGGTCACTGGCAGTTCTCACTTTTAACTATAAGAAGTCACGTGGAACTTAGTGATTTTTAACAAATACTAAATTCATCCTC ACTTTATGTTGATCTTCCAGCAACGTTTATATATTTAGTAATTTTAAAAATTCCTATTTGATACAATTAAATTTTATATAATGTGTTTTG CTTGAATAAAATGTTACATTTATACTTTGAGAACTCTGACCAAATCTTTCATACAGATACAGGTAAAGAAATATTTTTCTAGGTTGTCGG AGCTGCGTACCTCCCGCTGCTTCTCCTTGTGGTCCAGCACCACCTGCTGGTCAGTGGCAGAACTGTCCTGACTTCGTGAACATGGAACTG CTAAGAGGGAAAACTGCATCTCAGCATTTTCATTTTTTCTGAATATTAAGTGAAAAGCAAGTTCTATTTTTAAGAAATAGCTTTTGCTGA GGAGTTGTGAATTCCTGCACCTGTGTTTGATGTTATTTGTATGCAATGGATTGCTGGCCCTTTGTTCCTAAGCAAGAGAAGACCAAAGAT GCTTCCCCATGACCTCTCCGGTGAGGATCCGCACCCCCCGGACTTGCTCTCTGATGAAGGGCATGTCGTTCAGTGCCCCAGCAGTCAGAA GCAGTTTTTGTGGAACTAAAACTAATAATGTTACCCTCACAAAGGAAATGTTCTTTTACTAAGTTATTGCTGATTCTTGAATGTCTGTTA TATAACTGGGTCGGCTATCAAGAAGGAAGGTTATATTTCTTATCACATTTTAAGTTGTTAAGCAAATGTGAGTTTTTTTTTAAGAAATGA GGCTTTCACTGAGATTTGTGTGCTATTCATAATTTTAGGAATTACTGCTTAGAACAGCTGCTTGGGTGTATTTTTCTACACAGGTCTGTA CTTCTGCCACTTTCTCCTTTTAAAAAAACAAAACTATGCATATTATGTATTAAAGTTTTCTTGGTTTTATATATCAGAGAATAAAAACTT TTTTGACTTTTGAGTAAACAAAA >12445_12445_1_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000331581_PARD6G_chr18_77960815_ENST00000353265_length(amino acids)=343AA_BP= MDRHKPGKFEDFYKLVVHTHHISNSDVTIGYADVHGDLLPINNDDNFCKAVSSANPLLRVFIQKREEAERGSLGAGSLCRRRRALGALRD EGPRRRAHLDIGLPRDFRPVSSIIDVDLVPETHRRVRLHRHGCEKPLGFYIRDGASVRVTPHGLEKVPGIFISRMVPGGLAESTGLLAVN DEVLEVNGIEVAGKTLDQVTDMMIANSHNLIVTVKPANQRNNVVRGGRALGSSGPPSDGTAGFVGPPAPRVLQNFHPDEAESDEDNDVVI EGTLEPARPPQTPGAPAGSLSRVNGAGLAQRLQRDLALDGGLQRLLSSLRADPRHSLALPPGGVEEHGPAVTL -------------------------------------------------------------- >12445_12445_2_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000331581_PARD6G_chr18_77960815_ENST00000470488_length(transcript)=819nt_BP=295nt AGGGGGCGGGGTGGGGGAGGGAGCGAGGCCCTCCGAGAGCCGGGTTGGGCTCGCGGCGCTGTGATTGGTCTGCCCGGACTCCGCCTCCAG CGCATGTCATTAGCATCTCATTAGCTGTCCGCTCGGGCTCCGGAGGCAGCCAACGCCGCCAGTCTGAGGCAGGTGCCCGACATGGCGAGT GTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGGCTCCGGCTCCGGCTTCTGCTGTTGCTCTTC TCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCGTCATAAGCCTGGGAAGTTTGAAGATTTCTA CAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGATGTGCACGGAGACCTGCTGCCCATCAACAA TGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCAGAAACGAGATGACGGAGCCCTGCGACCAGG AACCTGACTCTTCAGCTCAAATCTTGGTTTGAAAGGGGGACTTCTCTGTGCCCCTGTTGTCGGCATGCCATGGTAGAGAATGGATGGCCC ACGCAGCTGGCCCCTCATGAGACCCCAGCCATCTCATTCCACTGGGAGCCACGTCAGGACGAAGGAGGACTGGGAGGGGCGTCTGGCTGC GTGGTGTGGGGCCTGCAGTGTTGTGGCACACGCAGCTTCAGCCAAACATCTAGCCCTGTACCTAGAACTAAGAAATAAAACTATAACATC AGATTTAAA >12445_12445_2_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000331581_PARD6G_chr18_77960815_ENST00000470488_length(amino acids)=119AA_BP=83 MGSRRCDWSARTPPPAHVISISLAVRSGSGGSQRRQSEAGARHGECSAAERIPVCGGSGGGGASRAPAPASAVALLRRGTDPHSLGRNSE GSLWTVISLGSLKISTSWLCTPTISPTVM -------------------------------------------------------------- >12445_12445_3_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000452722_PARD6G_chr18_77960815_ENST00000353265_length(transcript)=3743nt_BP=145nt AGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGG CTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCG TCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGA TGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCA GAAACGAGAGGAGGCCGAGCGTGGCAGCCTCGGCGCGGGCTCGCTGTGCAGGCGGAGGCGGGCGCTGGGCGCGCTGCGTGATGAAGGACC CCGGCGGCGTGCACACCTGGACATCGGCCTCCCGCGCGACTTCCGCCCCGTATCATCCATCATCGATGTGGACCTGGTCCCCGAGACGCA CCGGCGAGTGCGGCTGCACCGGCACGGCTGCGAGAAGCCGCTGGGCTTCTACATCCGCGATGGCGCCAGCGTGCGCGTGACCCCGCACGG GCTGGAGAAGGTGCCCGGCATCTTCATCTCGCGCATGGTACCCGGGGGCCTGGCGGAGAGCACCGGGCTGCTGGCTGTGAATGACGAGGT CCTGGAGGTGAACGGCATTGAGGTGGCCGGGAAGACGCTGGACCAGGTCACGGACATGATGATCGCCAACAGCCACAACCTCATCGTCAC CGTCAAGCCCGCCAACCAGCGCAACAACGTGGTGCGCGGCGGCCGCGCGTTGGGCAGCTCGGGACCGCCCTCGGACGGCACCGCGGGCTT CGTGGGTCCCCCCGCCCCGCGCGTCCTGCAGAACTTCCACCCCGACGAGGCGGAGAGCGATGAGGACAACGACGTCGTCATCGAGGGCAC ACTGGAGCCTGCACGTCCCCCCCAGACCCCGGGCGCGCCCGCAGGCAGCCTCTCCCGGGTCAATGGCGCGGGCCTGGCGCAGCGGCTGCA GCGGGACCTGGCCCTGGACGGCGGCCTCCAGCGGCTGCTCAGCTCCCTGCGGGCCGACCCCCGTCACAGCCTGGCGCTGCCGCCAGGCGG CGTGGAGGAGCACGGGCCCGCGGTCACGCTCTAGACTCCCGAGAGGCCCCCAAATCCTAGCTCCAGTTCCCCGGTAAGGACAGGGACAGG ACCTGCAGACTGCATCCGCTCATTTTTTGTTGTTTTTGTGACCACAAAAACAACGCTGCTCTTTGTTTCAACTTCCGGATATAAAAACAC AAGTATTGCCTATTTTTATAGAATTTCGCCACAGAACTTAAAATGAACGCCTGTAGAGACACTCTATACCAGGCCTGAGTGCAGGTCTTG GCGCCTTTGCAAAGTGATAGAAAAGATCTGACCAAGTGCAAAAAAATGCTTTTTAAAATTTTTACAAATACTTTGTTTTTAAAGTGAAGC TTTAAAATTACTTTTTTAAAATAGTCAAATTCATGTTTTTAAGTAAGTTGGCCCATGCAACATATCTCAGCATTCTTGCTGTGCTCTCTG CTGACTTGCAGGCGAGGCCGACTGTGCTGATGTACGTGACAAGAGGCTGGTTTTTAATAACGAATGCAGCCAGGTGCAGTGGCTCACGTC TGTAATCCCAGCACTTTGGGAGGCCTACGTGGGCTGATCACGAGGTCAGGAGGTCGAGACCAGCATGGCCAACATGGTGAAACCCTGTCT CTACTAAAAATATAAAAATTAGCCAGGTGTGGTGGCAGGCGCCTGTAATCCCAGCTACCTTGGGAGGCTGAGGCAGGAGAATCGCTTGAA CCCGGGAGGCAGAGGTTGCAGTGAGCTGAGATCGCACCACTGCACTCCAGCCTGGGTGACAGAGCAAGACTCTGTCTCGGGAAAAAAAAA AAATACACCCAAACCTGTCCATGTACAACCATGGAGATGAATTCCAGATTTTCACACTGTTCATAGTCATAATTCGGTTGGGACAGGAGA GCTCCCACCAGCACACTTATGGCACCCGCAGCTGTGGAATTGTTCCGGGGCTTTTTTTTGTTTGTTTTGAGACATAGTGTCTTGTTCTGT CATCCAAGCTGGAGGGATCTCAGCTCACTGCAACCTCCGCCTCCTGGGTTCAAGCAATTCTCCTGCCTCAGCCTCCCAAGGAGCTGGGAC AGGCACATGCCACCATACCCAGCTAATTTTTGTATTTTTAGTAGAGATGGGATTTCACCACGTTGGTCAGGCTGGTCTTGAACTCCTGAC CTCAGGTGATACGGTCCCCCCTCAGCCTCCAAAAATGCTAGGATTATAGACGTGAGCCACAGCACCAGACCAGGGGCTATTTTTGTGACC CACATAGGTGGCCCATTCAGCCTCAAGGGGAAATCCAGGGTAGGAAGCAGCAGCTCTGCTGGACTGGGCTCAGCAGTCCCATTTGGGATG TAAGCCTCTATTGCCTCTAAGATGACCCTTTTCCTGTGGGTTGACTGACTCTGAGGGCGATTTCGGAAATGTTTTCAGCAGAAATGAAGG ATAAGCATCGCACATGTCCAGCAATGTCTTTGAGAATGCAGTCATTTGCACGTGAGTTCTCATCTTTGTTTTCTGAAACAGACGTTTCTT TTAGTAAGTCACAATACATGCTAATTTTCATTTCTAGAATGGTCACTGGCAGTTCTCACTTTTAACTATAAGAAGTCACGTGGAACTTAG TGATTTTTAACAAATACTAAATTCATCCTCACTTTATGTTGATCTTCCAGCAACGTTTATATATTTAGTAATTTTAAAAATTCCTATTTG ATACAATTAAATTTTATATAATGTGTTTTGCTTGAATAAAATGTTACATTTATACTTTGAGAACTCTGACCAAATCTTTCATACAGATAC AGGTAAAGAAATATTTTTCTAGGTTGTCGGAGCTGCGTACCTCCCGCTGCTTCTCCTTGTGGTCCAGCACCACCTGCTGGTCAGTGGCAG AACTGTCCTGACTTCGTGAACATGGAACTGCTAAGAGGGAAAACTGCATCTCAGCATTTTCATTTTTTCTGAATATTAAGTGAAAAGCAA GTTCTATTTTTAAGAAATAGCTTTTGCTGAGGAGTTGTGAATTCCTGCACCTGTGTTTGATGTTATTTGTATGCAATGGATTGCTGGCCC TTTGTTCCTAAGCAAGAGAAGACCAAAGATGCTTCCCCATGACCTCTCCGGTGAGGATCCGCACCCCCCGGACTTGCTCTCTGATGAAGG GCATGTCGTTCAGTGCCCCAGCAGTCAGAAGCAGTTTTTGTGGAACTAAAACTAATAATGTTACCCTCACAAAGGAAATGTTCTTTTACT AAGTTATTGCTGATTCTTGAATGTCTGTTATATAACTGGGTCGGCTATCAAGAAGGAAGGTTATATTTCTTATCACATTTTAAGTTGTTA AGCAAATGTGAGTTTTTTTTTAAGAAATGAGGCTTTCACTGAGATTTGTGTGCTATTCATAATTTTAGGAATTACTGCTTAGAACAGCTG CTTGGGTGTATTTTTCTACACAGGTCTGTACTTCTGCCACTTTCTCCTTTTAAAAAAACAAAACTATGCATATTATGTATTAAAGTTTTC TTGGTTTTATATATCAGAGAATAAAAACTTTTTTGACTTTTGAGTAAACAAAA >12445_12445_3_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000452722_PARD6G_chr18_77960815_ENST00000353265_length(amino acids)=343AA_BP= MDRHKPGKFEDFYKLVVHTHHISNSDVTIGYADVHGDLLPINNDDNFCKAVSSANPLLRVFIQKREEAERGSLGAGSLCRRRRALGALRD EGPRRRAHLDIGLPRDFRPVSSIIDVDLVPETHRRVRLHRHGCEKPLGFYIRDGASVRVTPHGLEKVPGIFISRMVPGGLAESTGLLAVN DEVLEVNGIEVAGKTLDQVTDMMIANSHNLIVTVKPANQRNNVVRGGRALGSSGPPSDGTAGFVGPPAPRVLQNFHPDEAESDEDNDVVI EGTLEPARPPQTPGAPAGSLSRVNGAGLAQRLQRDLALDGGLQRLLSSLRADPRHSLALPPGGVEEHGPAVTL -------------------------------------------------------------- >12445_12445_4_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000452722_PARD6G_chr18_77960815_ENST00000470488_length(transcript)=669nt_BP=145nt AGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGG CTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCG TCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGA TGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCA GAAACGAGATGACGGAGCCCTGCGACCAGGAACCTGACTCTTCAGCTCAAATCTTGGTTTGAAAGGGGGACTTCTCTGTGCCCCTGTTGT CGGCATGCCATGGTAGAGAATGGATGGCCCACGCAGCTGGCCCCTCATGAGACCCCAGCCATCTCATTCCACTGGGAGCCACGTCAGGAC GAAGGAGGACTGGGAGGGGCGTCTGGCTGCGTGGTGTGGGGCCTGCAGTGTTGTGGCACACGCAGCTTCAGCCAAACATCTAGCCCTGTA CCTAGAACTAAGAAATAAAACTATAACATCAGATTTAAA >12445_12445_4_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000452722_PARD6G_chr18_77960815_ENST00000470488_length(amino acids)=84AA_BP= MTLQLKSWFERGTSLCPCCRHAMVENGWPTQLAPHETPAISFHWEPRQDEGGLGGASGCVVWGLQCCGTRSFSQTSSPVPRTKK -------------------------------------------------------------- >12445_12445_5_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000536727_PARD6G_chr18_77960815_ENST00000353265_length(transcript)=3743nt_BP=145nt AGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGG CTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCG TCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGA TGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCA GAAACGAGAGGAGGCCGAGCGTGGCAGCCTCGGCGCGGGCTCGCTGTGCAGGCGGAGGCGGGCGCTGGGCGCGCTGCGTGATGAAGGACC CCGGCGGCGTGCACACCTGGACATCGGCCTCCCGCGCGACTTCCGCCCCGTATCATCCATCATCGATGTGGACCTGGTCCCCGAGACGCA CCGGCGAGTGCGGCTGCACCGGCACGGCTGCGAGAAGCCGCTGGGCTTCTACATCCGCGATGGCGCCAGCGTGCGCGTGACCCCGCACGG GCTGGAGAAGGTGCCCGGCATCTTCATCTCGCGCATGGTACCCGGGGGCCTGGCGGAGAGCACCGGGCTGCTGGCTGTGAATGACGAGGT CCTGGAGGTGAACGGCATTGAGGTGGCCGGGAAGACGCTGGACCAGGTCACGGACATGATGATCGCCAACAGCCACAACCTCATCGTCAC CGTCAAGCCCGCCAACCAGCGCAACAACGTGGTGCGCGGCGGCCGCGCGTTGGGCAGCTCGGGACCGCCCTCGGACGGCACCGCGGGCTT CGTGGGTCCCCCCGCCCCGCGCGTCCTGCAGAACTTCCACCCCGACGAGGCGGAGAGCGATGAGGACAACGACGTCGTCATCGAGGGCAC ACTGGAGCCTGCACGTCCCCCCCAGACCCCGGGCGCGCCCGCAGGCAGCCTCTCCCGGGTCAATGGCGCGGGCCTGGCGCAGCGGCTGCA GCGGGACCTGGCCCTGGACGGCGGCCTCCAGCGGCTGCTCAGCTCCCTGCGGGCCGACCCCCGTCACAGCCTGGCGCTGCCGCCAGGCGG CGTGGAGGAGCACGGGCCCGCGGTCACGCTCTAGACTCCCGAGAGGCCCCCAAATCCTAGCTCCAGTTCCCCGGTAAGGACAGGGACAGG ACCTGCAGACTGCATCCGCTCATTTTTTGTTGTTTTTGTGACCACAAAAACAACGCTGCTCTTTGTTTCAACTTCCGGATATAAAAACAC AAGTATTGCCTATTTTTATAGAATTTCGCCACAGAACTTAAAATGAACGCCTGTAGAGACACTCTATACCAGGCCTGAGTGCAGGTCTTG GCGCCTTTGCAAAGTGATAGAAAAGATCTGACCAAGTGCAAAAAAATGCTTTTTAAAATTTTTACAAATACTTTGTTTTTAAAGTGAAGC TTTAAAATTACTTTTTTAAAATAGTCAAATTCATGTTTTTAAGTAAGTTGGCCCATGCAACATATCTCAGCATTCTTGCTGTGCTCTCTG CTGACTTGCAGGCGAGGCCGACTGTGCTGATGTACGTGACAAGAGGCTGGTTTTTAATAACGAATGCAGCCAGGTGCAGTGGCTCACGTC TGTAATCCCAGCACTTTGGGAGGCCTACGTGGGCTGATCACGAGGTCAGGAGGTCGAGACCAGCATGGCCAACATGGTGAAACCCTGTCT CTACTAAAAATATAAAAATTAGCCAGGTGTGGTGGCAGGCGCCTGTAATCCCAGCTACCTTGGGAGGCTGAGGCAGGAGAATCGCTTGAA CCCGGGAGGCAGAGGTTGCAGTGAGCTGAGATCGCACCACTGCACTCCAGCCTGGGTGACAGAGCAAGACTCTGTCTCGGGAAAAAAAAA AAATACACCCAAACCTGTCCATGTACAACCATGGAGATGAATTCCAGATTTTCACACTGTTCATAGTCATAATTCGGTTGGGACAGGAGA GCTCCCACCAGCACACTTATGGCACCCGCAGCTGTGGAATTGTTCCGGGGCTTTTTTTTGTTTGTTTTGAGACATAGTGTCTTGTTCTGT CATCCAAGCTGGAGGGATCTCAGCTCACTGCAACCTCCGCCTCCTGGGTTCAAGCAATTCTCCTGCCTCAGCCTCCCAAGGAGCTGGGAC AGGCACATGCCACCATACCCAGCTAATTTTTGTATTTTTAGTAGAGATGGGATTTCACCACGTTGGTCAGGCTGGTCTTGAACTCCTGAC CTCAGGTGATACGGTCCCCCCTCAGCCTCCAAAAATGCTAGGATTATAGACGTGAGCCACAGCACCAGACCAGGGGCTATTTTTGTGACC CACATAGGTGGCCCATTCAGCCTCAAGGGGAAATCCAGGGTAGGAAGCAGCAGCTCTGCTGGACTGGGCTCAGCAGTCCCATTTGGGATG TAAGCCTCTATTGCCTCTAAGATGACCCTTTTCCTGTGGGTTGACTGACTCTGAGGGCGATTTCGGAAATGTTTTCAGCAGAAATGAAGG ATAAGCATCGCACATGTCCAGCAATGTCTTTGAGAATGCAGTCATTTGCACGTGAGTTCTCATCTTTGTTTTCTGAAACAGACGTTTCTT TTAGTAAGTCACAATACATGCTAATTTTCATTTCTAGAATGGTCACTGGCAGTTCTCACTTTTAACTATAAGAAGTCACGTGGAACTTAG TGATTTTTAACAAATACTAAATTCATCCTCACTTTATGTTGATCTTCCAGCAACGTTTATATATTTAGTAATTTTAAAAATTCCTATTTG ATACAATTAAATTTTATATAATGTGTTTTGCTTGAATAAAATGTTACATTTATACTTTGAGAACTCTGACCAAATCTTTCATACAGATAC AGGTAAAGAAATATTTTTCTAGGTTGTCGGAGCTGCGTACCTCCCGCTGCTTCTCCTTGTGGTCCAGCACCACCTGCTGGTCAGTGGCAG AACTGTCCTGACTTCGTGAACATGGAACTGCTAAGAGGGAAAACTGCATCTCAGCATTTTCATTTTTTCTGAATATTAAGTGAAAAGCAA GTTCTATTTTTAAGAAATAGCTTTTGCTGAGGAGTTGTGAATTCCTGCACCTGTGTTTGATGTTATTTGTATGCAATGGATTGCTGGCCC TTTGTTCCTAAGCAAGAGAAGACCAAAGATGCTTCCCCATGACCTCTCCGGTGAGGATCCGCACCCCCCGGACTTGCTCTCTGATGAAGG GCATGTCGTTCAGTGCCCCAGCAGTCAGAAGCAGTTTTTGTGGAACTAAAACTAATAATGTTACCCTCACAAAGGAAATGTTCTTTTACT AAGTTATTGCTGATTCTTGAATGTCTGTTATATAACTGGGTCGGCTATCAAGAAGGAAGGTTATATTTCTTATCACATTTTAAGTTGTTA AGCAAATGTGAGTTTTTTTTTAAGAAATGAGGCTTTCACTGAGATTTGTGTGCTATTCATAATTTTAGGAATTACTGCTTAGAACAGCTG CTTGGGTGTATTTTTCTACACAGGTCTGTACTTCTGCCACTTTCTCCTTTTAAAAAAACAAAACTATGCATATTATGTATTAAAGTTTTC TTGGTTTTATATATCAGAGAATAAAAACTTTTTTGACTTTTGAGTAAACAAAA >12445_12445_5_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000536727_PARD6G_chr18_77960815_ENST00000353265_length(amino acids)=343AA_BP= MDRHKPGKFEDFYKLVVHTHHISNSDVTIGYADVHGDLLPINNDDNFCKAVSSANPLLRVFIQKREEAERGSLGAGSLCRRRRALGALRD EGPRRRAHLDIGLPRDFRPVSSIIDVDLVPETHRRVRLHRHGCEKPLGFYIRDGASVRVTPHGLEKVPGIFISRMVPGGLAESTGLLAVN DEVLEVNGIEVAGKTLDQVTDMMIANSHNLIVTVKPANQRNNVVRGGRALGSSGPPSDGTAGFVGPPAPRVLQNFHPDEAESDEDNDVVI EGTLEPARPPQTPGAPAGSLSRVNGAGLAQRLQRDLALDGGLQRLLSSLRADPRHSLALPPGGVEEHGPAVTL -------------------------------------------------------------- >12445_12445_6_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000536727_PARD6G_chr18_77960815_ENST00000470488_length(transcript)=669nt_BP=145nt AGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGG CTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCG TCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGA TGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCA GAAACGAGATGACGGAGCCCTGCGACCAGGAACCTGACTCTTCAGCTCAAATCTTGGTTTGAAAGGGGGACTTCTCTGTGCCCCTGTTGT CGGCATGCCATGGTAGAGAATGGATGGCCCACGCAGCTGGCCCCTCATGAGACCCCAGCCATCTCATTCCACTGGGAGCCACGTCAGGAC GAAGGAGGACTGGGAGGGGCGTCTGGCTGCGTGGTGTGGGGCCTGCAGTGTTGTGGCACACGCAGCTTCAGCCAAACATCTAGCCCTGTA CCTAGAACTAAGAAATAAAACTATAACATCAGATTTAAA >12445_12445_6_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000536727_PARD6G_chr18_77960815_ENST00000470488_length(amino acids)=84AA_BP= MTLQLKSWFERGTSLCPCCRHAMVENGWPTQLAPHETPAISFHWEPRQDEGGLGGASGCVVWGLQCCGTRSFSQTSSPVPRTKK -------------------------------------------------------------- >12445_12445_7_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000537058_PARD6G_chr18_77960815_ENST00000353265_length(transcript)=3743nt_BP=145nt AGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGG CTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCG TCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGA TGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCA GAAACGAGAGGAGGCCGAGCGTGGCAGCCTCGGCGCGGGCTCGCTGTGCAGGCGGAGGCGGGCGCTGGGCGCGCTGCGTGATGAAGGACC CCGGCGGCGTGCACACCTGGACATCGGCCTCCCGCGCGACTTCCGCCCCGTATCATCCATCATCGATGTGGACCTGGTCCCCGAGACGCA CCGGCGAGTGCGGCTGCACCGGCACGGCTGCGAGAAGCCGCTGGGCTTCTACATCCGCGATGGCGCCAGCGTGCGCGTGACCCCGCACGG GCTGGAGAAGGTGCCCGGCATCTTCATCTCGCGCATGGTACCCGGGGGCCTGGCGGAGAGCACCGGGCTGCTGGCTGTGAATGACGAGGT CCTGGAGGTGAACGGCATTGAGGTGGCCGGGAAGACGCTGGACCAGGTCACGGACATGATGATCGCCAACAGCCACAACCTCATCGTCAC CGTCAAGCCCGCCAACCAGCGCAACAACGTGGTGCGCGGCGGCCGCGCGTTGGGCAGCTCGGGACCGCCCTCGGACGGCACCGCGGGCTT CGTGGGTCCCCCCGCCCCGCGCGTCCTGCAGAACTTCCACCCCGACGAGGCGGAGAGCGATGAGGACAACGACGTCGTCATCGAGGGCAC ACTGGAGCCTGCACGTCCCCCCCAGACCCCGGGCGCGCCCGCAGGCAGCCTCTCCCGGGTCAATGGCGCGGGCCTGGCGCAGCGGCTGCA GCGGGACCTGGCCCTGGACGGCGGCCTCCAGCGGCTGCTCAGCTCCCTGCGGGCCGACCCCCGTCACAGCCTGGCGCTGCCGCCAGGCGG CGTGGAGGAGCACGGGCCCGCGGTCACGCTCTAGACTCCCGAGAGGCCCCCAAATCCTAGCTCCAGTTCCCCGGTAAGGACAGGGACAGG ACCTGCAGACTGCATCCGCTCATTTTTTGTTGTTTTTGTGACCACAAAAACAACGCTGCTCTTTGTTTCAACTTCCGGATATAAAAACAC AAGTATTGCCTATTTTTATAGAATTTCGCCACAGAACTTAAAATGAACGCCTGTAGAGACACTCTATACCAGGCCTGAGTGCAGGTCTTG GCGCCTTTGCAAAGTGATAGAAAAGATCTGACCAAGTGCAAAAAAATGCTTTTTAAAATTTTTACAAATACTTTGTTTTTAAAGTGAAGC TTTAAAATTACTTTTTTAAAATAGTCAAATTCATGTTTTTAAGTAAGTTGGCCCATGCAACATATCTCAGCATTCTTGCTGTGCTCTCTG CTGACTTGCAGGCGAGGCCGACTGTGCTGATGTACGTGACAAGAGGCTGGTTTTTAATAACGAATGCAGCCAGGTGCAGTGGCTCACGTC TGTAATCCCAGCACTTTGGGAGGCCTACGTGGGCTGATCACGAGGTCAGGAGGTCGAGACCAGCATGGCCAACATGGTGAAACCCTGTCT CTACTAAAAATATAAAAATTAGCCAGGTGTGGTGGCAGGCGCCTGTAATCCCAGCTACCTTGGGAGGCTGAGGCAGGAGAATCGCTTGAA CCCGGGAGGCAGAGGTTGCAGTGAGCTGAGATCGCACCACTGCACTCCAGCCTGGGTGACAGAGCAAGACTCTGTCTCGGGAAAAAAAAA AAATACACCCAAACCTGTCCATGTACAACCATGGAGATGAATTCCAGATTTTCACACTGTTCATAGTCATAATTCGGTTGGGACAGGAGA GCTCCCACCAGCACACTTATGGCACCCGCAGCTGTGGAATTGTTCCGGGGCTTTTTTTTGTTTGTTTTGAGACATAGTGTCTTGTTCTGT CATCCAAGCTGGAGGGATCTCAGCTCACTGCAACCTCCGCCTCCTGGGTTCAAGCAATTCTCCTGCCTCAGCCTCCCAAGGAGCTGGGAC AGGCACATGCCACCATACCCAGCTAATTTTTGTATTTTTAGTAGAGATGGGATTTCACCACGTTGGTCAGGCTGGTCTTGAACTCCTGAC CTCAGGTGATACGGTCCCCCCTCAGCCTCCAAAAATGCTAGGATTATAGACGTGAGCCACAGCACCAGACCAGGGGCTATTTTTGTGACC CACATAGGTGGCCCATTCAGCCTCAAGGGGAAATCCAGGGTAGGAAGCAGCAGCTCTGCTGGACTGGGCTCAGCAGTCCCATTTGGGATG TAAGCCTCTATTGCCTCTAAGATGACCCTTTTCCTGTGGGTTGACTGACTCTGAGGGCGATTTCGGAAATGTTTTCAGCAGAAATGAAGG ATAAGCATCGCACATGTCCAGCAATGTCTTTGAGAATGCAGTCATTTGCACGTGAGTTCTCATCTTTGTTTTCTGAAACAGACGTTTCTT TTAGTAAGTCACAATACATGCTAATTTTCATTTCTAGAATGGTCACTGGCAGTTCTCACTTTTAACTATAAGAAGTCACGTGGAACTTAG TGATTTTTAACAAATACTAAATTCATCCTCACTTTATGTTGATCTTCCAGCAACGTTTATATATTTAGTAATTTTAAAAATTCCTATTTG ATACAATTAAATTTTATATAATGTGTTTTGCTTGAATAAAATGTTACATTTATACTTTGAGAACTCTGACCAAATCTTTCATACAGATAC AGGTAAAGAAATATTTTTCTAGGTTGTCGGAGCTGCGTACCTCCCGCTGCTTCTCCTTGTGGTCCAGCACCACCTGCTGGTCAGTGGCAG AACTGTCCTGACTTCGTGAACATGGAACTGCTAAGAGGGAAAACTGCATCTCAGCATTTTCATTTTTTCTGAATATTAAGTGAAAAGCAA GTTCTATTTTTAAGAAATAGCTTTTGCTGAGGAGTTGTGAATTCCTGCACCTGTGTTTGATGTTATTTGTATGCAATGGATTGCTGGCCC TTTGTTCCTAAGCAAGAGAAGACCAAAGATGCTTCCCCATGACCTCTCCGGTGAGGATCCGCACCCCCCGGACTTGCTCTCTGATGAAGG GCATGTCGTTCAGTGCCCCAGCAGTCAGAAGCAGTTTTTGTGGAACTAAAACTAATAATGTTACCCTCACAAAGGAAATGTTCTTTTACT AAGTTATTGCTGATTCTTGAATGTCTGTTATATAACTGGGTCGGCTATCAAGAAGGAAGGTTATATTTCTTATCACATTTTAAGTTGTTA AGCAAATGTGAGTTTTTTTTTAAGAAATGAGGCTTTCACTGAGATTTGTGTGCTATTCATAATTTTAGGAATTACTGCTTAGAACAGCTG CTTGGGTGTATTTTTCTACACAGGTCTGTACTTCTGCCACTTTCTCCTTTTAAAAAAACAAAACTATGCATATTATGTATTAAAGTTTTC TTGGTTTTATATATCAGAGAATAAAAACTTTTTTGACTTTTGAGTAAACAAAA >12445_12445_7_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000537058_PARD6G_chr18_77960815_ENST00000353265_length(amino acids)=343AA_BP= MDRHKPGKFEDFYKLVVHTHHISNSDVTIGYADVHGDLLPINNDDNFCKAVSSANPLLRVFIQKREEAERGSLGAGSLCRRRRALGALRD EGPRRRAHLDIGLPRDFRPVSSIIDVDLVPETHRRVRLHRHGCEKPLGFYIRDGASVRVTPHGLEKVPGIFISRMVPGGLAESTGLLAVN DEVLEVNGIEVAGKTLDQVTDMMIANSHNLIVTVKPANQRNNVVRGGRALGSSGPPSDGTAGFVGPPAPRVLQNFHPDEAESDEDNDVVI EGTLEPARPPQTPGAPAGSLSRVNGAGLAQRLQRDLALDGGLQRLLSSLRADPRHSLALPPGGVEEHGPAVTL -------------------------------------------------------------- >12445_12445_8_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000537058_PARD6G_chr18_77960815_ENST00000470488_length(transcript)=669nt_BP=145nt AGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCGGCGGCGGCGCCTCCCGGG CTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCGAAGGTTCTCTCTGGACCG TCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGTAACTATTGGCTATGCAGA TGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCTGCTCAGGGTCTTCATCCA GAAACGAGATGACGGAGCCCTGCGACCAGGAACCTGACTCTTCAGCTCAAATCTTGGTTTGAAAGGGGGACTTCTCTGTGCCCCTGTTGT CGGCATGCCATGGTAGAGAATGGATGGCCCACGCAGCTGGCCCCTCATGAGACCCCAGCCATCTCATTCCACTGGGAGCCACGTCAGGAC GAAGGAGGACTGGGAGGGGCGTCTGGCTGCGTGGTGTGGGGCCTGCAGTGTTGTGGCACACGCAGCTTCAGCCAAACATCTAGCCCTGTA CCTAGAACTAAGAAATAAAACTATAACATCAGATTTAAA >12445_12445_8_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000537058_PARD6G_chr18_77960815_ENST00000470488_length(amino acids)=84AA_BP= MTLQLKSWFERGTSLCPCCRHAMVENGWPTQLAPHETPAISFHWEPRQDEGGLGGASGCVVWGLQCCGTRSFSQTSSPVPRTKK -------------------------------------------------------------- >12445_12445_9_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000542447_PARD6G_chr18_77960815_ENST00000353265_length(transcript)=3851nt_BP=253nt GGTTGGGCTCGCGGCGCTGTGATTGGTCTGCCCGGACTCCGCCTCCAGCGCATGTCATTAGCATCTCATTAGCTGTCCGCTCGGGCTCCG GAGGCAGCCAACGCCGCCAGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCG GCGGCGGCGCCTCCCGGGCTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCG AAGGTTCTCTCTGGACCGTCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGT AACTATTGGCTATGCAGATGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCT GCTCAGGGTCTTCATCCAGAAACGAGAGGAGGCCGAGCGTGGCAGCCTCGGCGCGGGCTCGCTGTGCAGGCGGAGGCGGGCGCTGGGCGC GCTGCGTGATGAAGGACCCCGGCGGCGTGCACACCTGGACATCGGCCTCCCGCGCGACTTCCGCCCCGTATCATCCATCATCGATGTGGA CCTGGTCCCCGAGACGCACCGGCGAGTGCGGCTGCACCGGCACGGCTGCGAGAAGCCGCTGGGCTTCTACATCCGCGATGGCGCCAGCGT GCGCGTGACCCCGCACGGGCTGGAGAAGGTGCCCGGCATCTTCATCTCGCGCATGGTACCCGGGGGCCTGGCGGAGAGCACCGGGCTGCT GGCTGTGAATGACGAGGTCCTGGAGGTGAACGGCATTGAGGTGGCCGGGAAGACGCTGGACCAGGTCACGGACATGATGATCGCCAACAG CCACAACCTCATCGTCACCGTCAAGCCCGCCAACCAGCGCAACAACGTGGTGCGCGGCGGCCGCGCGTTGGGCAGCTCGGGACCGCCCTC GGACGGCACCGCGGGCTTCGTGGGTCCCCCCGCCCCGCGCGTCCTGCAGAACTTCCACCCCGACGAGGCGGAGAGCGATGAGGACAACGA CGTCGTCATCGAGGGCACACTGGAGCCTGCACGTCCCCCCCAGACCCCGGGCGCGCCCGCAGGCAGCCTCTCCCGGGTCAATGGCGCGGG CCTGGCGCAGCGGCTGCAGCGGGACCTGGCCCTGGACGGCGGCCTCCAGCGGCTGCTCAGCTCCCTGCGGGCCGACCCCCGTCACAGCCT GGCGCTGCCGCCAGGCGGCGTGGAGGAGCACGGGCCCGCGGTCACGCTCTAGACTCCCGAGAGGCCCCCAAATCCTAGCTCCAGTTCCCC GGTAAGGACAGGGACAGGACCTGCAGACTGCATCCGCTCATTTTTTGTTGTTTTTGTGACCACAAAAACAACGCTGCTCTTTGTTTCAAC TTCCGGATATAAAAACACAAGTATTGCCTATTTTTATAGAATTTCGCCACAGAACTTAAAATGAACGCCTGTAGAGACACTCTATACCAG GCCTGAGTGCAGGTCTTGGCGCCTTTGCAAAGTGATAGAAAAGATCTGACCAAGTGCAAAAAAATGCTTTTTAAAATTTTTACAAATACT TTGTTTTTAAAGTGAAGCTTTAAAATTACTTTTTTAAAATAGTCAAATTCATGTTTTTAAGTAAGTTGGCCCATGCAACATATCTCAGCA TTCTTGCTGTGCTCTCTGCTGACTTGCAGGCGAGGCCGACTGTGCTGATGTACGTGACAAGAGGCTGGTTTTTAATAACGAATGCAGCCA GGTGCAGTGGCTCACGTCTGTAATCCCAGCACTTTGGGAGGCCTACGTGGGCTGATCACGAGGTCAGGAGGTCGAGACCAGCATGGCCAA CATGGTGAAACCCTGTCTCTACTAAAAATATAAAAATTAGCCAGGTGTGGTGGCAGGCGCCTGTAATCCCAGCTACCTTGGGAGGCTGAG GCAGGAGAATCGCTTGAACCCGGGAGGCAGAGGTTGCAGTGAGCTGAGATCGCACCACTGCACTCCAGCCTGGGTGACAGAGCAAGACTC TGTCTCGGGAAAAAAAAAAAATACACCCAAACCTGTCCATGTACAACCATGGAGATGAATTCCAGATTTTCACACTGTTCATAGTCATAA TTCGGTTGGGACAGGAGAGCTCCCACCAGCACACTTATGGCACCCGCAGCTGTGGAATTGTTCCGGGGCTTTTTTTTGTTTGTTTTGAGA CATAGTGTCTTGTTCTGTCATCCAAGCTGGAGGGATCTCAGCTCACTGCAACCTCCGCCTCCTGGGTTCAAGCAATTCTCCTGCCTCAGC CTCCCAAGGAGCTGGGACAGGCACATGCCACCATACCCAGCTAATTTTTGTATTTTTAGTAGAGATGGGATTTCACCACGTTGGTCAGGC TGGTCTTGAACTCCTGACCTCAGGTGATACGGTCCCCCCTCAGCCTCCAAAAATGCTAGGATTATAGACGTGAGCCACAGCACCAGACCA GGGGCTATTTTTGTGACCCACATAGGTGGCCCATTCAGCCTCAAGGGGAAATCCAGGGTAGGAAGCAGCAGCTCTGCTGGACTGGGCTCA GCAGTCCCATTTGGGATGTAAGCCTCTATTGCCTCTAAGATGACCCTTTTCCTGTGGGTTGACTGACTCTGAGGGCGATTTCGGAAATGT TTTCAGCAGAAATGAAGGATAAGCATCGCACATGTCCAGCAATGTCTTTGAGAATGCAGTCATTTGCACGTGAGTTCTCATCTTTGTTTT CTGAAACAGACGTTTCTTTTAGTAAGTCACAATACATGCTAATTTTCATTTCTAGAATGGTCACTGGCAGTTCTCACTTTTAACTATAAG AAGTCACGTGGAACTTAGTGATTTTTAACAAATACTAAATTCATCCTCACTTTATGTTGATCTTCCAGCAACGTTTATATATTTAGTAAT TTTAAAAATTCCTATTTGATACAATTAAATTTTATATAATGTGTTTTGCTTGAATAAAATGTTACATTTATACTTTGAGAACTCTGACCA AATCTTTCATACAGATACAGGTAAAGAAATATTTTTCTAGGTTGTCGGAGCTGCGTACCTCCCGCTGCTTCTCCTTGTGGTCCAGCACCA CCTGCTGGTCAGTGGCAGAACTGTCCTGACTTCGTGAACATGGAACTGCTAAGAGGGAAAACTGCATCTCAGCATTTTCATTTTTTCTGA ATATTAAGTGAAAAGCAAGTTCTATTTTTAAGAAATAGCTTTTGCTGAGGAGTTGTGAATTCCTGCACCTGTGTTTGATGTTATTTGTAT GCAATGGATTGCTGGCCCTTTGTTCCTAAGCAAGAGAAGACCAAAGATGCTTCCCCATGACCTCTCCGGTGAGGATCCGCACCCCCCGGA CTTGCTCTCTGATGAAGGGCATGTCGTTCAGTGCCCCAGCAGTCAGAAGCAGTTTTTGTGGAACTAAAACTAATAATGTTACCCTCACAA AGGAAATGTTCTTTTACTAAGTTATTGCTGATTCTTGAATGTCTGTTATATAACTGGGTCGGCTATCAAGAAGGAAGGTTATATTTCTTA TCACATTTTAAGTTGTTAAGCAAATGTGAGTTTTTTTTTAAGAAATGAGGCTTTCACTGAGATTTGTGTGCTATTCATAATTTTAGGAAT TACTGCTTAGAACAGCTGCTTGGGTGTATTTTTCTACACAGGTCTGTACTTCTGCCACTTTCTCCTTTTAAAAAAACAAAACTATGCATA TTATGTATTAAAGTTTTCTTGGTTTTATATATCAGAGAATAAAAACTTTTTTGACTTTTGAGTAAACAAAA >12445_12445_9_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000542447_PARD6G_chr18_77960815_ENST00000353265_length(amino acids)=343AA_BP= MDRHKPGKFEDFYKLVVHTHHISNSDVTIGYADVHGDLLPINNDDNFCKAVSSANPLLRVFIQKREEAERGSLGAGSLCRRRRALGALRD EGPRRRAHLDIGLPRDFRPVSSIIDVDLVPETHRRVRLHRHGCEKPLGFYIRDGASVRVTPHGLEKVPGIFISRMVPGGLAESTGLLAVN DEVLEVNGIEVAGKTLDQVTDMMIANSHNLIVTVKPANQRNNVVRGGRALGSSGPPSDGTAGFVGPPAPRVLQNFHPDEAESDEDNDVVI EGTLEPARPPQTPGAPAGSLSRVNGAGLAQRLQRDLALDGGLQRLLSSLRADPRHSLALPPGGVEEHGPAVTL -------------------------------------------------------------- >12445_12445_10_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000542447_PARD6G_chr18_77960815_ENST00000470488_length(transcript)=777nt_BP=253nt GGTTGGGCTCGCGGCGCTGTGATTGGTCTGCCCGGACTCCGCCTCCAGCGCATGTCATTAGCATCTCATTAGCTGTCCGCTCGGGCTCCG GAGGCAGCCAACGCCGCCAGTCTGAGGCAGGTGCCCGACATGGCGAGTGTAGTGCTGCCGAGCGGATCCCAGTGTGCGGCGGCAGCGGCG GCGGCGGCGCCTCCCGGGCTCCGGCTCCGGCTTCTGCTGTTGCTCTTCTCCGCCGCGGCACTGATCCCCACAGTTTGGGGCGGAATTCCG AAGGTTCTCTCTGGACCGTCATAAGCCTGGGAAGTTTGAAGATTTCTACAAGCTGGTTGTGCACACCCACCATATCTCCAACAGTGATGT AACTATTGGCTATGCAGATGTGCACGGAGACCTGCTGCCCATCAACAATGATGACAACTTCTGCAAGGCGGTTTCTAGTGCAAATCCCCT GCTCAGGGTCTTCATCCAGAAACGAGATGACGGAGCCCTGCGACCAGGAACCTGACTCTTCAGCTCAAATCTTGGTTTGAAAGGGGGACT TCTCTGTGCCCCTGTTGTCGGCATGCCATGGTAGAGAATGGATGGCCCACGCAGCTGGCCCCTCATGAGACCCCAGCCATCTCATTCCAC TGGGAGCCACGTCAGGACGAAGGAGGACTGGGAGGGGCGTCTGGCTGCGTGGTGTGGGGCCTGCAGTGTTGTGGCACACGCAGCTTCAGC CAAACATCTAGCCCTGTACCTAGAACTAAGAAATAAAACTATAACATCAGATTTAAA >12445_12445_10_CADM1-PARD6G_CADM1_chr11_115374988_ENST00000542447_PARD6G_chr18_77960815_ENST00000470488_length(amino acids)=119AA_BP=83 LGSRRCDWSARTPPPAHVISISLAVRSGSGGSQRRQSEAGARHGECSAAERIPVCGGSGGGGASRAPAPASAVALLRRGTDPHSLGRNSE GSLWTVISLGSLKISTSWLCTPTISPTVM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CADM1-PARD6G |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000353265 | 0 | 3 | 127_254 | 24.0 | 377.0 | PARD3 and CDC42 | |
| Tgene | PARD6G | chr11:115374988 | chr18:77960815 | ENST00000470488 | 0 | 3 | 127_254 | 24.0 | 108.0 | PARD3 and CDC42 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CADM1-PARD6G |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CADM1-PARD6G |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CADM1 | C0004352 | Autistic Disorder | 1 | CTD_human |
| Hgene | CADM1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Hgene | CADM1 | C0011573 | Endogenous depression | 1 | CTD_human |
| Hgene | CADM1 | C0011581 | Depressive disorder | 1 | CTD_human |
| Hgene | CADM1 | C0025193 | Melancholia | 1 | CTD_human |
| Hgene | CADM1 | C0041696 | Unipolar Depression | 1 | CTD_human |
| Hgene | CADM1 | C0086133 | Depressive Syndrome | 1 | CTD_human |
| Hgene | CADM1 | C0282126 | Depression, Neurotic | 1 | CTD_human |
| Hgene | CADM1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Hgene | CADM1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Hgene | CADM1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Hgene | CADM1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
