
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ABCF1-SHC2 (FusionGDB2 ID:HG23TG25759) |
Fusion Gene Summary for ABCF1-SHC2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ABCF1-SHC2 | Fusion gene ID: hg23tg25759 | Hgene | Tgene | Gene symbol | ABCF1 | SHC2 | Gene ID | 23 | 25759 |
| Gene name | ATP binding cassette subfamily F member 1 | SHC adaptor protein 2 | |
| Synonyms | ABC27|ABC50 | SCK|SHCB|SLI | |
| Cytomap | ('ABCF1')('SHC2') 6p21.33 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ATP-binding cassette sub-family F member 1ATP-binding cassette 50 (TNF-alpha stimulated)ATP-binding cassette, sub-family F (GCN20), member 1TNF-alpha-stimulated ABC proteinTNFalpha-inducible ATP-binding protein | SHC-transforming protein 2SH2 domain protein C2SHC (Src homology 2 domain containing) transforming protein 2SHC-transforming protein Bneuronal Shc adaptor homologprotein Scksrc homology 2 domain-containing-transforming protein C2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000326195, ENST00000376545, ENST00000383587, ENST00000383588, ENST00000396515, ENST00000400609, ENST00000412443, ENST00000416932, ENST00000419893, ENST00000420257, ENST00000421042, ENST00000421608, ENST00000423247, ENST00000426219, ENST00000448939, ENST00000452530, ENST00000454094, ENST00000456791, ENST00000457078, ENST00000457111, ENST00000457970, ENST00000470464, ENST00000475528, ENST00000485585, ENST00000486105, ENST00000494413, ENST00000495194, ENST00000546690, ENST00000552867, | ||
| Fusion gene scores | * DoF score | 6 X 5 X 4=120 | 7 X 4 X 7=196 |
| # samples | 6 | 7 | |
| ** MAII score | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/196*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ABCF1 [Title/Abstract] AND SHC2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ABCF1(30554488)-SHC2(440932), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ABCF1-SHC2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ABCF1-SHC2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ABCF1-SHC2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ABCF1-SHC2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ABCF1 (5'-gene) Fusion gene breakpoints across ABCF1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across SHC2 (3'-gene) Fusion gene breakpoints across SHC2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
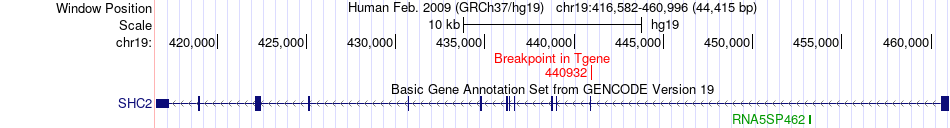 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-TM-A7CF-01A | ABCF1 | chr6 | 30554488 | - | SHC2 | chr19 | 440932 | - |
| ChimerDB4 | LGG | TCGA-TM-A7CF-01A | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
Top |
Fusion Gene ORF analysis for ABCF1-SHC2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000326195 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| In-frame | ENST00000376545 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000383587 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000383588 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000396515 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000400609 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000412443 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000416932 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000419893 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000420257 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000421042 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000421608 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000423247 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000426219 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000448939 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000452530 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000454094 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000456791 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000457078 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000457111 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000457970 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000470464 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000475528 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000485585 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000486105 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000494413 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000495194 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000546690 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
| intron-3CDS | ENST00000552867 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000326195 | ABCF1 | chr6 | 30554488 | + | ENST00000264554 | SHC2 | chr19 | 440932 | - | 4169 | 2143 | 25 | 3423 | 1132 |
| ENST00000376545 | ABCF1 | chr6 | 30554488 | + | ENST00000264554 | SHC2 | chr19 | 440932 | - | 4038 | 2012 | 8 | 3292 | 1094 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000326195 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - | 0.005918701 | 0.9940813 |
| ENST00000376545 | ENST00000264554 | ABCF1 | chr6 | 30554488 | + | SHC2 | chr19 | 440932 | - | 0.00810428 | 0.99189574 |
Top |
Fusion Genomic Features for ABCF1-SHC2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
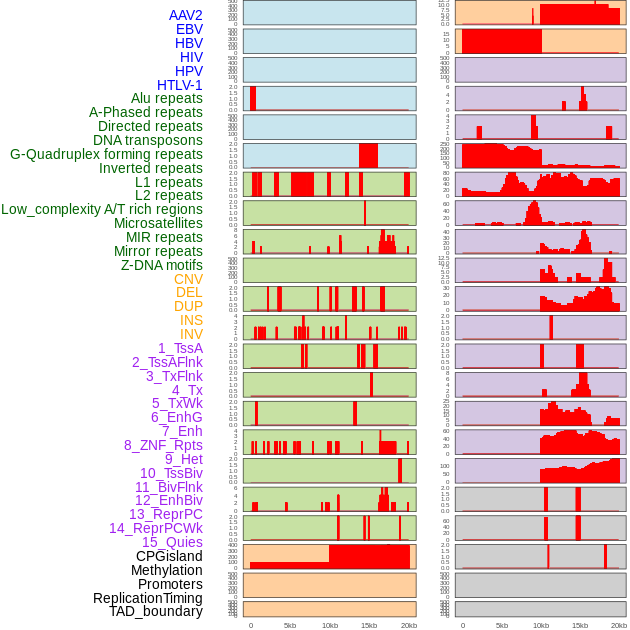 |
Top |
Fusion Protein Features for ABCF1-SHC2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:30554488/chr19:440932) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000326195 | + | 20 | 25 | 141_243 | 677 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000376545 | + | 19 | 24 | 141_243 | 639 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000326195 | + | 20 | 25 | 304_548 | 677 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000376545 | + | 19 | 24 | 304_548 | 639 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000326195 | + | 20 | 25 | 336_343 | 677 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000326195 | + | 20 | 25 | 658_665 | 677 | 846.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000376545 | + | 19 | 24 | 336_343 | 639 | 808.0 | Nucleotide binding | ATP 1 |
| Tgene | SHC2 | chr6:30554488 | chr19:440932 | ENST00000264554 | 0 | 13 | 487_578 | 156 | 694.6666666666666 | Domain | SH2 | |
| Tgene | SHC2 | chr6:30554488 | chr19:440932 | ENST00000264554 | 0 | 13 | 330_486 | 156 | 694.6666666666666 | Region | Note=CH1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383587 | + | 1 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383588 | + | 1 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000412443 | + | 1 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000419893 | + | 1 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000420257 | + | 1 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000421042 | + | 1 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000423247 | + | 1 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000426219 | + | 1 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000448939 | + | 1 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000452530 | + | 1 | 24 | 141_243 | 0 | 808.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457078 | + | 1 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457111 | + | 1 | 25 | 141_243 | 0 | 846.0 | Compositional bias | Note=Glu-rich |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000326195 | + | 20 | 25 | 625_840 | 677 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000376545 | + | 19 | 24 | 625_840 | 639 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383587 | + | 1 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383587 | + | 1 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383588 | + | 1 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383588 | + | 1 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000412443 | + | 1 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000412443 | + | 1 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000419893 | + | 1 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000419893 | + | 1 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000420257 | + | 1 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000420257 | + | 1 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000421042 | + | 1 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000421042 | + | 1 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000423247 | + | 1 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000423247 | + | 1 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000426219 | + | 1 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000426219 | + | 1 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000448939 | + | 1 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000448939 | + | 1 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000452530 | + | 1 | 24 | 304_548 | 0 | 808.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000452530 | + | 1 | 24 | 625_840 | 0 | 808.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457078 | + | 1 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457078 | + | 1 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457111 | + | 1 | 25 | 304_548 | 0 | 846.0 | Domain | ABC transporter 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457111 | + | 1 | 25 | 625_840 | 0 | 846.0 | Domain | ABC transporter 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000376545 | + | 19 | 24 | 658_665 | 639 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383587 | + | 1 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383587 | + | 1 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383588 | + | 1 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000383588 | + | 1 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000412443 | + | 1 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000412443 | + | 1 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000419893 | + | 1 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000419893 | + | 1 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000420257 | + | 1 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000420257 | + | 1 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000421042 | + | 1 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000421042 | + | 1 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000423247 | + | 1 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000423247 | + | 1 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000426219 | + | 1 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000426219 | + | 1 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000448939 | + | 1 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000448939 | + | 1 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000452530 | + | 1 | 24 | 336_343 | 0 | 808.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000452530 | + | 1 | 24 | 658_665 | 0 | 808.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457078 | + | 1 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457078 | + | 1 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457111 | + | 1 | 25 | 336_343 | 0 | 846.0 | Nucleotide binding | ATP 1 |
| Hgene | ABCF1 | chr6:30554488 | chr19:440932 | ENST00000457111 | + | 1 | 25 | 658_665 | 0 | 846.0 | Nucleotide binding | ATP 2 |
| Tgene | SHC2 | chr6:30554488 | chr19:440932 | ENST00000264554 | 0 | 13 | 147_329 | 156 | 694.6666666666666 | Domain | PID |
Top |
Fusion Gene Sequence for ABCF1-SHC2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >343_343_1_ABCF1-SHC2_ABCF1_chr6_30554488_ENST00000326195_SHC2_chr19_440932_ENST00000264554_length(transcript)=4169nt_BP=2143nt CGGGGGCGGGGTTGGCCGCGCCAGCTTGGAGAGCCAGCCCCATCGGGGTTCCCCGCCGCCGGAAGCGGAAATAGCACCGGGCGCCGCCAC AGTAGCTGTAACTGCCACCGCGATGCCGAAGGCGCCCAAGCAGCAGCCGCCGGAGCCCGAGTGGATCGGGGACGGAGAGAGCACGAGCCC ATCAGACAAAGTGGTGAAGAAAGGGAAGAAGGACAAGAAGATCAAAAAAACGTTCTTTGAAGAGCTGGCAGTAGAAGATAAACAGGCTGG GGAAGAAGAGAAAGTGCTCAAGGAGAAGGAGCAGCAGCAGCAGCAACAGCAACAGCAGCAAAAAAAAAAGCGAGATACCCGAAAAGGCAG GCGGAAGAAGGATGTGGATGATGATGGAGAAGAGAAAGAGCTCATGGAGCGTCTTAAGAAGCTCTCAGTGCCAACCAGTGATGAGGAGGA TGAAGTACCCGCCCCAAAACCCCGCGGAGGGAAGAAAACCAAGGGTGGTAATGTTTTTGCAGCCCTGATTCAGGATCAGAGTGAGGAAGA GGAGGAGGAAGAAAAACATCCTCCTAAGCCTGCCAAGCCGGAGAAGAATCGGATCAATAAGGCCGTATCTGAGGAACAGCAGCCTGCACT CAAGGGCAAAAAGGGAAAGGAAGAGAAGTCAAAAGGGAAGGCTAAGCCTCAAAATAAATTCGCTGCTCTGGACAATGAAGAGGAGGATAA AGAAGAAGAAATTATAAAGGAAAAGGAGCCTCCCAAACAAGGGAAGGAGAAGGCCAAGAAGGCAGAGCAGGGTTCAGAGGAAGAAGGAGA AGGGGAAGAAGAGGAGGAGGAAGGAGGAGAGTCTAAGGCAGATGATCCCTATGCTCATCTTAGCAAAAAGGAGAAGAAAAAGCTGAAAAA ACAGATGGAGTATGAGCGCCAAGTGGCTTCATTAAAAGCAGCCAATGCAGCTGAAAATGACTTCTCCGTGTCCCAGGCGGAGATGTCCTC CCGCCAAGCCATGTTAGAAAATGCATCTGACATCAAGCTGGAGAAGTTCAGCATCTCCGCTCATGGCAAGGAGCTGTTCGTCAATGCAGA CCTGTACATTGTAGCCGGCCGCCGCTACGGGCTGGTAGGACCCAATGGCAAGGGCAAGACCACACTCCTCAAGCACATTGCCAACCGAGC CCTGAGCATCCCTCCCAACATTGATGTGTTGCTGTGTGAGCAGGAGGTGGTAGCAGATGAGACACCAGCAGTCCAGGCTGTTCTTCGAGC TGACACCAAGCGATTGAAGCTGCTGGAAGAGGAGCGGCGGCTTCAGGGACAGCTGGAACAAGGGGATGACACAGCTGCTGAGAGGCTAGA GAAGGTGTATGAGGAATTGCGGGCCACTGGGGCGGCAGCTGCAGAGGCCAAAGCACGGCGGATCCTGGCTGGCCTGGGCTTTGACCCTGA AATGCAGAATCGACCCACACAGAAGTTCTCAGGGGGCTGGCGCATGCGTGTCTCCCTGGCCAGGGCACTGTTCATGGAGCCCACACTGCT GATGCTGGATGAGCCCACCAACCACCTGGACCTCAACGCTGTCATCTGGCTTAATAACTACCTCCAGGGCTGGCGGAAGACCTTGCTGAT CGTCTCCCATGACCAGGGCTTCTTGGATGATGTCTGCACTGATATCATCCACCTCGATGCCCAGCGGCTCCACTACTATAGGGGCAATTA CATGACCTTCAAAAAGATGTACCAGCAGAAGCAGAAAGAACTGCTGAAACAGTATGAGAAGCAAGAGAAAAAGCTGAAGGAGCTGAAGGC AGGCGGGAAGTCCACCAAGCAGGCGGAAAAACAAACGAAGGAAGCCCTGACTCGGAAGCAGCAGAAATGCCGACGGAAAAACCAAGATGA GGAATCCCAGGAGGCCCCTGAGCTCCTGAAGCGCCCTAAGGAGTACACTGTGCGCTTCACTTTTCCAGACCCCCCACCACTCAGCCCTCC AGTGCTGGGTCTGCATGGTGTGACATTCGGCTACCAGGGACAGAAACCACTCTTTAAGAACTTGGATTTTGGCATCGACATGGATTCAAG GATTTGCATTGTGGGCCCTAATGGTGTGGGGAAGAGTACGCTACTCCTGCTGCTGACTGGCAAGCTGACACCGTACATGGGCTGCATCGA GGTTCTCCGCTCTATGCGCTCCCTGGACTTTAACACGCGCACGCAGGTGACCAGGGAAGCCATCAACCGGCTCCATGAGGCCGTGCCTGG CGTCCGGGGATCCTGGAAGAAAAAGGCCCCCAACAAGGCCCTGGCGTCCGTCCTGGGCAAGAGCAACCTTCGCTTTGCCGGCATGAGCAT CTCCATCCACATCTCCACTGATGGCCTCAGCCTCTCCGTGCCTGCCACGCGCCAGGTCATCGCCAACCACCACATGCCGTCCATCTCCTT CGCGTCAGGCGGAGACACGGACATGACGGATTACGTGGCCTACGTCGCCAAGGACCCCATCAACCAGAGAGCCTGCCACATCCTGGAGTG CTGTGAGGGCCTGGCACAGAGCATCATCAGCACCGTGGGCCAAGCTTTCGAGCTGCGCTTCAAGCAGTACCTGCACAGCCCGCCCAAGGT GGCGCTGCCCCCAGAAAGGCTGGCAGGGCCGGAGGAGTCGGCCTGGGGGGACGAGGAGGACTCTTTGGAGCACAATTACTACAACAGCAT CCCGGGGAAGGAGCCGCCGCTGGGCGGGCTAGTGGACTCCAGGCTGGCCCTGACACAGCCCTGCGCCCTCACGGCCCTCGACCAGGGCCC ATCTCCTTCTCTAAGAGATGCCTGCAGCCTGCCATGGGACGTGGGGTCCACCGGTACAGCTCCACCGGGGGACGGCTACGTGCAGGCGGA CGCCCGGGGCCCCCCGGACCACGAGGAGCACCTGTATGTCAACACCCAGGGTCTGGACGCCCCCGAGCCGGAGGACAGCCCCAAAAAGGA TCTGTTTGACATGCGACCCTTTGAGGATGCCCTGAAGTTGCATGAGTGCTCAGTGGCGGCAGGCGTGACAGCAGCCCCTCTTCCCTTGGA GGACCAGTGGCCCAGCCCCCCTACCCGCCGGGCCCCTGTGGCCCCCACGGAGGAACAGCTGCGTCAGGAGCCCTGGTACCACGGCCGGAT GAGCCGCCGGGCGGCAGAGAGGATGCTTCGAGCTGACGGGGACTTCCTTGTGCGAGACAGCGTCACCAACCCCGGGCAGTATGTCCTCAC CGGCATGCACGCCGGGCAGCCCAAGCACCTGCTGCTCGTGGACCCCGAGGGCGTGGTACGGACGAAGGACGTGCTGTTTGAGAGCATCAG CCACCTGATCGACCACCACCTGCAGAACGGGCAGCCCATCGTGGCCGCCGAGAGTGAGCTGCACCTGCGTGGCGTGGTCTCACGGGAGCC CTGAGCCAGGTGACCGTTCTCAGCCCTGGCTCCTGCCTGTTCTCAGCCCCGGCTCCTGCCTGTCATGGCCTTGGGGCTCTCAGCCCTCCT CCTGGATCCTGGTCAGGCCGAACCACCGCTGCCTCTCCTTCCTCCGCATGAGCCTCTGGCATGGTCCTTCCTCCAGCTGGCCCCGGGCTG GGCAGAGCCTCCTCCTGCCGGGGCCCCTGCCCACCCCCTCCTTTGCCTGGAGTGAGGGTGTTCATACCAAAGACGGAACCATTTCGCCTT TAAAGAAAATATATCCAGAAGCAGCCGCTGCCTCGGAGCCCTGGCCCTTGGGTCCCCCTCTCGCCTGGCTGGTTCGGTCTAACGCCCCGG AGAGTCAGGGCTCCCAGGACCCTGGGGAGGAGGCGGATTCCGGGCCTGGCTGGGCTTCCTCTTCCCACCTGAGGACTGGGTGCACAGTTG TCTTTGAGGGGGGACGCTTAAGGTGCTTTGGGGTTCTCAGGCCAGGATACATGCTGGCGCTGAAGTGCAGGCAGCCTTGAGGTCACCTGG GATCTCGGGGTAGCCAGGCTGCTCCAAGACAGTGGATATCGAGGCAGTCGTGAGGCCCCTACTCCACCTGGGAGCAGGGGAAGGACGTGG TTGCCTGGCCTGGGCTGCGCCCAGCAGCTTCCCCCCAGTCCTGCCCTCCCAGCTGTCGACCCAGATGGGATGTTCGGATCGGTTTGTAAT TAAACCTGGGAATGGCCACAAGAGCACAA >343_343_1_ABCF1-SHC2_ABCF1_chr6_30554488_ENST00000326195_SHC2_chr19_440932_ENST00000264554_length(amino acids)=1132AA_BP=706 MESQPHRGSPPPEAEIAPGAATVAVTATAMPKAPKQQPPEPEWIGDGESTSPSDKVVKKGKKDKKIKKTFFEELAVEDKQAGEEEKVLKE KEQQQQQQQQQQKKKRDTRKGRRKKDVDDDGEEKELMERLKKLSVPTSDEEDEVPAPKPRGGKKTKGGNVFAALIQDQSEEEEEEEKHPP KPAKPEKNRINKAVSEEQQPALKGKKGKEEKSKGKAKPQNKFAALDNEEEDKEEEIIKEKEPPKQGKEKAKKAEQGSEEEGEGEEEEEEG GESKADDPYAHLSKKEKKKLKKQMEYERQVASLKAANAAENDFSVSQAEMSSRQAMLENASDIKLEKFSISAHGKELFVNADLYIVAGRR YGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEVVADETPAVQAVLRADTKRLKLLEEERRLQGQLEQGDDTAAERLEKVYEELRA TGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGWRMRVSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFL DDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLKQYEKQEKKLKELKAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPEL LKRPKEYTVRFTFPDPPPLSPPVLGLHGVTFGYQGQKPLFKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPYMGCIEVLRSMRSL DFNTRTQVTREAINRLHEAVPGVRGSWKKKAPNKALASVLGKSNLRFAGMSISIHISTDGLSLSVPATRQVIANHHMPSISFASGGDTDM TDYVAYVAKDPINQRACHILECCEGLAQSIISTVGQAFELRFKQYLHSPPKVALPPERLAGPEESAWGDEEDSLEHNYYNSIPGKEPPLG GLVDSRLALTQPCALTALDQGPSPSLRDACSLPWDVGSTGTAPPGDGYVQADARGPPDHEEHLYVNTQGLDAPEPEDSPKKDLFDMRPFE DALKLHECSVAAGVTAAPLPLEDQWPSPPTRRAPVAPTEEQLRQEPWYHGRMSRRAAERMLRADGDFLVRDSVTNPGQYVLTGMHAGQPK HLLLVDPEGVVRTKDVLFESISHLIDHHLQNGQPIVAAESELHLRGVVSREP -------------------------------------------------------------- >343_343_2_ABCF1-SHC2_ABCF1_chr6_30554488_ENST00000376545_SHC2_chr19_440932_ENST00000264554_length(transcript)=4038nt_BP=2012nt GCGCCAGCTTGGAGAGCCAGCCCCATCGGGGTTCCCCGCCGCCGGAAGCGGAAATAGCACCGGGCGCCGCCACAGTAGCTGTAACTGCCA CCGCGATGCCGAAGGCGCCCAAGCAGCAGCCGCCGGAGCCCGAGTGGATCGGGGACGGAGAGAGCACGAGCCCATCAGACAAAGTGGTGA AGAAAGGGAAGAAGGACAAGAAGATCAAAAAAACGTTCTTTGAAGAGCTGGCAGTAGAAGATAAACAGGCTGGGGAAGAAGAGAAAGTGC TCAAGGAGAAGGAGCAGCAGCAGCAGCAACAGCAACAGCAGCAAAAAAAAAAGCGAGATACCCGAAAAGGCAGGCGGAAGAAGGATGTGG ATGATGATGGAGAAGAGAAAGAGCTCATGGAGCGTCTTAAGAAGCTCTCAGTGCCAACCAGTGATGAGGAGGATGAAGTACCCGCCCCAA AACCCCGCGGAGGGAAGAAAACCAAGGGTGGTAATGTTTTTGCAGCCCTGATTCAGGATCAGAGTGAGGAAGAGGAGGAGGAAGAAAAAC ATCCTCCTAAGCCTGCCAAGCCGGAGAAGAATCGGATCAATAAGGCCGTATCTGAGGAACAGCAGCCTGCACTCAAGGGCAAAAAGGGAA AGGAAGAGAAGTCAAAAGGGAAGGCTAAGCCTCAAAATAAATTCGCTGCTCTGGACAATGAAGAGGAGGATAAAGAAGAAGAAATTATAA AGGAAAAGGAGCCTCCCAAACAAGGGAAGGAGAAGGCCAAGAAGGCAGAGCAGATGGAGTATGAGCGCCAAGTGGCTTCATTAAAAGCAG CCAATGCAGCTGAAAATGACTTCTCCGTGTCCCAGGCGGAGATGTCCTCCCGCCAAGCCATGTTAGAAAATGCATCTGACATCAAGCTGG AGAAGTTCAGCATCTCCGCTCATGGCAAGGAGCTGTTCGTCAATGCAGACCTGTACATTGTAGCCGGCCGCCGCTACGGGCTGGTAGGAC CCAATGGCAAGGGCAAGACCACACTCCTCAAGCACATTGCCAACCGAGCCCTGAGCATCCCTCCCAACATTGATGTGTTGCTGTGTGAGC AGGAGGTGGTAGCAGATGAGACACCAGCAGTCCAGGCTGTTCTTCGAGCTGACACCAAGCGATTGAAGCTGCTGGAAGAGGAGCGGCGGC TTCAGGGACAGCTGGAACAAGGGGATGACACAGCTGCTGAGAGGCTAGAGAAGGTGTATGAGGAATTGCGGGCCACTGGGGCGGCAGCTG CAGAGGCCAAAGCACGGCGGATCCTGGCTGGCCTGGGCTTTGACCCTGAAATGCAGAATCGACCCACACAGAAGTTCTCAGGGGGCTGGC GCATGCGTGTCTCCCTGGCCAGGGCACTGTTCATGGAGCCCACACTGCTGATGCTGGATGAGCCCACCAACCACCTGGACCTCAACGCTG TCATCTGGCTTAATAACTACCTCCAGGGCTGGCGGAAGACCTTGCTGATCGTCTCCCATGACCAGGGCTTCTTGGATGATGTCTGCACTG ATATCATCCACCTCGATGCCCAGCGGCTCCACTACTATAGGGGCAATTACATGACCTTCAAAAAGATGTACCAGCAGAAGCAGAAAGAAC TGCTGAAACAGTATGAGAAGCAAGAGAAAAAGCTGAAGGAGCTGAAGGCAGGCGGGAAGTCCACCAAGCAGGCGGAAAAACAAACGAAGG AAGCCCTGACTCGGAAGCAGCAGAAATGCCGACGGAAAAACCAAGATGAGGAATCCCAGGAGGCCCCTGAGCTCCTGAAGCGCCCTAAGG AGTACACTGTGCGCTTCACTTTTCCAGACCCCCCACCACTCAGCCCTCCAGTGCTGGGTCTGCATGGTGTGACATTCGGCTACCAGGGAC AGAAACCACTCTTTAAGAACTTGGATTTTGGCATCGACATGGATTCAAGGATTTGCATTGTGGGCCCTAATGGTGTGGGGAAGAGTACGC TACTCCTGCTGCTGACTGGCAAGCTGACACCGTACATGGGCTGCATCGAGGTTCTCCGCTCTATGCGCTCCCTGGACTTTAACACGCGCA CGCAGGTGACCAGGGAAGCCATCAACCGGCTCCATGAGGCCGTGCCTGGCGTCCGGGGATCCTGGAAGAAAAAGGCCCCCAACAAGGCCC TGGCGTCCGTCCTGGGCAAGAGCAACCTTCGCTTTGCCGGCATGAGCATCTCCATCCACATCTCCACTGATGGCCTCAGCCTCTCCGTGC CTGCCACGCGCCAGGTCATCGCCAACCACCACATGCCGTCCATCTCCTTCGCGTCAGGCGGAGACACGGACATGACGGATTACGTGGCCT ACGTCGCCAAGGACCCCATCAACCAGAGAGCCTGCCACATCCTGGAGTGCTGTGAGGGCCTGGCACAGAGCATCATCAGCACCGTGGGCC AAGCTTTCGAGCTGCGCTTCAAGCAGTACCTGCACAGCCCGCCCAAGGTGGCGCTGCCCCCAGAAAGGCTGGCAGGGCCGGAGGAGTCGG CCTGGGGGGACGAGGAGGACTCTTTGGAGCACAATTACTACAACAGCATCCCGGGGAAGGAGCCGCCGCTGGGCGGGCTAGTGGACTCCA GGCTGGCCCTGACACAGCCCTGCGCCCTCACGGCCCTCGACCAGGGCCCATCTCCTTCTCTAAGAGATGCCTGCAGCCTGCCATGGGACG TGGGGTCCACCGGTACAGCTCCACCGGGGGACGGCTACGTGCAGGCGGACGCCCGGGGCCCCCCGGACCACGAGGAGCACCTGTATGTCA ACACCCAGGGTCTGGACGCCCCCGAGCCGGAGGACAGCCCCAAAAAGGATCTGTTTGACATGCGACCCTTTGAGGATGCCCTGAAGTTGC ATGAGTGCTCAGTGGCGGCAGGCGTGACAGCAGCCCCTCTTCCCTTGGAGGACCAGTGGCCCAGCCCCCCTACCCGCCGGGCCCCTGTGG CCCCCACGGAGGAACAGCTGCGTCAGGAGCCCTGGTACCACGGCCGGATGAGCCGCCGGGCGGCAGAGAGGATGCTTCGAGCTGACGGGG ACTTCCTTGTGCGAGACAGCGTCACCAACCCCGGGCAGTATGTCCTCACCGGCATGCACGCCGGGCAGCCCAAGCACCTGCTGCTCGTGG ACCCCGAGGGCGTGGTACGGACGAAGGACGTGCTGTTTGAGAGCATCAGCCACCTGATCGACCACCACCTGCAGAACGGGCAGCCCATCG TGGCCGCCGAGAGTGAGCTGCACCTGCGTGGCGTGGTCTCACGGGAGCCCTGAGCCAGGTGACCGTTCTCAGCCCTGGCTCCTGCCTGTT CTCAGCCCCGGCTCCTGCCTGTCATGGCCTTGGGGCTCTCAGCCCTCCTCCTGGATCCTGGTCAGGCCGAACCACCGCTGCCTCTCCTTC CTCCGCATGAGCCTCTGGCATGGTCCTTCCTCCAGCTGGCCCCGGGCTGGGCAGAGCCTCCTCCTGCCGGGGCCCCTGCCCACCCCCTCC TTTGCCTGGAGTGAGGGTGTTCATACCAAAGACGGAACCATTTCGCCTTTAAAGAAAATATATCCAGAAGCAGCCGCTGCCTCGGAGCCC TGGCCCTTGGGTCCCCCTCTCGCCTGGCTGGTTCGGTCTAACGCCCCGGAGAGTCAGGGCTCCCAGGACCCTGGGGAGGAGGCGGATTCC GGGCCTGGCTGGGCTTCCTCTTCCCACCTGAGGACTGGGTGCACAGTTGTCTTTGAGGGGGGACGCTTAAGGTGCTTTGGGGTTCTCAGG CCAGGATACATGCTGGCGCTGAAGTGCAGGCAGCCTTGAGGTCACCTGGGATCTCGGGGTAGCCAGGCTGCTCCAAGACAGTGGATATCG AGGCAGTCGTGAGGCCCCTACTCCACCTGGGAGCAGGGGAAGGACGTGGTTGCCTGGCCTGGGCTGCGCCCAGCAGCTTCCCCCCAGTCC TGCCCTCCCAGCTGTCGACCCAGATGGGATGTTCGGATCGGTTTGTAATTAAACCTGGGAATGGCCACAAGAGCACAA >343_343_2_ABCF1-SHC2_ABCF1_chr6_30554488_ENST00000376545_SHC2_chr19_440932_ENST00000264554_length(amino acids)=1094AA_BP=668 MESQPHRGSPPPEAEIAPGAATVAVTATAMPKAPKQQPPEPEWIGDGESTSPSDKVVKKGKKDKKIKKTFFEELAVEDKQAGEEEKVLKE KEQQQQQQQQQQKKKRDTRKGRRKKDVDDDGEEKELMERLKKLSVPTSDEEDEVPAPKPRGGKKTKGGNVFAALIQDQSEEEEEEEKHPP KPAKPEKNRINKAVSEEQQPALKGKKGKEEKSKGKAKPQNKFAALDNEEEDKEEEIIKEKEPPKQGKEKAKKAEQMEYERQVASLKAANA AENDFSVSQAEMSSRQAMLENASDIKLEKFSISAHGKELFVNADLYIVAGRRYGLVGPNGKGKTTLLKHIANRALSIPPNIDVLLCEQEV VADETPAVQAVLRADTKRLKLLEEERRLQGQLEQGDDTAAERLEKVYEELRATGAAAAEAKARRILAGLGFDPEMQNRPTQKFSGGWRMR VSLARALFMEPTLLMLDEPTNHLDLNAVIWLNNYLQGWRKTLLIVSHDQGFLDDVCTDIIHLDAQRLHYYRGNYMTFKKMYQQKQKELLK QYEKQEKKLKELKAGGKSTKQAEKQTKEALTRKQQKCRRKNQDEESQEAPELLKRPKEYTVRFTFPDPPPLSPPVLGLHGVTFGYQGQKP LFKNLDFGIDMDSRICIVGPNGVGKSTLLLLLTGKLTPYMGCIEVLRSMRSLDFNTRTQVTREAINRLHEAVPGVRGSWKKKAPNKALAS VLGKSNLRFAGMSISIHISTDGLSLSVPATRQVIANHHMPSISFASGGDTDMTDYVAYVAKDPINQRACHILECCEGLAQSIISTVGQAF ELRFKQYLHSPPKVALPPERLAGPEESAWGDEEDSLEHNYYNSIPGKEPPLGGLVDSRLALTQPCALTALDQGPSPSLRDACSLPWDVGS TGTAPPGDGYVQADARGPPDHEEHLYVNTQGLDAPEPEDSPKKDLFDMRPFEDALKLHECSVAAGVTAAPLPLEDQWPSPPTRRAPVAPT EEQLRQEPWYHGRMSRRAAERMLRADGDFLVRDSVTNPGQYVLTGMHAGQPKHLLLVDPEGVVRTKDVLFESISHLIDHHLQNGQPIVAA ESELHLRGVVSREP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ABCF1-SHC2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ABCF1-SHC2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ABCF1-SHC2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ABCF1 | C0020517 | Hypersensitivity | 1 | CTD_human |
| Hgene | ABCF1 | C0032285 | Pneumonia | 1 | CTD_human |
| Hgene | ABCF1 | C0032300 | Lobar Pneumonia | 1 | CTD_human |
| Hgene | ABCF1 | C0235874 | Disease Exacerbation | 1 | CTD_human |
| Hgene | ABCF1 | C0887898 | Experimental Lung Inflammation | 1 | CTD_human |
| Hgene | ABCF1 | C1527304 | Allergic Reaction | 1 | CTD_human |
| Hgene | ABCF1 | C3714636 | Pneumonitis | 1 | CTD_human |
