
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AUTS2-ACTR1A (FusionGDB2 ID:HG26053TG10121) |
Fusion Gene Summary for AUTS2-ACTR1A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AUTS2-ACTR1A | Fusion gene ID: hg26053tg10121 | Hgene | Tgene | Gene symbol | AUTS2 | ACTR1A | Gene ID | 26053 | 10121 |
| Gene name | activator of transcription and developmental regulator AUTS2 | actin related protein 1A | |
| Synonyms | FBRSL2|MRD26 | ARP1|Arp1A|CTRN1 | |
| Cytomap | ('AUTS2')('ACTR1A') 7q11.22 | 10q24.32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | autism susceptibility gene 2 proteinAUTS2, activator of transcription and developmental regulatorautism susceptibility candidate 2autism-related protein 1 | alpha-centractinARP1 actin related protein 1 homolog AARP1 actin-related protein 1 homolog A, centractin alphaactin-RPVcentractincentrosome-associated actin homologepididymis secretory sperm binding protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000342771, ENST00000403018, ENST00000406775, ENST00000489774, | ||
| Fusion gene scores | * DoF score | 29 X 23 X 8=5336 | 5 X 5 X 4=100 |
| # samples | 30 | 5 | |
| ** MAII score | log2(30/5336*10)=-4.1527242606887 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AUTS2 [Title/Abstract] AND ACTR1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AUTS2(69364484)-ACTR1A(104250370), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | AUTS2-ACTR1A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AUTS2-ACTR1A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AUTS2 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 25519132 |
| Hgene | AUTS2 | GO:0051571 | positive regulation of histone H3-K4 methylation | 25519132 |
| Hgene | AUTS2 | GO:2000620 | positive regulation of histone H4-K16 acetylation | 25519132 |
 Fusion gene breakpoints across AUTS2 (5'-gene) Fusion gene breakpoints across AUTS2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
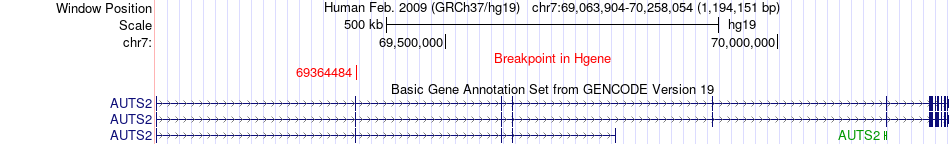 |
 Fusion gene breakpoints across ACTR1A (3'-gene) Fusion gene breakpoints across ACTR1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
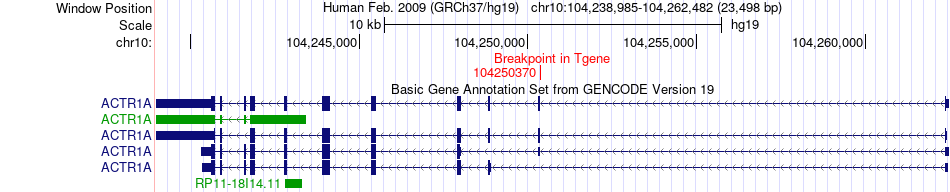 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-LI-A67I-01A | AUTS2 | chr7 | 69364484 | - | ACTR1A | chr10 | 104250370 | - |
| ChimerDB4 | SARC | TCGA-LI-A67I-01A | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
Top |
Fusion Gene ORF analysis for AUTS2-ACTR1A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000342771 | ENST00000545684 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-5UTR | ENST00000403018 | ENST00000545684 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-5UTR | ENST00000406775 | ENST00000545684 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000342771 | ENST00000446605 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000342771 | ENST00000470322 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000342771 | ENST00000487599 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000403018 | ENST00000446605 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000403018 | ENST00000470322 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000403018 | ENST00000487599 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000406775 | ENST00000446605 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000406775 | ENST00000470322 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| 5CDS-intron | ENST00000406775 | ENST00000487599 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| In-frame | ENST00000342771 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| In-frame | ENST00000403018 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| In-frame | ENST00000406775 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| intron-3CDS | ENST00000489774 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| intron-5UTR | ENST00000489774 | ENST00000545684 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| intron-intron | ENST00000489774 | ENST00000446605 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| intron-intron | ENST00000489774 | ENST00000470322 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
| intron-intron | ENST00000489774 | ENST00000487599 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000406775 | AUTS2 | chr7 | 69364484 | + | ENST00000369905 | ACTR1A | chr10 | 104250370 | - | 3974 | 1257 | 735 | 2339 | 534 |
| ENST00000342771 | AUTS2 | chr7 | 69364484 | + | ENST00000369905 | ACTR1A | chr10 | 104250370 | - | 3560 | 843 | 321 | 1925 | 534 |
| ENST00000403018 | AUTS2 | chr7 | 69364484 | + | ENST00000369905 | ACTR1A | chr10 | 104250370 | - | 3282 | 565 | 43 | 1647 | 534 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000406775 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - | 0.004293852 | 0.99570614 |
| ENST00000342771 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - | 0.00441218 | 0.9955878 |
| ENST00000403018 | ENST00000369905 | AUTS2 | chr7 | 69364484 | + | ACTR1A | chr10 | 104250370 | - | 0.004612472 | 0.9953875 |
Top |
Fusion Genomic Features for AUTS2-ACTR1A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
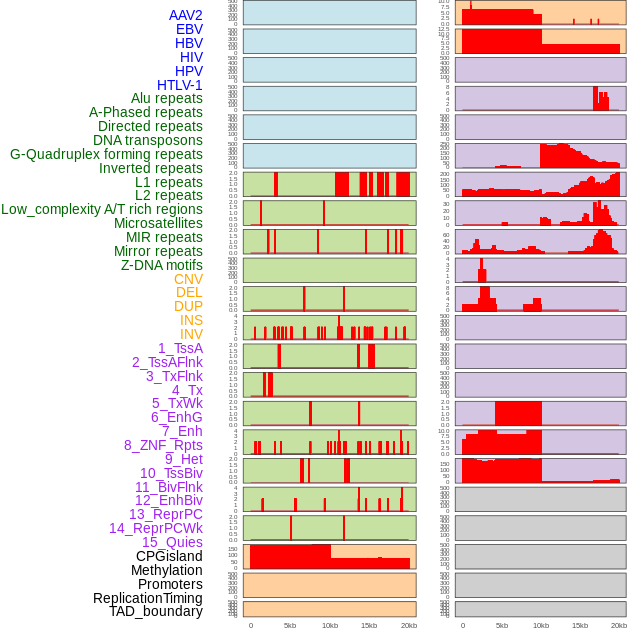 |
Top |
Fusion Protein Features for AUTS2-ACTR1A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:69364484/chr10:104250370) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000342771 | + | 2 | 19 | 1122_1181 | 174 | 1260.0 | Compositional bias | His-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000342771 | + | 2 | 19 | 288_471 | 174 | 1260.0 | Compositional bias | Pro-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000342771 | + | 2 | 19 | 525_548 | 174 | 1260.0 | Compositional bias | His-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000342771 | + | 2 | 19 | 545_646 | 174 | 1260.0 | Compositional bias | Pro-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000403018 | + | 2 | 5 | 1122_1181 | 174 | 267.0 | Compositional bias | His-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000403018 | + | 2 | 5 | 288_471 | 174 | 267.0 | Compositional bias | Pro-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000403018 | + | 2 | 5 | 525_548 | 174 | 267.0 | Compositional bias | His-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000403018 | + | 2 | 5 | 545_646 | 174 | 267.0 | Compositional bias | Pro-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000406775 | + | 2 | 18 | 1122_1181 | 174 | 1236.0 | Compositional bias | His-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000406775 | + | 2 | 18 | 288_471 | 174 | 1236.0 | Compositional bias | Pro-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000406775 | + | 2 | 18 | 525_548 | 174 | 1236.0 | Compositional bias | His-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000406775 | + | 2 | 18 | 545_646 | 174 | 1236.0 | Compositional bias | Pro-rich |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000342771 | + | 2 | 19 | 289_472 | 174 | 1260.0 | Region | Important for regulation of lamellipodia formation |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000403018 | + | 2 | 5 | 289_472 | 174 | 267.0 | Region | Important for regulation of lamellipodia formation |
| Hgene | AUTS2 | chr7:69364484 | chr10:104250370 | ENST00000406775 | + | 2 | 18 | 289_472 | 174 | 1236.0 | Region | Important for regulation of lamellipodia formation |
Top |
Fusion Gene Sequence for AUTS2-ACTR1A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >8493_8493_1_AUTS2-ACTR1A_AUTS2_chr7_69364484_ENST00000342771_ACTR1A_chr10_104250370_ENST00000369905_length(transcript)=3560nt_BP=843nt CTGTGTGTTCAGCCATTACTTTGCTCGGCGCTGCTCCCAGGCATCTCCGACCCTCGGTGCTGTGGGGAGCCCCACACTTGGGCTCCTCGC CTCTCGCCCTCGCTCCCCGTCCCTCCTCCCCTCTCTCCGCCCCTTCCCCCTTTTCTTTCTCCTCTCTTTCTTCCCCTCTCTCCCTTCTTT CGGCCGCCGTCTCCCCCGCGCCCTCCTCGGGGCGGAGGGAAGCCGTGAAGGGGGAGGGAGGGCTCGGTGTCAATTTTTTTTTGTGTGGCT GCGGCCGTAGCCTGTGGCGGGCAAGCGGGGAGACCCCGGCGCAGCAGAACCATGGATGGCCCGACGCGGGGCCATGGACTCCGCAAAAAG CGGCGGTCGCGGTCGCAGCGAGACCGGGAGAGGCGCTCCCGGGGCGGGCTGGGGGCCGGCGCGGCCGGCGGCGGCGGGGCTGGCCGGACC CGGGCGCTCTCACTCGCCTCGTCGTCGGGCTCCGACAAGGAAGACAATGGGAAGCCCCCGTCCTCCGCCCCGTCCCGGCCCAGACCCCCG CGGAGGAAGCGGAGAGAGTCCACCTCGGCAGAAGAGGACATCATTGATGGATTTGCCATGACCAGCTTTGTCACTTTTGAAGCGCTGGAG AAAGATGTAGCACTTAAGCCTCAGGAACGTGTGGAGAAACGCCAGACGCCCCTGACCAAGAAGAAACGAGAAGCACTTACCAATGGCTTG TCCTTTCATTCAAAGAAGAGCAGACTCAGCCACCCACACCACTACAGCTCAGATCGAGAAAATGACCGCAATCTCTGCCAGCACCTTGGG AAGAGAAAGAAAATGCCGAAGGCACTCAGACAGGGATCCGGTGTGATTAAAGCTGGTTTTGCTGGTGATCAGATCCCCAAATACTGCTTT CCAAACTATGTGGGCCGACCCAAGCACGTTCGTGTCATGGCAGGAGCCCTTGAAGGCGACATCTTCATTGGCCCCAAAGCTGAGGAGCAC CGAGGGCTGCTTTCAATCCGCTATCCCATGGAGCATGGCATCGTCAAGGATTGGAACGACATGGAACGCATTTGGCAATATGTCTATTCT AAGGACCAGCTGCAGACTTTCTCAGAGGAGCATCCTGTGCTCCTGACTGAGGCGCCTTTAAACCCACGAAAAAACCGGGAACGAGCTGCC GAAGTTTTCTTCGAGACCTTCAATGTGCCCGCTCTTTTCATCTCCATGCAAGCTGTACTCAGCCTTTACGCTACAGGCAGGACCACAGGG GTGGTGCTGGATTCTGGGGATGGAGTCACCCATGCTGTGCCCATCTATGAGGGCTTTGCCATGCCCCACTCCATCATGCGCATCGACATC GCGGGCCGGGACGTCTCTCGCTTCCTGCGCCTCTACCTGCGTAAGGAGGGCTACGACTTCCACTCATCCTCTGAGTTTGAGATTGTCAAG GCCATAAAAGAAAGAGCCTGTTACCTATCCATAAACCCCCAAAAGGATGAGACGCTAGAGACAGAGAAAGCTCAGTACTACCTGCCTGAT GGCAGCACCATTGAGATTGGTCCTTCCCGATTCCGGGCCCCTGAGTTGCTCTTCAGGCCAGATTTGATTGGAGAGGAGAGTGAAGGCATC CACGAGGTCCTGGTGTTCGCCATTCAGAAGTCAGACATGGACCTGCGGCGCACGCTTTTCTCTAACATTGTCCTCTCAGGAGGCTCTACC CTGTTCAAAGGTTTTGGTGACAGGCTCCTGAGTGAAGTGAAGAAACTAGCTCCAAAAGATGTGAAGATCAGGATATCTGCACCTCAGGAG AGACTGTATTCCACGTGGATTGGGGGCTCCATCCTTGCCTCCCTGGACACCTTTAAGAAGATGTGGGTCTCCAAAAAGGAATATGAGGAA GACGGTGCCCGATCCATCCACAGAAAAACCTTCTAATGTCGGGACATCATCTTCACCTCTCTCTGAAGTTAACTCCACTTTAAAACTCGC TTTCTTGAGTCGGAGTGTTTGCGAGGAACTGCCTGTGTGTGAGTGCGTGTGTGGATATGAGTGTGTGTGCACATGCGAGTGCCGTGTGGC CCTGGGACCCTGGGCCCAGAAAGGACGATGAACTACCTGCAGTGGTGATGGCCTGAGGCCTGGGGTTGACCACTAACTGGCTCCTGACAG GGAAGAGCGCTGGCAGAGGCTGTGCTCCCTCCTCAGGTGGCCTCTGGCTGGCTGTGGGGGACTCCGTTTACTACCACAGGGAGACAGAGG GAGGTAAGCCATCCCCCGGGAGACCTTGCTGCTGACCATCCTAGGCTGGGCTGGCCCCACCCTCACCCCCACCCCCAGGGTGCCCTGAGG CCCCAGGCAGCTGCTGCCTCCACTATCGATGCCTCCTGACTGCACACTGAGGACTGGGACTGGGGTTGAGTTCTGTCTGGTTTTGTTGCC ATTTTGGTTTGGGAGGCTGGAAAAGCACCCCAAGAGCTATTACAGAGACTGGAGTCAGGAGAGAGCAGGAGGCCCTCATGTTCACCAGGG AACAGGACCACACCGGCCACTGGAGGAGGGCAGGAGCAGTCCTCACTCTGAATGGCTGCAGAGTTAATGTTCCCAGCCCAGTCCCCTTTC GGGGGCCTTGGGAGAGTTTAAGGCACCTGCTGGTTCCAGGACCTCGCTTTCCATCTGTTCTTGTTGCAATGCCATCTTCAAACCGTTTTA TTTATTGAAGTGTTTGTTCAGTTAGGGGCTGGAGAGAGGGAGCTTGCTGCCTCCTGCCTTGCTACACTAATGTTTACAGCACCTAAGCTT AGCCTCCAGGGCCCCACCTCTCCCAGCTGATGGTGAGCTGACAGTGTCCACAGGTTCCAGGACCATTTGAGATTGGAAGCTACACTCAAA GACACTCCCACCAGGCTCTTTCTCCCTTTTCCTCTTGCTCACTGCCCTGGAATCAACAGGCTGGTTGCTGGTTAGATTTTCTGAAACAGG AGGTAAAATTTTTCTTTGGCAGAGGCCCCTAAGCAAGGGAGGGGTGTTGGAGAGCCAGTGCCCTTAAGACTGGAGAAAGCTGCAATTTAC CAAGTTGCCTTTTGCCACTGTAGCTGACCAGGGGACTAGGTTGTAGAGGTGGGAAGGCCCCCTCTGGGCTGATCTTGTGCCATTCTTGAC CTTGGACCTGCTTGGTTAAGGAGGGAGTGGGCCAGACCAGAGTGCCAGGAGCTAATGGAGCCAGGCCTGACTCCTAGGAGTGGTCCAAAG GCCTTCAGCCTAGATGGTGCAAAGCTGGGGCCAGCCTGTCTTCACCGGCACCCTCACCTGTGACACCAAGACCCACCCCAATCCCAGACT TCACACAGTATTCTCCCCCACGCCGTCCTATGACCAAAGGCCCCTGCCAGGTGTGGGCCACAGCAGCAGGTATGTGTGAAAGCAACGTAG CGCCCCGCGGACTGCAGTGCGCTTAACCAACTCACCTCCCTTCTCTTAGCCCAAGCCTGTCCCTCGCACAGCCTCGCACAAACCACATTG CCTGGTGGGGCCCAGTGTACTGAAATAAAGTCGTTCCGATAGACACGTCA >8493_8493_1_AUTS2-ACTR1A_AUTS2_chr7_69364484_ENST00000342771_ACTR1A_chr10_104250370_ENST00000369905_length(amino acids)=534AA_BP=173 MDGPTRGHGLRKKRRSRSQRDRERRSRGGLGAGAAGGGGAGRTRALSLASSSGSDKEDNGKPPSSAPSRPRPPRRKRRESTSAEEDIIDG FAMTSFVTFEALEKDVALKPQERVEKRQTPLTKKKREALTNGLSFHSKKSRLSHPHHYSSDRENDRNLCQHLGKRKKMPKALRQGSGVIK AGFAGDQIPKYCFPNYVGRPKHVRVMAGALEGDIFIGPKAEEHRGLLSIRYPMEHGIVKDWNDMERIWQYVYSKDQLQTFSEEHPVLLTE APLNPRKNRERAAEVFFETFNVPALFISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFAMPHSIMRIDIAGRDVSRFLRLYLRKEG YDFHSSSEFEIVKAIKERACYLSINPQKDETLETEKAQYYLPDGSTIEIGPSRFRAPELLFRPDLIGEESEGIHEVLVFAIQKSDMDLRR TLFSNIVLSGGSTLFKGFGDRLLSEVKKLAPKDVKIRISAPQERLYSTWIGGSILASLDTFKKMWVSKKEYEEDGARSIHRKTF -------------------------------------------------------------- >8493_8493_2_AUTS2-ACTR1A_AUTS2_chr7_69364484_ENST00000403018_ACTR1A_chr10_104250370_ENST00000369905_length(transcript)=3282nt_BP=565nt AGCCTGTGGCGGGCAAGCGGGGAGACCCCGGCGCAGCAGAACCATGGATGGCCCGACGCGGGGCCATGGACTCCGCAAAAAGCGGCGGTC GCGGTCGCAGCGAGACCGGGAGAGGCGCTCCCGGGGCGGGCTGGGGGCCGGCGCGGCCGGCGGCGGCGGGGCTGGCCGGACCCGGGCGCT CTCACTCGCCTCGTCGTCGGGCTCCGACAAGGAAGACAATGGGAAGCCCCCGTCCTCCGCCCCGTCCCGGCCCAGACCCCCGCGGAGGAA GCGGAGAGAGTCCACCTCGGCAGAAGAGGACATCATTGATGGATTTGCCATGACCAGCTTTGTCACTTTTGAAGCGCTGGAGAAAGATGT AGCACTTAAGCCTCAGGAACGTGTGGAGAAACGCCAGACGCCCCTGACCAAGAAGAAACGAGAAGCACTTACCAATGGCTTGTCCTTTCA TTCAAAGAAGAGCAGACTCAGCCACCCACACCACTACAGCTCAGATCGAGAAAATGACCGCAATCTCTGCCAGCACCTTGGGAAGAGAAA GAAAATGCCGAAGGCACTCAGACAGGGATCCGGTGTGATTAAAGCTGGTTTTGCTGGTGATCAGATCCCCAAATACTGCTTTCCAAACTA TGTGGGCCGACCCAAGCACGTTCGTGTCATGGCAGGAGCCCTTGAAGGCGACATCTTCATTGGCCCCAAAGCTGAGGAGCACCGAGGGCT GCTTTCAATCCGCTATCCCATGGAGCATGGCATCGTCAAGGATTGGAACGACATGGAACGCATTTGGCAATATGTCTATTCTAAGGACCA GCTGCAGACTTTCTCAGAGGAGCATCCTGTGCTCCTGACTGAGGCGCCTTTAAACCCACGAAAAAACCGGGAACGAGCTGCCGAAGTTTT CTTCGAGACCTTCAATGTGCCCGCTCTTTTCATCTCCATGCAAGCTGTACTCAGCCTTTACGCTACAGGCAGGACCACAGGGGTGGTGCT GGATTCTGGGGATGGAGTCACCCATGCTGTGCCCATCTATGAGGGCTTTGCCATGCCCCACTCCATCATGCGCATCGACATCGCGGGCCG GGACGTCTCTCGCTTCCTGCGCCTCTACCTGCGTAAGGAGGGCTACGACTTCCACTCATCCTCTGAGTTTGAGATTGTCAAGGCCATAAA AGAAAGAGCCTGTTACCTATCCATAAACCCCCAAAAGGATGAGACGCTAGAGACAGAGAAAGCTCAGTACTACCTGCCTGATGGCAGCAC CATTGAGATTGGTCCTTCCCGATTCCGGGCCCCTGAGTTGCTCTTCAGGCCAGATTTGATTGGAGAGGAGAGTGAAGGCATCCACGAGGT CCTGGTGTTCGCCATTCAGAAGTCAGACATGGACCTGCGGCGCACGCTTTTCTCTAACATTGTCCTCTCAGGAGGCTCTACCCTGTTCAA AGGTTTTGGTGACAGGCTCCTGAGTGAAGTGAAGAAACTAGCTCCAAAAGATGTGAAGATCAGGATATCTGCACCTCAGGAGAGACTGTA TTCCACGTGGATTGGGGGCTCCATCCTTGCCTCCCTGGACACCTTTAAGAAGATGTGGGTCTCCAAAAAGGAATATGAGGAAGACGGTGC CCGATCCATCCACAGAAAAACCTTCTAATGTCGGGACATCATCTTCACCTCTCTCTGAAGTTAACTCCACTTTAAAACTCGCTTTCTTGA GTCGGAGTGTTTGCGAGGAACTGCCTGTGTGTGAGTGCGTGTGTGGATATGAGTGTGTGTGCACATGCGAGTGCCGTGTGGCCCTGGGAC CCTGGGCCCAGAAAGGACGATGAACTACCTGCAGTGGTGATGGCCTGAGGCCTGGGGTTGACCACTAACTGGCTCCTGACAGGGAAGAGC GCTGGCAGAGGCTGTGCTCCCTCCTCAGGTGGCCTCTGGCTGGCTGTGGGGGACTCCGTTTACTACCACAGGGAGACAGAGGGAGGTAAG CCATCCCCCGGGAGACCTTGCTGCTGACCATCCTAGGCTGGGCTGGCCCCACCCTCACCCCCACCCCCAGGGTGCCCTGAGGCCCCAGGC AGCTGCTGCCTCCACTATCGATGCCTCCTGACTGCACACTGAGGACTGGGACTGGGGTTGAGTTCTGTCTGGTTTTGTTGCCATTTTGGT TTGGGAGGCTGGAAAAGCACCCCAAGAGCTATTACAGAGACTGGAGTCAGGAGAGAGCAGGAGGCCCTCATGTTCACCAGGGAACAGGAC CACACCGGCCACTGGAGGAGGGCAGGAGCAGTCCTCACTCTGAATGGCTGCAGAGTTAATGTTCCCAGCCCAGTCCCCTTTCGGGGGCCT TGGGAGAGTTTAAGGCACCTGCTGGTTCCAGGACCTCGCTTTCCATCTGTTCTTGTTGCAATGCCATCTTCAAACCGTTTTATTTATTGA AGTGTTTGTTCAGTTAGGGGCTGGAGAGAGGGAGCTTGCTGCCTCCTGCCTTGCTACACTAATGTTTACAGCACCTAAGCTTAGCCTCCA GGGCCCCACCTCTCCCAGCTGATGGTGAGCTGACAGTGTCCACAGGTTCCAGGACCATTTGAGATTGGAAGCTACACTCAAAGACACTCC CACCAGGCTCTTTCTCCCTTTTCCTCTTGCTCACTGCCCTGGAATCAACAGGCTGGTTGCTGGTTAGATTTTCTGAAACAGGAGGTAAAA TTTTTCTTTGGCAGAGGCCCCTAAGCAAGGGAGGGGTGTTGGAGAGCCAGTGCCCTTAAGACTGGAGAAAGCTGCAATTTACCAAGTTGC CTTTTGCCACTGTAGCTGACCAGGGGACTAGGTTGTAGAGGTGGGAAGGCCCCCTCTGGGCTGATCTTGTGCCATTCTTGACCTTGGACC TGCTTGGTTAAGGAGGGAGTGGGCCAGACCAGAGTGCCAGGAGCTAATGGAGCCAGGCCTGACTCCTAGGAGTGGTCCAAAGGCCTTCAG CCTAGATGGTGCAAAGCTGGGGCCAGCCTGTCTTCACCGGCACCCTCACCTGTGACACCAAGACCCACCCCAATCCCAGACTTCACACAG TATTCTCCCCCACGCCGTCCTATGACCAAAGGCCCCTGCCAGGTGTGGGCCACAGCAGCAGGTATGTGTGAAAGCAACGTAGCGCCCCGC GGACTGCAGTGCGCTTAACCAACTCACCTCCCTTCTCTTAGCCCAAGCCTGTCCCTCGCACAGCCTCGCACAAACCACATTGCCTGGTGG GGCCCAGTGTACTGAAATAAAGTCGTTCCGATAGACACGTCA >8493_8493_2_AUTS2-ACTR1A_AUTS2_chr7_69364484_ENST00000403018_ACTR1A_chr10_104250370_ENST00000369905_length(amino acids)=534AA_BP=173 MDGPTRGHGLRKKRRSRSQRDRERRSRGGLGAGAAGGGGAGRTRALSLASSSGSDKEDNGKPPSSAPSRPRPPRRKRRESTSAEEDIIDG FAMTSFVTFEALEKDVALKPQERVEKRQTPLTKKKREALTNGLSFHSKKSRLSHPHHYSSDRENDRNLCQHLGKRKKMPKALRQGSGVIK AGFAGDQIPKYCFPNYVGRPKHVRVMAGALEGDIFIGPKAEEHRGLLSIRYPMEHGIVKDWNDMERIWQYVYSKDQLQTFSEEHPVLLTE APLNPRKNRERAAEVFFETFNVPALFISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFAMPHSIMRIDIAGRDVSRFLRLYLRKEG YDFHSSSEFEIVKAIKERACYLSINPQKDETLETEKAQYYLPDGSTIEIGPSRFRAPELLFRPDLIGEESEGIHEVLVFAIQKSDMDLRR TLFSNIVLSGGSTLFKGFGDRLLSEVKKLAPKDVKIRISAPQERLYSTWIGGSILASLDTFKKMWVSKKEYEEDGARSIHRKTF -------------------------------------------------------------- >8493_8493_3_AUTS2-ACTR1A_AUTS2_chr7_69364484_ENST00000406775_ACTR1A_chr10_104250370_ENST00000369905_length(transcript)=3974nt_BP=1257nt CGCTCGCAGTTTCGCCCTCTCTTCCGCTAATGATTGCATTATTATGCTCCCCTCTCTGGGGGGTCTCGCCCCTCTTGGGTCGCTCCGGAG CCCCGGCCTCCCCTGGCTGCATTTCTTAAAAATTTGGGAGCCTGGGAGTGAGTTTTCTCCGAGGCGTGTGTGAGAGGCGGCGGGGGTGTT TTCCTGCGCGAGGGGCGGGTGAAGTTCATTGCCCCCACTTTTCCCGCGACCTTTTTCGGACCCGATTTTGGATCGAGTTGAGGGGGGCGC GGGCGTTTTCGGGGGGCGGGGGGCGCGGCGGAGAATGGCCGCGGGGAGGGCTCCCCGGAGCCTCCCAGTCTCTTGATCAAAGCATTCCGC TATTCTGATTTATTGCTTGCTTGGTGAGTTATTTTTTTTTCCTCTAAAGGAGACCTGTGTGTTCAGCCATTACTTTGCTCGGCGCTGCTC CCAGGCATCTCCGACCCTCGGTGCTGTGGGGAGCCCCACACTTGGGCTCCTCGCCTCTCGCCCTCGCTCCCCGTCCCTCCTCCCCTCTCT CCGCCCCTTCCCCCTTTTCTTTCTCCTCTCTTTCTTCCCCTCTCTCCCTTCTTTCGGCCGCCGTCTCCCCCGCGCCCTCCTCGGGGCGGA GGGAAGCCGTGAAGGGGGAGGGAGGGCTCGGTGTCAATTTTTTTTTGTGTGGCTGCGGCCGTAGCCTGTGGCGGGCAAGCGGGGAGACCC CGGCGCAGCAGAACCATGGATGGCCCGACGCGGGGCCATGGACTCCGCAAAAAGCGGCGGTCGCGGTCGCAGCGAGACCGGGAGAGGCGC TCCCGGGGCGGGCTGGGGGCCGGCGCGGCCGGCGGCGGCGGGGCTGGCCGGACCCGGGCGCTCTCACTCGCCTCGTCGTCGGGCTCCGAC AAGGAAGACAATGGGAAGCCCCCGTCCTCCGCCCCGTCCCGGCCCAGACCCCCGCGGAGGAAGCGGAGAGAGTCCACCTCGGCAGAAGAG GACATCATTGATGGATTTGCCATGACCAGCTTTGTCACTTTTGAAGCGCTGGAGAAAGATGTAGCACTTAAGCCTCAGGAACGTGTGGAG AAACGCCAGACGCCCCTGACCAAGAAGAAACGAGAAGCACTTACCAATGGCTTGTCCTTTCATTCAAAGAAGAGCAGACTCAGCCACCCA CACCACTACAGCTCAGATCGAGAAAATGACCGCAATCTCTGCCAGCACCTTGGGAAGAGAAAGAAAATGCCGAAGGCACTCAGACAGGGA TCCGGTGTGATTAAAGCTGGTTTTGCTGGTGATCAGATCCCCAAATACTGCTTTCCAAACTATGTGGGCCGACCCAAGCACGTTCGTGTC ATGGCAGGAGCCCTTGAAGGCGACATCTTCATTGGCCCCAAAGCTGAGGAGCACCGAGGGCTGCTTTCAATCCGCTATCCCATGGAGCAT GGCATCGTCAAGGATTGGAACGACATGGAACGCATTTGGCAATATGTCTATTCTAAGGACCAGCTGCAGACTTTCTCAGAGGAGCATCCT GTGCTCCTGACTGAGGCGCCTTTAAACCCACGAAAAAACCGGGAACGAGCTGCCGAAGTTTTCTTCGAGACCTTCAATGTGCCCGCTCTT TTCATCTCCATGCAAGCTGTACTCAGCCTTTACGCTACAGGCAGGACCACAGGGGTGGTGCTGGATTCTGGGGATGGAGTCACCCATGCT GTGCCCATCTATGAGGGCTTTGCCATGCCCCACTCCATCATGCGCATCGACATCGCGGGCCGGGACGTCTCTCGCTTCCTGCGCCTCTAC CTGCGTAAGGAGGGCTACGACTTCCACTCATCCTCTGAGTTTGAGATTGTCAAGGCCATAAAAGAAAGAGCCTGTTACCTATCCATAAAC CCCCAAAAGGATGAGACGCTAGAGACAGAGAAAGCTCAGTACTACCTGCCTGATGGCAGCACCATTGAGATTGGTCCTTCCCGATTCCGG GCCCCTGAGTTGCTCTTCAGGCCAGATTTGATTGGAGAGGAGAGTGAAGGCATCCACGAGGTCCTGGTGTTCGCCATTCAGAAGTCAGAC ATGGACCTGCGGCGCACGCTTTTCTCTAACATTGTCCTCTCAGGAGGCTCTACCCTGTTCAAAGGTTTTGGTGACAGGCTCCTGAGTGAA GTGAAGAAACTAGCTCCAAAAGATGTGAAGATCAGGATATCTGCACCTCAGGAGAGACTGTATTCCACGTGGATTGGGGGCTCCATCCTT GCCTCCCTGGACACCTTTAAGAAGATGTGGGTCTCCAAAAAGGAATATGAGGAAGACGGTGCCCGATCCATCCACAGAAAAACCTTCTAA TGTCGGGACATCATCTTCACCTCTCTCTGAAGTTAACTCCACTTTAAAACTCGCTTTCTTGAGTCGGAGTGTTTGCGAGGAACTGCCTGT GTGTGAGTGCGTGTGTGGATATGAGTGTGTGTGCACATGCGAGTGCCGTGTGGCCCTGGGACCCTGGGCCCAGAAAGGACGATGAACTAC CTGCAGTGGTGATGGCCTGAGGCCTGGGGTTGACCACTAACTGGCTCCTGACAGGGAAGAGCGCTGGCAGAGGCTGTGCTCCCTCCTCAG GTGGCCTCTGGCTGGCTGTGGGGGACTCCGTTTACTACCACAGGGAGACAGAGGGAGGTAAGCCATCCCCCGGGAGACCTTGCTGCTGAC CATCCTAGGCTGGGCTGGCCCCACCCTCACCCCCACCCCCAGGGTGCCCTGAGGCCCCAGGCAGCTGCTGCCTCCACTATCGATGCCTCC TGACTGCACACTGAGGACTGGGACTGGGGTTGAGTTCTGTCTGGTTTTGTTGCCATTTTGGTTTGGGAGGCTGGAAAAGCACCCCAAGAG CTATTACAGAGACTGGAGTCAGGAGAGAGCAGGAGGCCCTCATGTTCACCAGGGAACAGGACCACACCGGCCACTGGAGGAGGGCAGGAG CAGTCCTCACTCTGAATGGCTGCAGAGTTAATGTTCCCAGCCCAGTCCCCTTTCGGGGGCCTTGGGAGAGTTTAAGGCACCTGCTGGTTC CAGGACCTCGCTTTCCATCTGTTCTTGTTGCAATGCCATCTTCAAACCGTTTTATTTATTGAAGTGTTTGTTCAGTTAGGGGCTGGAGAG AGGGAGCTTGCTGCCTCCTGCCTTGCTACACTAATGTTTACAGCACCTAAGCTTAGCCTCCAGGGCCCCACCTCTCCCAGCTGATGGTGA GCTGACAGTGTCCACAGGTTCCAGGACCATTTGAGATTGGAAGCTACACTCAAAGACACTCCCACCAGGCTCTTTCTCCCTTTTCCTCTT GCTCACTGCCCTGGAATCAACAGGCTGGTTGCTGGTTAGATTTTCTGAAACAGGAGGTAAAATTTTTCTTTGGCAGAGGCCCCTAAGCAA GGGAGGGGTGTTGGAGAGCCAGTGCCCTTAAGACTGGAGAAAGCTGCAATTTACCAAGTTGCCTTTTGCCACTGTAGCTGACCAGGGGAC TAGGTTGTAGAGGTGGGAAGGCCCCCTCTGGGCTGATCTTGTGCCATTCTTGACCTTGGACCTGCTTGGTTAAGGAGGGAGTGGGCCAGA CCAGAGTGCCAGGAGCTAATGGAGCCAGGCCTGACTCCTAGGAGTGGTCCAAAGGCCTTCAGCCTAGATGGTGCAAAGCTGGGGCCAGCC TGTCTTCACCGGCACCCTCACCTGTGACACCAAGACCCACCCCAATCCCAGACTTCACACAGTATTCTCCCCCACGCCGTCCTATGACCA AAGGCCCCTGCCAGGTGTGGGCCACAGCAGCAGGTATGTGTGAAAGCAACGTAGCGCCCCGCGGACTGCAGTGCGCTTAACCAACTCACC TCCCTTCTCTTAGCCCAAGCCTGTCCCTCGCACAGCCTCGCACAAACCACATTGCCTGGTGGGGCCCAGTGTACTGAAATAAAGTCGTTC CGATAGACACGTCA >8493_8493_3_AUTS2-ACTR1A_AUTS2_chr7_69364484_ENST00000406775_ACTR1A_chr10_104250370_ENST00000369905_length(amino acids)=534AA_BP=173 MDGPTRGHGLRKKRRSRSQRDRERRSRGGLGAGAAGGGGAGRTRALSLASSSGSDKEDNGKPPSSAPSRPRPPRRKRRESTSAEEDIIDG FAMTSFVTFEALEKDVALKPQERVEKRQTPLTKKKREALTNGLSFHSKKSRLSHPHHYSSDRENDRNLCQHLGKRKKMPKALRQGSGVIK AGFAGDQIPKYCFPNYVGRPKHVRVMAGALEGDIFIGPKAEEHRGLLSIRYPMEHGIVKDWNDMERIWQYVYSKDQLQTFSEEHPVLLTE APLNPRKNRERAAEVFFETFNVPALFISMQAVLSLYATGRTTGVVLDSGDGVTHAVPIYEGFAMPHSIMRIDIAGRDVSRFLRLYLRKEG YDFHSSSEFEIVKAIKERACYLSINPQKDETLETEKAQYYLPDGSTIEIGPSRFRAPELLFRPDLIGEESEGIHEVLVFAIQKSDMDLRR TLFSNIVLSGGSTLFKGFGDRLLSEVKKLAPKDVKIRISAPQERLYSTWIGGSILASLDTFKKMWVSKKEYEEDGARSIHRKTF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AUTS2-ACTR1A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AUTS2-ACTR1A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AUTS2-ACTR1A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | AUTS2 | C0036341 | Schizophrenia | 2 | PSYGENET |
| Hgene | AUTS2 | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Hgene | AUTS2 | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Hgene | AUTS2 | C0014544 | Epilepsy | 1 | CTD_human |
| Hgene | AUTS2 | C0086237 | Epilepsy, Cryptogenic | 1 | CTD_human |
| Hgene | AUTS2 | C0236018 | Aura | 1 | CTD_human |
| Hgene | AUTS2 | C0751111 | Awakening Epilepsy | 1 | CTD_human |
| Hgene | AUTS2 | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Hgene | AUTS2 | C2677504 | AUTISM, SUSCEPTIBILITY TO, 15 | 1 | GENOMICS_ENGLAND |
| Hgene | AUTS2 | C4014435 | MENTAL RETARDATION, AUTOSOMAL DOMINANT 26 | 1 | CTD_human;ORPHANET |
