
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BLOC1S6-DYM (FusionGDB2 ID:HG26258TG54808) |
Fusion Gene Summary for BLOC1S6-DYM |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BLOC1S6-DYM | Fusion gene ID: hg26258tg54808 | Hgene | Tgene | Gene symbol | BLOC1S6 | DYM | Gene ID | 26258 | 54808 |
| Gene name | biogenesis of lysosomal organelles complex 1 subunit 6 | dymeclin | |
| Synonyms | BLOS6|HPS9|PA|PALLID|PLDN | DMC|SMC | |
| Cytomap | ('BLOC1S6')('DYM') 15q21.1 | 18q21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | biogenesis of lysosome-related organelles complex 1 subunit 6BLOC-1 subunit 6BLOC-1 subunit pallidinbiogenesis of lysosomal organelles complex-1, subunit 5, pallidinbiogenesis of lysosomal organelles complex-1, subunit 6, pallidinpallid protein homol | dymeclindyggve-Melchior-Clausen syndrome protein | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | . | Q7RTS9 | |
| Ensembl transtripts involved in fusion gene | ENST00000220531, ENST00000565323, ENST00000566753, ENST00000564765, ENST00000565409, ENST00000568816, ENST00000562384, ENST00000565216, ENST00000567461, ENST00000567740, | ||
| Fusion gene scores | * DoF score | 4 X 3 X 3=36 | 19 X 11 X 13=2717 |
| # samples | 5 | 24 | |
| ** MAII score | log2(5/36*10)=0.473931188332412 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(24/2717*10)=-3.50090825461346 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: BLOC1S6 [Title/Abstract] AND DYM [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BLOC1S6(45884474)-DYM(46918015), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BLOC1S6 | GO:0032402 | melanosome transport | 17182842 |
| Hgene | BLOC1S6 | GO:0035646 | endosome to melanosome transport | 17182842 |
| Hgene | BLOC1S6 | GO:0050942 | positive regulation of pigment cell differentiation | 17182842 |
 Fusion gene breakpoints across BLOC1S6 (5'-gene) Fusion gene breakpoints across BLOC1S6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
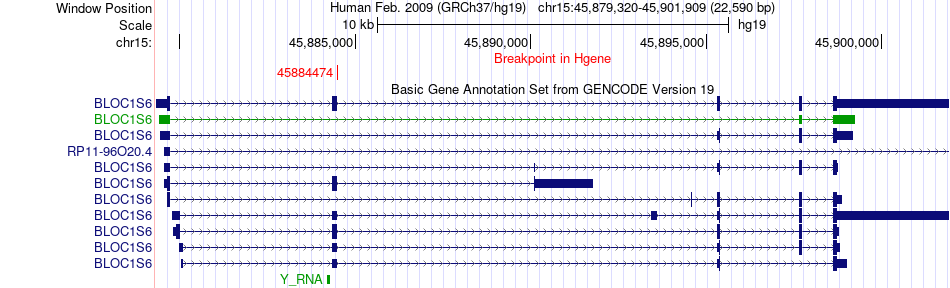 |
 Fusion gene breakpoints across DYM (3'-gene) Fusion gene breakpoints across DYM (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
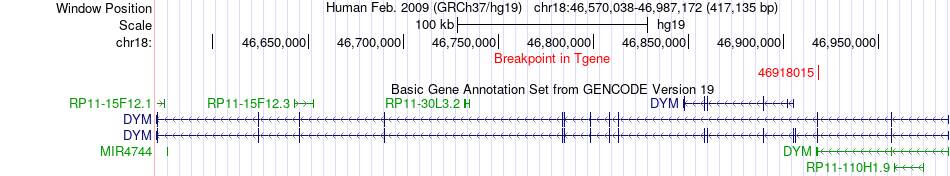 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
Top |
Fusion Gene ORF analysis for BLOC1S6-DYM |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000220531 | ENST00000442713 | BLOC1S6 | chr15 | 45884474 | + | DYM | chr18 | 46918015 | - | 0.000635072 | 0.9993649 |
| ENST00000220531 | ENST00000269445 | BLOC1S6 | chr15 | 45884474 | + | DYM | chr18 | 46918015 | - | 0.000863248 | 0.99913675 |
| ENST00000566753 | ENST00000442713 | BLOC1S6 | chr15 | 45884474 | + | DYM | chr18 | 46918015 | - | 0.000510646 | 0.9994893 |
| ENST00000566753 | ENST00000269445 | BLOC1S6 | chr15 | 45884474 | + | DYM | chr18 | 46918015 | - | 0.000751531 | 0.99924845 |
| ENST00000565323 | ENST00000442713 | BLOC1S6 | chr15 | 45884474 | + | DYM | chr18 | 46918015 | - | 0.000685329 | 0.99931467 |
| ENST00000565323 | ENST00000269445 | BLOC1S6 | chr15 | 45884474 | + | DYM | chr18 | 46918015 | - | 0.001556732 | 0.9984433 |
Top |
Fusion Genomic Features for BLOC1S6-DYM |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
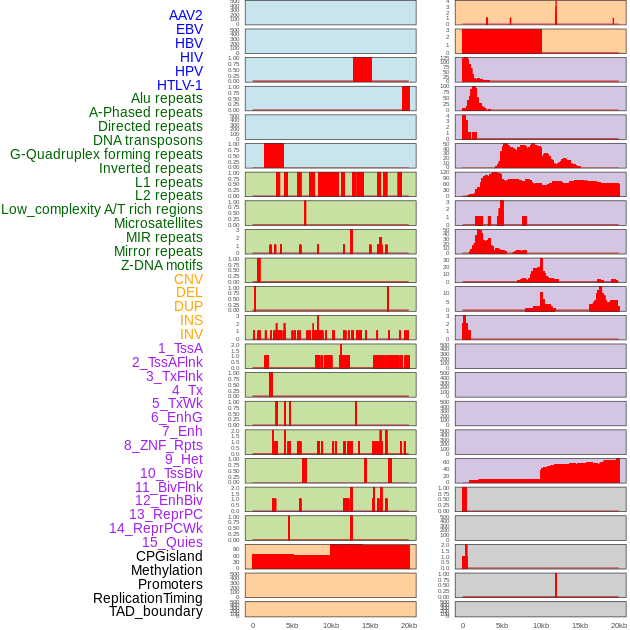 |
Top |
Fusion Protein Features for BLOC1S6-DYM |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:45884474/chr18:46918015) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | DYM |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Necessary for correct organization of Golgi apparatus. Involved in bone development. {ECO:0000269|PubMed:21280149}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BLOC1S6 | chr15:45884474 | chr18:46918015 | ENST00000220531 | + | 2 | 5 | 63_167 | 74 | 173.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for BLOC1S6-DYM |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >9833_9833_1_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000220531_DYM_chr18_46918015_ENST00000269445_length(transcript)=2650nt_BP=545nt ACCCACCCCGTGAGCAGCCCAAACCGACCACCGTCCTCCCCCAGCTCCCACCAATCGGATCGCAGGGACTCGAGCCCCACACTGCTGAGT CCGCTGACACTGCGTCCGGGGCCAGACGACGATATCAGCGCGGGGTCCCCACAACGCCATGGGGCAGAGCCAACTCTCGAGCGCGTGATC GAAGCCCGCAGTTTTTTCGCCCCCGTCACTTCCGGGTGCGACAATCTCTTCTGTCCGGCCAGCCGCTGGAGTCGTTAGGTGCCGCCTTGC TTCTGACGAGCCACACGTTTGCTTCTTCCCTGTGTTCCCAGCTGGAGGGACATGAGTGTCCCTGGGCCGTCGTCTCCGGACGGGGCCCTG ACACGGCCACCCTACTGCCTGGAGGCCGGGGAGCCGACGCCTGGTTTAAGTGACACTTCTCCAGATGAAGGGTTAATAGAGGACTTGACT ATAGAAGACAAAGCAGTGGAGCAACTGGCAGAAGGATTGCTTTCTCATTATTTGCCAGATCTGCAGAGATCAAAACAAGCCCTCCAGGAA CTCACTAGTGAGTTGAAACTCTTGGAGGAAGCAACCATTTCAGTCTGCAGGTCATTAGTTGAAAACAATCCTCGAACAGGAAATCTTGGT GCACTAATTAAGGTCTTCCTTTCTAGAACCAAAGAACTAAAACTTTCAGCAGAATGTCAGAACCACATCTTCATTTGGCAGACACACAAT GCTTTGTTTATTATTTGCTGTTTGCTGAAAGTGTTCATCTGTCAGATGTCAGAGGAGGAATTACAACTTCATTTTACTTATGAAGAAAAA TCTCCTGGCAATTACAGTTCTGACTCAGAAGATCTTTTGGAAGAATTGCTGTGCTGTTTGATGCAGTTGATCACTGATATTCCACTCTTA GATATTACATATGAAATATCAGTAGAAGCTATATCAACAATGGTTGTTTTCCTTTCCTGCCAACTCTTCCACAAAGAAGTTTTGCGACAG AGCATCAGCCACAAGTATTTGATGCGAGGTCCATGTCTTCCATACACCAGCAAACTTGTGAAGACCTTATTATATAACTTTATCAGACAA GAAAAGCCACCTCCTCCAGGGGCCCATGTTTTCCCTCAGCAGTCGGATGGGGGAGGACTGCTTTATGGACTTGCATCAGGAGTAGCAACA GGACTCTGGACTGTCTTCACACTAGGTGGTGTGGGCAGCAAAGCGGCTGCCTCTCCAGAGCTTTCTTCCCCTCTGGCCAACCAGAGTCTC CTGCTTCTGCTGGTGTTGGCCAATCTGACAGATGCCTCAGATGCGCCAAACCCCTACAGACAAGCCATTATGTCCTTCAAGAACACACAA GATAGCAGTCCTTTCCCCTCATCAATTCCACATGCCTTCCAGATCAACTTTAATAGTTTGTACACAGCTCTTTGTGAACAGCAGACATCT GATCAAGCAACTCTCCTCTTGTATACCTTGCTCCATCAAAATAGTAATATTAGAACATACATGTTGGCTCGCACAGATATGGAAAATCTT GTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGAATTCACACCATGTGTATATGGCCCTTATAATATTGTTGATCCTTACG GAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAAAAAATATTACTTGGTATTCAGAACGAGTTTTAACTGAAATCTCCTTG GGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACAACATGACTAGGACACGAGACAAGTACCTTCACACAAATTGTTTGGCA GCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGTATGCTGCCCAGAGGATCATCAGTTTATTTTCTTTGCTGTCTAAAAAA CACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTTCGCTGAGTTCTAATGATGTTCCTCTACCAGATTATGCACAAGACCTA AATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCAACTCCTGCCTGACAAATTCCCTTCACCACAACCCAAACTTGGTATAC GCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTCATCCTTCATTTCAGGATATAATGCAAAATATTGATCTGGTGATCTCC TTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAGTGGAACGGGTCCTGGAAATCATTAAGCAAGGCGTCGTTGCGCTGCCC AAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATGTGGAAGAGGAGCAGCCCGAGGAGTTTTTTATCCCCTATGTCTGGTCT CTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGGACATCCAGCTGTTCACCATGGATTCCGACTGAGGGCAGGATGCTCTC CCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTATTTCTGGGTAACAGAAGTAGACAGACAGGTTACTTGGTGTATCTTCT GTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGATTTGGAGAATTGGTGCTAGTTGGCAATAGATAACTCAGCGTAGATAG TATTGCAAAAAGGGGAGGAAATACACAACAATAATAAATG >9833_9833_1_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000220531_DYM_chr18_46918015_ENST00000269445_length(amino acids)=708AA_BP=72 MLLPCVPSWRDMSVPGPSSPDGALTRPPYCLEAGEPTPGLSDTSPDEGLIEDLTIEDKAVEQLAEGLLSHYLPDLQRSKQALQELTSELK LLEEATISVCRSLVENNPRTGNLGALIKVFLSRTKELKLSAECQNHIFIWQTHNALFIICCLLKVFICQMSEEELQLHFTYEEKSPGNYS SDSEDLLEELLCCLMQLITDIPLLDITYEISVEAISTMVVFLSCQLFHKEVLRQSISHKYLMRGPCLPYTSKLVKTLLYNFIRQEKPPPP GAHVFPQQSDGGGLLYGLASGVATGLWTVFTLGGVGSKAAASPELSSPLANQSLLLLLVLANLTDASDAPNPYRQAIMSFKNTQDSSPFP SSIPHAFQINFNSLYTALCEQQTSDQATLLLYTLLHQNSNIRTYMLARTDMENLVLPILEILYHVEERNSHHVYMALIILLILTEDDGFN RSIHEVILKNITWYSERVLTEISLGSLLILVVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVLE QATQSLRGSLSSNDVPLPDYAQDLNVIEEVIRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSRL LQAGAELSVERVLEIIKQGVVALPKDRLKKFPELKFKYVEEEQPEEFFIPYVWSLVYNSAVGLYWNPQDIQLFTMDSD -------------------------------------------------------------- >9833_9833_2_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000220531_DYM_chr18_46918015_ENST00000442713_length(transcript)=2231nt_BP=545nt ACCCACCCCGTGAGCAGCCCAAACCGACCACCGTCCTCCCCCAGCTCCCACCAATCGGATCGCAGGGACTCGAGCCCCACACTGCTGAGT CCGCTGACACTGCGTCCGGGGCCAGACGACGATATCAGCGCGGGGTCCCCACAACGCCATGGGGCAGAGCCAACTCTCGAGCGCGTGATC GAAGCCCGCAGTTTTTTCGCCCCCGTCACTTCCGGGTGCGACAATCTCTTCTGTCCGGCCAGCCGCTGGAGTCGTTAGGTGCCGCCTTGC TTCTGACGAGCCACACGTTTGCTTCTTCCCTGTGTTCCCAGCTGGAGGGACATGAGTGTCCCTGGGCCGTCGTCTCCGGACGGGGCCCTG ACACGGCCACCCTACTGCCTGGAGGCCGGGGAGCCGACGCCTGGTTTAAGTGACACTTCTCCAGATGAAGGGTTAATAGAGGACTTGACT ATAGAAGACAAAGCAGTGGAGCAACTGGCAGAAGGATTGCTTTCTCATTATTTGCCAGATCTGCAGAGATCAAAACAAGCCCTCCAGGAA CTCACTAGTGAGTTGAAACTCTTGGAGGAAGCAACCATTTCAGTCTGCAGGTCATTAGCAGGACTCTGGACTGTCTTCACACTAGGTGGT GTGGGCAGCAAAGCGGCTGCCTCTCCAGAGCTTTCTTCCCCTCTGGCCAACCAGAGTCTCCTGCTTCTGCTGGTGTTGGCCAATCTGACA GATGCCTCAGATGCGCCAAACCCCTACAGACAAGCCATTATGTCCTTCAAGAACACACAAGATAGCAGTCCTTTCCCCTCATCAATTCCA CATGCCTTCCAGATCAACTTTAATAGTTTGTACACAGCTCTTTGTGAACAGCAGACATCTGATCAAGCAACTCTCCTCTTGTATACCTTG CTCCATCAAAATAGTAATATTAGAACATACATGTTGGCTCGCACAGATATGGAAAATCTTGTTTTACCAATTCTTGAGATTCTGTATCAT GTTGAAGAAAGGAATTCACACCATGTGTATATGGCCCTTATAATATTGTTGATCCTTACGGAAGATGATGGCTTCAACAGATCCATTCAT GAAGTGATACTAAAAAATATTACTTGGTATTCAGAACGAGTTTTAACTGAAATCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGA ACCATTCAATACAACATGACTAGGACACGAGACAAGTACCTTCACACAAATTGTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGT TCTCTCCATCAGTATGCTGCCCAGAGGATCATCAGTTTATTTTCTTTGCTGTCTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAG TCCTTGAGAGGTTCGCTGAGTTCTAATGATGTTCCTCTACCAGATTATGCACAAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATG TTAGAGATCATCAACTCCTGCCTGACAAATTCCCTTCACCACAACCCAAACTTGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAA CAATTTCGAACTCATCCTTCATTTCAGGATATAATGCAAAATATTGATCTGGTGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGA GCTGAGCTGTCAGTGGAACGGGTCCTGGAAATCATTAAGCAAGGCGTCGTTGCGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTG AAATTCAAATATGTGGAAGAGGAGCAGCCCGAGGAGTTTTTTATCCCCTATGTCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTAC TGGAATCCACAGGACATCCAGCTGTTCACCATGGATTCCGACTGAGGGCAGGATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCT TCAAGTTCTTTTATTTCTGGGTAACAGAAGTAGACAGACAGGTTACTTGGTGTATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCC TCACACACTTTGATTTGGAGAATTGGTGCTAGTTGGCAATAGATAACTCAGCGTAGATAGTATTGCAAAAAGGGGAGGAAATACACAACA ATAATAAATGTAAAAACCTGCTATTCAACATGCAGTTTTATTTCGAAGCCAAAAATCTAGAGCTTTCCCAAGATCCTGTTGCCTTAGGCA CATCACACTTCAACAGTGCACACTATCCAACAGTGCACACTATTCAACAGTGCACACTATTCAAAAGCGTA >9833_9833_2_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000220531_DYM_chr18_46918015_ENST00000442713_length(amino acids)=518AA_BP=72 MLLPCVPSWRDMSVPGPSSPDGALTRPPYCLEAGEPTPGLSDTSPDEGLIEDLTIEDKAVEQLAEGLLSHYLPDLQRSKQALQELTSELK LLEEATISVCRSLAGLWTVFTLGGVGSKAAASPELSSPLANQSLLLLLVLANLTDASDAPNPYRQAIMSFKNTQDSSPFPSSIPHAFQIN FNSLYTALCEQQTSDQATLLLYTLLHQNSNIRTYMLARTDMENLVLPILEILYHVEERNSHHVYMALIILLILTEDDGFNRSIHEVILKN ITWYSERVLTEISLGSLLILVVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVLEQATQSLRGSL SSNDVPLPDYAQDLNVIEEVIRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSRLLQAGAELSVE RVLEIIKQGVVALPKDRLKKFPELKFKYVEEEQPEEFFIPYVWSLVYNSAVGLYWNPQDIQLFTMDSD -------------------------------------------------------------- >9833_9833_3_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000565323_DYM_chr18_46918015_ENST00000269445_length(transcript)=2427nt_BP=322nt AAGCGGTTTTTGGCGGCTGCGGCCCCCGGGCCCTGCCCCGAGTGCAGAAATGGGGAACCGCGCCGGCCCCTCTGCCCAGCGCAATGGGCG GAACTGAAGTCCCGAGGGGAAGCTTTGTATTGGGGGTGCAATTGAAGGCGGTTGATGGTGAAGCCATTCTGAGTTTATGTGTTGACTCCG GTTTAAGTGACACTTCTCCAGATGAAGGGTTAATAGAGGACTTGACTATAGAAGACAAAGCAGTGGAGCAACTGGCAGAAGGATTGCTTT CTCATTATTTGCCAGATCTGCAGAGATCAAAACAAGCCCTCCAGGAACTCACTAGTGAGTTGAAACTCTTGGAGGAAGCAACCATTTCAG TCTGCAGGTCATTAGTTGAAAACAATCCTCGAACAGGAAATCTTGGTGCACTAATTAAGGTCTTCCTTTCTAGAACCAAAGAACTAAAAC TTTCAGCAGAATGTCAGAACCACATCTTCATTTGGCAGACACACAATGCTTTGTTTATTATTTGCTGTTTGCTGAAAGTGTTCATCTGTC AGATGTCAGAGGAGGAATTACAACTTCATTTTACTTATGAAGAAAAATCTCCTGGCAATTACAGTTCTGACTCAGAAGATCTTTTGGAAG AATTGCTGTGCTGTTTGATGCAGTTGATCACTGATATTCCACTCTTAGATATTACATATGAAATATCAGTAGAAGCTATATCAACAATGG TTGTTTTCCTTTCCTGCCAACTCTTCCACAAAGAAGTTTTGCGACAGAGCATCAGCCACAAGTATTTGATGCGAGGTCCATGTCTTCCAT ACACCAGCAAACTTGTGAAGACCTTATTATATAACTTTATCAGACAAGAAAAGCCACCTCCTCCAGGGGCCCATGTTTTCCCTCAGCAGT CGGATGGGGGAGGACTGCTTTATGGACTTGCATCAGGAGTAGCAACAGGACTCTGGACTGTCTTCACACTAGGTGGTGTGGGCAGCAAAG CGGCTGCCTCTCCAGAGCTTTCTTCCCCTCTGGCCAACCAGAGTCTCCTGCTTCTGCTGGTGTTGGCCAATCTGACAGATGCCTCAGATG CGCCAAACCCCTACAGACAAGCCATTATGTCCTTCAAGAACACACAAGATAGCAGTCCTTTCCCCTCATCAATTCCACATGCCTTCCAGA TCAACTTTAATAGTTTGTACACAGCTCTTTGTGAACAGCAGACATCTGATCAAGCAACTCTCCTCTTGTATACCTTGCTCCATCAAAATA GTAATATTAGAACATACATGTTGGCTCGCACAGATATGGAAAATCTTGTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGA ATTCACACCATGTGTATATGGCCCTTATAATATTGTTGATCCTTACGGAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAA AAAATATTACTTGGTATTCAGAACGAGTTTTAACTGAAATCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACA ACATGACTAGGACACGAGACAAGTACCTTCACACAAATTGTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGT ATGCTGCCCAGAGGATCATCAGTTTATTTTCTTTGCTGTCTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTT CGCTGAGTTCTAATGATGTTCCTCTACCAGATTATGCACAAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCA ACTCCTGCCTGACAAATTCCCTTCACCACAACCCAAACTTGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTC ATCCTTCATTTCAGGATATAATGCAAAATATTGATCTGGTGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAG TGGAACGGGTCCTGGAAATCATTAAGCAAGGCGTCGTTGCGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATG TGGAAGAGGAGCAGCCCGAGGAGTTTTTTATCCCCTATGTCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGG ACATCCAGCTGTTCACCATGGATTCCGACTGAGGGCAGGATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTA TTTCTGGGTAACAGAAGTAGACAGACAGGTTACTTGGTGTATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGA TTTGGAGAATTGGTGCTAGTTGGCAATAGATAACTCAGCGTAGATAGTATTGCAAAAAGGGGAGGAAATACACAACAATAATAAATG >9833_9833_3_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000565323_DYM_chr18_46918015_ENST00000269445_length(amino acids)=719AA_BP=83 MPRVQKWGTAPAPLPSAMGGTEVPRGSFVLGVQLKAVDGEAILSLCVDSGLSDTSPDEGLIEDLTIEDKAVEQLAEGLLSHYLPDLQRSK QALQELTSELKLLEEATISVCRSLVENNPRTGNLGALIKVFLSRTKELKLSAECQNHIFIWQTHNALFIICCLLKVFICQMSEEELQLHF TYEEKSPGNYSSDSEDLLEELLCCLMQLITDIPLLDITYEISVEAISTMVVFLSCQLFHKEVLRQSISHKYLMRGPCLPYTSKLVKTLLY NFIRQEKPPPPGAHVFPQQSDGGGLLYGLASGVATGLWTVFTLGGVGSKAAASPELSSPLANQSLLLLLVLANLTDASDAPNPYRQAIMS FKNTQDSSPFPSSIPHAFQINFNSLYTALCEQQTSDQATLLLYTLLHQNSNIRTYMLARTDMENLVLPILEILYHVEERNSHHVYMALII LLILTEDDGFNRSIHEVILKNITWYSERVLTEISLGSLLILVVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFS LLSKKHNKVLEQATQSLRGSLSSNDVPLPDYAQDLNVIEEVIRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNI DLVISFFSSRLLQAGAELSVERVLEIIKQGVVALPKDRLKKFPELKFKYVEEEQPEEFFIPYVWSLVYNSAVGLYWNPQDIQLFTMDSD -------------------------------------------------------------- >9833_9833_4_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000565323_DYM_chr18_46918015_ENST00000442713_length(transcript)=2008nt_BP=322nt AAGCGGTTTTTGGCGGCTGCGGCCCCCGGGCCCTGCCCCGAGTGCAGAAATGGGGAACCGCGCCGGCCCCTCTGCCCAGCGCAATGGGCG GAACTGAAGTCCCGAGGGGAAGCTTTGTATTGGGGGTGCAATTGAAGGCGGTTGATGGTGAAGCCATTCTGAGTTTATGTGTTGACTCCG GTTTAAGTGACACTTCTCCAGATGAAGGGTTAATAGAGGACTTGACTATAGAAGACAAAGCAGTGGAGCAACTGGCAGAAGGATTGCTTT CTCATTATTTGCCAGATCTGCAGAGATCAAAACAAGCCCTCCAGGAACTCACTAGTGAGTTGAAACTCTTGGAGGAAGCAACCATTTCAG TCTGCAGGTCATTAGCAGGACTCTGGACTGTCTTCACACTAGGTGGTGTGGGCAGCAAAGCGGCTGCCTCTCCAGAGCTTTCTTCCCCTC TGGCCAACCAGAGTCTCCTGCTTCTGCTGGTGTTGGCCAATCTGACAGATGCCTCAGATGCGCCAAACCCCTACAGACAAGCCATTATGT CCTTCAAGAACACACAAGATAGCAGTCCTTTCCCCTCATCAATTCCACATGCCTTCCAGATCAACTTTAATAGTTTGTACACAGCTCTTT GTGAACAGCAGACATCTGATCAAGCAACTCTCCTCTTGTATACCTTGCTCCATCAAAATAGTAATATTAGAACATACATGTTGGCTCGCA CAGATATGGAAAATCTTGTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGAATTCACACCATGTGTATATGGCCCTTATAA TATTGTTGATCCTTACGGAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAAAAAATATTACTTGGTATTCAGAACGAGTTT TAACTGAAATCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACAACATGACTAGGACACGAGACAAGTACCTTC ACACAAATTGTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGTATGCTGCCCAGAGGATCATCAGTTTATTTT CTTTGCTGTCTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTTCGCTGAGTTCTAATGATGTTCCTCTACCAG ATTATGCACAAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCAACTCCTGCCTGACAAATTCCCTTCACCACA ACCCAAACTTGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTCATCCTTCATTTCAGGATATAATGCAAAATA TTGATCTGGTGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAGTGGAACGGGTCCTGGAAATCATTAAGCAAG GCGTCGTTGCGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATGTGGAAGAGGAGCAGCCCGAGGAGTTTTTTA TCCCCTATGTCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGGACATCCAGCTGTTCACCATGGATTCCGACT GAGGGCAGGATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTATTTCTGGGTAACAGAAGTAGACAGACAGGT TACTTGGTGTATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGATTTGGAGAATTGGTGCTAGTTGGCAATAGA TAACTCAGCGTAGATAGTATTGCAAAAAGGGGAGGAAATACACAACAATAATAAATGTAAAAACCTGCTATTCAACATGCAGTTTTATTT CGAAGCCAAAAATCTAGAGCTTTCCCAAGATCCTGTTGCCTTAGGCACATCACACTTCAACAGTGCACACTATCCAACAGTGCACACTAT TCAACAGTGCACACTATTCAAAAGCGTA >9833_9833_4_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000565323_DYM_chr18_46918015_ENST00000442713_length(amino acids)=529AA_BP=83 MPRVQKWGTAPAPLPSAMGGTEVPRGSFVLGVQLKAVDGEAILSLCVDSGLSDTSPDEGLIEDLTIEDKAVEQLAEGLLSHYLPDLQRSK QALQELTSELKLLEEATISVCRSLAGLWTVFTLGGVGSKAAASPELSSPLANQSLLLLLVLANLTDASDAPNPYRQAIMSFKNTQDSSPF PSSIPHAFQINFNSLYTALCEQQTSDQATLLLYTLLHQNSNIRTYMLARTDMENLVLPILEILYHVEERNSHHVYMALIILLILTEDDGF NRSIHEVILKNITWYSERVLTEISLGSLLILVVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVL EQATQSLRGSLSSNDVPLPDYAQDLNVIEEVIRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSR LLQAGAELSVERVLEIIKQGVVALPKDRLKKFPELKFKYVEEEQPEEFFIPYVWSLVYNSAVGLYWNPQDIQLFTMDSD -------------------------------------------------------------- >9833_9833_5_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000566753_DYM_chr18_46918015_ENST00000269445_length(transcript)=2418nt_BP=313nt GTCCGGCCAGCCGCTGGAGTCGTTAGGTGCCGCCTTGCTTCTGACGAGCCACACGTTTGCTTCTTCCCTGTGTTCCCAGCTGGAGGGACA TGAGTGTCCCTGGGCCGTCGTCTCCGGACGGGGCCCTGACACGGCCACCCTACTGCCTGGAGGCCGGGGAGCCGACGCCTGGTTTAAGTG ACACTTCTCCAGATGAAGGGTTAATAGAGGACTTGACTATAGAAGACAAAGCAGTGGAGCAACTGGCAGAAGGATTGCTTTCTCATTATT TGCCAGATCTGCAGAGATCAAAACAAGCCCTCCAGGAACTCACTAGTGAGTTGAAACTCTTGGAGGAAGCAACCATTTCAGTCTGCAGGT CATTAGTTGAAAACAATCCTCGAACAGGAAATCTTGGTGCACTAATTAAGGTCTTCCTTTCTAGAACCAAAGAACTAAAACTTTCAGCAG AATGTCAGAACCACATCTTCATTTGGCAGACACACAATGCTTTGTTTATTATTTGCTGTTTGCTGAAAGTGTTCATCTGTCAGATGTCAG AGGAGGAATTACAACTTCATTTTACTTATGAAGAAAAATCTCCTGGCAATTACAGTTCTGACTCAGAAGATCTTTTGGAAGAATTGCTGT GCTGTTTGATGCAGTTGATCACTGATATTCCACTCTTAGATATTACATATGAAATATCAGTAGAAGCTATATCAACAATGGTTGTTTTCC TTTCCTGCCAACTCTTCCACAAAGAAGTTTTGCGACAGAGCATCAGCCACAAGTATTTGATGCGAGGTCCATGTCTTCCATACACCAGCA AACTTGTGAAGACCTTATTATATAACTTTATCAGACAAGAAAAGCCACCTCCTCCAGGGGCCCATGTTTTCCCTCAGCAGTCGGATGGGG GAGGACTGCTTTATGGACTTGCATCAGGAGTAGCAACAGGACTCTGGACTGTCTTCACACTAGGTGGTGTGGGCAGCAAAGCGGCTGCCT CTCCAGAGCTTTCTTCCCCTCTGGCCAACCAGAGTCTCCTGCTTCTGCTGGTGTTGGCCAATCTGACAGATGCCTCAGATGCGCCAAACC CCTACAGACAAGCCATTATGTCCTTCAAGAACACACAAGATAGCAGTCCTTTCCCCTCATCAATTCCACATGCCTTCCAGATCAACTTTA ATAGTTTGTACACAGCTCTTTGTGAACAGCAGACATCTGATCAAGCAACTCTCCTCTTGTATACCTTGCTCCATCAAAATAGTAATATTA GAACATACATGTTGGCTCGCACAGATATGGAAAATCTTGTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGAATTCACACC ATGTGTATATGGCCCTTATAATATTGTTGATCCTTACGGAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAAAAAATATTA CTTGGTATTCAGAACGAGTTTTAACTGAAATCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACAACATGACTA GGACACGAGACAAGTACCTTCACACAAATTGTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGTATGCTGCCC AGAGGATCATCAGTTTATTTTCTTTGCTGTCTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTTCGCTGAGTT CTAATGATGTTCCTCTACCAGATTATGCACAAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCAACTCCTGCC TGACAAATTCCCTTCACCACAACCCAAACTTGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTCATCCTTCAT TTCAGGATATAATGCAAAATATTGATCTGGTGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAGTGGAACGGG TCCTGGAAATCATTAAGCAAGGCGTCGTTGCGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATGTGGAAGAGG AGCAGCCCGAGGAGTTTTTTATCCCCTATGTCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGGACATCCAGC TGTTCACCATGGATTCCGACTGAGGGCAGGATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTATTTCTGGGT AACAGAAGTAGACAGACAGGTTACTTGGTGTATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGATTTGGAGAA TTGGTGCTAGTTGGCAATAGATAACTCAGCGTAGATAGTATTGCAAAAAGGGGAGGAAATACACAACAATAATAAATG >9833_9833_5_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000566753_DYM_chr18_46918015_ENST00000269445_length(amino acids)=708AA_BP=72 MLLPCVPSWRDMSVPGPSSPDGALTRPPYCLEAGEPTPGLSDTSPDEGLIEDLTIEDKAVEQLAEGLLSHYLPDLQRSKQALQELTSELK LLEEATISVCRSLVENNPRTGNLGALIKVFLSRTKELKLSAECQNHIFIWQTHNALFIICCLLKVFICQMSEEELQLHFTYEEKSPGNYS SDSEDLLEELLCCLMQLITDIPLLDITYEISVEAISTMVVFLSCQLFHKEVLRQSISHKYLMRGPCLPYTSKLVKTLLYNFIRQEKPPPP GAHVFPQQSDGGGLLYGLASGVATGLWTVFTLGGVGSKAAASPELSSPLANQSLLLLLVLANLTDASDAPNPYRQAIMSFKNTQDSSPFP SSIPHAFQINFNSLYTALCEQQTSDQATLLLYTLLHQNSNIRTYMLARTDMENLVLPILEILYHVEERNSHHVYMALIILLILTEDDGFN RSIHEVILKNITWYSERVLTEISLGSLLILVVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVLE QATQSLRGSLSSNDVPLPDYAQDLNVIEEVIRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSRL LQAGAELSVERVLEIIKQGVVALPKDRLKKFPELKFKYVEEEQPEEFFIPYVWSLVYNSAVGLYWNPQDIQLFTMDSD -------------------------------------------------------------- >9833_9833_6_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000566753_DYM_chr18_46918015_ENST00000442713_length(transcript)=1999nt_BP=313nt GTCCGGCCAGCCGCTGGAGTCGTTAGGTGCCGCCTTGCTTCTGACGAGCCACACGTTTGCTTCTTCCCTGTGTTCCCAGCTGGAGGGACA TGAGTGTCCCTGGGCCGTCGTCTCCGGACGGGGCCCTGACACGGCCACCCTACTGCCTGGAGGCCGGGGAGCCGACGCCTGGTTTAAGTG ACACTTCTCCAGATGAAGGGTTAATAGAGGACTTGACTATAGAAGACAAAGCAGTGGAGCAACTGGCAGAAGGATTGCTTTCTCATTATT TGCCAGATCTGCAGAGATCAAAACAAGCCCTCCAGGAACTCACTAGTGAGTTGAAACTCTTGGAGGAAGCAACCATTTCAGTCTGCAGGT CATTAGCAGGACTCTGGACTGTCTTCACACTAGGTGGTGTGGGCAGCAAAGCGGCTGCCTCTCCAGAGCTTTCTTCCCCTCTGGCCAACC AGAGTCTCCTGCTTCTGCTGGTGTTGGCCAATCTGACAGATGCCTCAGATGCGCCAAACCCCTACAGACAAGCCATTATGTCCTTCAAGA ACACACAAGATAGCAGTCCTTTCCCCTCATCAATTCCACATGCCTTCCAGATCAACTTTAATAGTTTGTACACAGCTCTTTGTGAACAGC AGACATCTGATCAAGCAACTCTCCTCTTGTATACCTTGCTCCATCAAAATAGTAATATTAGAACATACATGTTGGCTCGCACAGATATGG AAAATCTTGTTTTACCAATTCTTGAGATTCTGTATCATGTTGAAGAAAGGAATTCACACCATGTGTATATGGCCCTTATAATATTGTTGA TCCTTACGGAAGATGATGGCTTCAACAGATCCATTCATGAAGTGATACTAAAAAATATTACTTGGTATTCAGAACGAGTTTTAACTGAAA TCTCCTTGGGGAGTCTCCTGATCCTGGTGGTAATAAGAACCATTCAATACAACATGACTAGGACACGAGACAAGTACCTTCACACAAATT GTTTGGCAGCTTTAGCAAATATGTCGGCACAGTTTCGTTCTCTCCATCAGTATGCTGCCCAGAGGATCATCAGTTTATTTTCTTTGCTGT CTAAAAAACACAACAAAGTTCTGGAACAAGCCACACAGTCCTTGAGAGGTTCGCTGAGTTCTAATGATGTTCCTCTACCAGATTATGCAC AAGACCTAAATGTCATTGAAGAAGTGATTCGAATGATGTTAGAGATCATCAACTCCTGCCTGACAAATTCCCTTCACCACAACCCAAACT TGGTATACGCCCTGCTTTACAAACGCGATCTCTTTGAACAATTTCGAACTCATCCTTCATTTCAGGATATAATGCAAAATATTGATCTGG TGATCTCCTTCTTTAGCTCAAGGTTGCTGCAAGCTGGAGCTGAGCTGTCAGTGGAACGGGTCCTGGAAATCATTAAGCAAGGCGTCGTTG CGCTGCCCAAAGACAGACTGAAGAAATTTCCAGAATTGAAATTCAAATATGTGGAAGAGGAGCAGCCCGAGGAGTTTTTTATCCCCTATG TCTGGTCTCTTGTCTACAACTCAGCAGTCGGCCTGTACTGGAATCCACAGGACATCCAGCTGTTCACCATGGATTCCGACTGAGGGCAGG ATGCTCTCCCACCCGGACCCCTCCAGCCAAGCAGCCCTTCAAGTTCTTTTATTTCTGGGTAACAGAAGTAGACAGACAGGTTACTTGGTG TATCTTCTGTTAAAGAGGATTGCACGAGTGTGTTTTCCTCACACACTTTGATTTGGAGAATTGGTGCTAGTTGGCAATAGATAACTCAGC GTAGATAGTATTGCAAAAAGGGGAGGAAATACACAACAATAATAAATGTAAAAACCTGCTATTCAACATGCAGTTTTATTTCGAAGCCAA AAATCTAGAGCTTTCCCAAGATCCTGTTGCCTTAGGCACATCACACTTCAACAGTGCACACTATCCAACAGTGCACACTATTCAACAGTG CACACTATTCAAAAGCGTA >9833_9833_6_BLOC1S6-DYM_BLOC1S6_chr15_45884474_ENST00000566753_DYM_chr18_46918015_ENST00000442713_length(amino acids)=518AA_BP=72 MLLPCVPSWRDMSVPGPSSPDGALTRPPYCLEAGEPTPGLSDTSPDEGLIEDLTIEDKAVEQLAEGLLSHYLPDLQRSKQALQELTSELK LLEEATISVCRSLAGLWTVFTLGGVGSKAAASPELSSPLANQSLLLLLVLANLTDASDAPNPYRQAIMSFKNTQDSSPFPSSIPHAFQIN FNSLYTALCEQQTSDQATLLLYTLLHQNSNIRTYMLARTDMENLVLPILEILYHVEERNSHHVYMALIILLILTEDDGFNRSIHEVILKN ITWYSERVLTEISLGSLLILVVIRTIQYNMTRTRDKYLHTNCLAALANMSAQFRSLHQYAAQRIISLFSLLSKKHNKVLEQATQSLRGSL SSNDVPLPDYAQDLNVIEEVIRMMLEIINSCLTNSLHHNPNLVYALLYKRDLFEQFRTHPSFQDIMQNIDLVISFFSSRLLQAGAELSVE RVLEIIKQGVVALPKDRLKKFPELKFKYVEEEQPEEFFIPYVWSLVYNSAVGLYWNPQDIQLFTMDSD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BLOC1S6-DYM |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BLOC1S6-DYM |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BLOC1S6-DYM |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | BLOC1S6 | C3280026 | HERMANSKY-PUDLAK SYNDROME 9 | 3 | GENOMICS_ENGLAND;ORPHANET |
| Hgene | BLOC1S6 | C1844666 | Immune dysregulation | 1 | GENOMICS_ENGLAND |
| Tgene | C0265286 | Dyggve-Melchior-Clausen syndrome | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C3888088 | SMITH-MCCORT DYSPLASIA 1 | 4 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C1846431 | SMITH-MCCORT DYSPLASIA | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C0036341 | Schizophrenia | 1 | PSYGENET |
