
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ARFIP1-FGB (FusionGDB2 ID:HG27236TG2244) |
Fusion Gene Summary for ARFIP1-FGB |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ARFIP1-FGB | Fusion gene ID: hg27236tg2244 | Hgene | Tgene | Gene symbol | ARFIP1 | FGB | Gene ID | 27236 | 2244 |
| Gene name | ADP ribosylation factor interacting protein 1 | fibrinogen beta chain | |
| Synonyms | HSU52521 | HEL-S-78p | |
| Cytomap | ('ARFIP1')('FGB') 4q31.3 | 4q31.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | arfaptin-1 | fibrinogen beta chainbeta-fibrinogenepididymis secretory sperm binding protein Li 78pfibrinogen, B beta polypeptide | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P53367 | P02675 | |
| Ensembl transtripts involved in fusion gene | ENST00000353617, ENST00000356064, ENST00000405727, ENST00000429148, ENST00000451320, ENST00000511289, | ||
| Fusion gene scores | * DoF score | 11 X 7 X 7=539 | 4 X 5 X 2=40 |
| # samples | 11 | 5 | |
| ** MAII score | log2(11/539*10)=-2.29278174922785 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/40*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: ARFIP1 [Title/Abstract] AND FGB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ARFIP1(153750878)-FGB(155486960), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | ARFIP1-FGB seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARFIP1-FGB seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARFIP1-FGB seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ARFIP1-FGB seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ARFIP1 | GO:0006886 | intracellular protein transport | 12606037 |
| Hgene | ARFIP1 | GO:0050708 | regulation of protein secretion | 12606037 |
| Tgene | FGB | GO:0007160 | cell-matrix adhesion | 10903502 |
| Tgene | FGB | GO:0031639 | plasminogen activation | 16846481 |
| Tgene | FGB | GO:0034116 | positive regulation of heterotypic cell-cell adhesion | 8100742 |
| Tgene | FGB | GO:0034622 | cellular protein-containing complex assembly | 8910396 |
| Tgene | FGB | GO:0042730 | fibrinolysis | 16846481 |
| Tgene | FGB | GO:0043152 | induction of bacterial agglutination | 24367264 |
| Tgene | FGB | GO:0045907 | positive regulation of vasoconstriction | 15739255 |
| Tgene | FGB | GO:0045921 | positive regulation of exocytosis | 19193866 |
| Tgene | FGB | GO:0050714 | positive regulation of protein secretion | 19193866 |
| Tgene | FGB | GO:0051258 | protein polymerization | 12706644 |
| Tgene | FGB | GO:0051592 | response to calcium ion | 6777381 |
| Tgene | FGB | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 10903502|19193866 |
| Tgene | FGB | GO:0070527 | platelet aggregation | 6281794 |
| Tgene | FGB | GO:0072378 | blood coagulation, fibrin clot formation | 16846481 |
| Tgene | FGB | GO:0090277 | positive regulation of peptide hormone secretion | 19193866 |
| Tgene | FGB | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 10903502 |
| Tgene | FGB | GO:2000352 | negative regulation of endothelial cell apoptotic process | 10903502 |
 Fusion gene breakpoints across ARFIP1 (5'-gene) Fusion gene breakpoints across ARFIP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
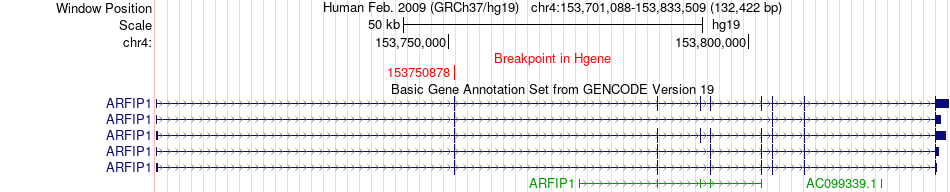 |
 Fusion gene breakpoints across FGB (3'-gene) Fusion gene breakpoints across FGB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
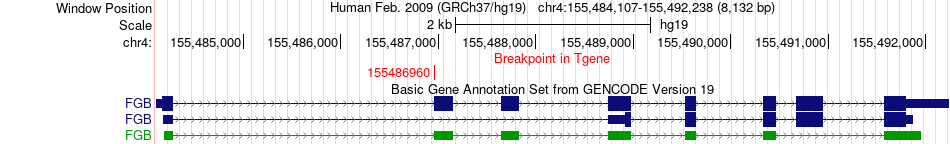 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A13Z-01A | ARFIP1 | chr4 | 153750878 | - | FGB | chr4 | 155486960 | + |
| ChimerDB4 | BRCA | TCGA-D8-A13Z-01A | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
Top |
Fusion Gene ORF analysis for ARFIP1-FGB |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000353617 | ENST00000502545 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-3UTR | ENST00000356064 | ENST00000502545 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-3UTR | ENST00000405727 | ENST00000502545 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-3UTR | ENST00000429148 | ENST00000502545 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-3UTR | ENST00000451320 | ENST00000502545 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-intron | ENST00000353617 | ENST00000509493 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-intron | ENST00000356064 | ENST00000509493 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-intron | ENST00000405727 | ENST00000509493 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-intron | ENST00000429148 | ENST00000509493 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| 5CDS-intron | ENST00000451320 | ENST00000509493 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| In-frame | ENST00000353617 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| In-frame | ENST00000356064 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| In-frame | ENST00000405727 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| In-frame | ENST00000429148 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| In-frame | ENST00000451320 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| intron-3CDS | ENST00000511289 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| intron-3UTR | ENST00000511289 | ENST00000502545 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
| intron-intron | ENST00000511289 | ENST00000509493 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000451320 | ARFIP1 | chr4 | 153750878 | + | ENST00000302068 | FGB | chr4 | 155486960 | + | 2055 | 257 | 86 | 1618 | 510 |
| ENST00000429148 | ARFIP1 | chr4 | 153750878 | + | ENST00000302068 | FGB | chr4 | 155486960 | + | 2053 | 255 | 84 | 1616 | 510 |
| ENST00000353617 | ARFIP1 | chr4 | 153750878 | + | ENST00000302068 | FGB | chr4 | 155486960 | + | 2167 | 369 | 63 | 1730 | 555 |
| ENST00000405727 | ARFIP1 | chr4 | 153750878 | + | ENST00000302068 | FGB | chr4 | 155486960 | + | 2021 | 223 | 52 | 1584 | 510 |
| ENST00000356064 | ARFIP1 | chr4 | 153750878 | + | ENST00000302068 | FGB | chr4 | 155486960 | + | 2142 | 344 | 38 | 1705 | 555 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000451320 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + | 0.00040122 | 0.9995988 |
| ENST00000429148 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + | 0.000392686 | 0.9996074 |
| ENST00000353617 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + | 0.000433676 | 0.99956626 |
| ENST00000405727 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + | 0.000427758 | 0.9995722 |
| ENST00000356064 | ENST00000302068 | ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486960 | + | 0.000469682 | 0.9995303 |
Top |
Fusion Genomic Features for ARFIP1-FGB |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486959 | + | 1.84E-07 | 0.99999976 |
| ARFIP1 | chr4 | 153750878 | + | FGB | chr4 | 155486959 | + | 1.84E-07 | 0.99999976 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
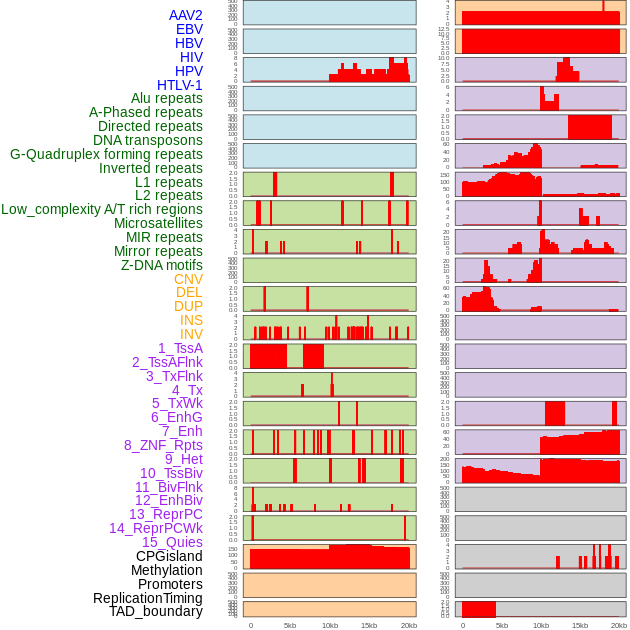 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
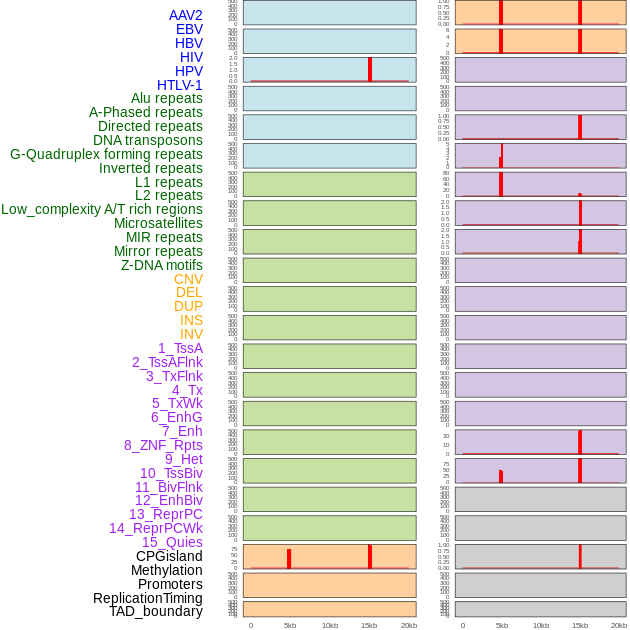 |
Top |
Fusion Protein Features for ARFIP1-FGB |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:153750878/chr4:155486960) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ARFIP1 | FGB |
| FUNCTION: Plays a role in controlling biogenesis of secretory granules at the trans-Golgi network (PubMed:22981988). Mechanisitically, binds ARF-GTP at the neck of a growing secretory granule precursor and forms a protective scaffold (PubMed:9038142, PubMed:22981988). Once the granule precursor has been completely loaded, active PRKD1 phosphorylates ARFIP1 and releases it from ARFs (PubMed:22981988). In turn, ARFs induce fission (PubMed:22981988). Through this mechanism, ensures proper secretory granule formation at the Golgi of pancreatic beta cells (PubMed:22981988). {ECO:0000269|PubMed:22981988, ECO:0000269|PubMed:9038142}. | FUNCTION: Cleaved by the protease thrombin to yield monomers which, together with fibrinogen alpha (FGA) and fibrinogen gamma (FGG), polymerize to form an insoluble fibrin matrix. Fibrin has a major function in hemostasis as one of the primary components of blood clots. In addition, functions during the early stages of wound repair to stabilize the lesion and guide cell migration during re-epithelialization. Was originally thought to be essential for platelet aggregation, based on in vitro studies using anticoagulated blood. However subsequent studies have shown that it is not absolutely required for thrombus formation in vivo. Enhances expression of SELP in activated platelets. Maternal fibrinogen is essential for successful pregnancy. Fibrin deposition is also associated with infection, where it protects against IFNG-mediated hemorrhage. May also facilitate the antibacterial immune response via both innate and T-cell mediated pathways. {ECO:0000250|UniProtKB:E9PV24}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FGB | chr4:153750878 | chr4:155486960 | ENST00000302068 | 0 | 8 | 157_222 | 38 | 492.0 | Coiled coil | Ontology_term=ECO:0000305,ECO:0000305,ECO:0000305 | |
| Tgene | FGB | chr4:153750878 | chr4:155486960 | ENST00000302068 | 0 | 8 | 232_488 | 38 | 492.0 | Domain | Fibrinogen C-terminal | |
| Tgene | FGB | chr4:153750878 | chr4:155486960 | ENST00000302068 | 0 | 8 | 45_47 | 38 | 492.0 | Region | Note=Beta-chain polymerization%2C binding distal domain of another fibrin |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARFIP1 | chr4:153750878 | chr4:155486960 | ENST00000353617 | + | 2 | 9 | 153_353 | 31 | 374.0 | Domain | AH |
| Hgene | ARFIP1 | chr4:153750878 | chr4:155486960 | ENST00000356064 | + | 2 | 8 | 153_353 | 31 | 342.0 | Domain | AH |
| Hgene | ARFIP1 | chr4:153750878 | chr4:155486960 | ENST00000405727 | + | 2 | 8 | 153_353 | 31 | 342.0 | Domain | AH |
| Hgene | ARFIP1 | chr4:153750878 | chr4:155486960 | ENST00000451320 | + | 2 | 9 | 153_353 | 31 | 374.0 | Domain | AH |
Top |
Fusion Gene Sequence for ARFIP1-FGB |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >5894_5894_1_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000353617_FGB_chr4_155486960_ENST00000302068_length(transcript)=2167nt_BP=369nt ACTTCCGGCTTCGCTGCTCTTGGTTCTGGTTCTGGAGGCTGGGTTGAGAGGTCGCCGGTCCGACTGTCCTCGGCGGTTGGTCAGTGTGAA TTTGTGACAGCTGCAGTTGCTCCCCGCCCCCGAGCAGCCGAGGTGCGTGGGGGAAGGGGAAGAAGGAAAAGGTCCGGGTCGCGTTTCCGC TCAGTTTTTGCCAGGGTTGAGGCGATTCCAGAGAGCGGCGGAAAGGATAAGCCTCGACTTAGAAGCTACCGAGTTACCCTAAGAAAGGAG TCTACCATGGCTCAAGAATCTCCCAAAAATTCAGCAGCAGAAATTCCAGTGACTAGTAATGGAGAAGTTGATGACTCTCGTGAACATAGC TTTAATAGGGGTTTCTTCAGTGCCCGTGGTCATCGACCCCTTGACAAGAAGAGAGAAGAGGCTCCCAGCCTGAGGCCTGCCCCACCGCCC ATCAGTGGAGGTGGCTATCGGGCTCGTCCAGCCAAAGCAGCTGCCACTCAAAAGAAAGTAGAAAGAAAAGCCCCTGATGCTGGAGGCTGT CTTCACGCTGACCCAGACCTGGGGGTGTTGTGTCCTACAGGATGTCAGTTGCAAGAGGCTTTGCTACAACAGGAAAGGCCAATCAGAAAT AGTGTTGATGAGTTAAATAACAATGTGGAAGCTGTTTCCCAGACCTCCTCTTCTTCCTTTCAGTACATGTATTTGCTGAAAGACCTGTGG CAAAAGAGGCAGAAGCAAGTAAAAGATAATGAAAATGTAGTCAATGAGTACTCCTCAGAACTGGAAAAGCACCAATTATATATAGATGAG ACTGTGAATAGCAATATCCCAACTAACCTTCGTGTGCTTCGTTCAATCCTGGAAAACCTGAGAAGCAAAATACAAAAGTTAGAATCTGAT GTCTCAGCTCAAATGGAATATTGTCGCACCCCATGCACTGTCAGTTGCAATATTCCTGTGGTGTCTGGCAAAGAATGTGAGGAAATTATC AGGAAAGGAGGTGAAACATCTGAAATGTATCTCATTCAACCTGACAGTTCTGTCAAACCGTATAGAGTATACTGTGACATGAATACAGAA AATGGAGGATGGACAGTGATTCAGAACCGTCAAGACGGTAGTGTTGACTTTGGCAGGAAATGGGATCCATATAAACAGGGATTTGGAAAT GTTGCAACCAACACAGATGGGAAGAATTACTGTGGCCTACCAGGTGAATATTGGCTTGGAAATGATAAAATTAGCCAGCTTACCAGGATG GGACCCACAGAACTTTTGATAGAAATGGAGGACTGGAAAGGAGACAAAGTAAAGGCTCACTATGGAGGATTCACTGTACAGAATGAAGCC AACAAATACCAGATCTCAGTGAACAAATACAGAGGAACAGCCGGTAATGCCCTCATGGATGGAGCATCTCAGCTGATGGGAGAAAACAGG ACCATGACCATTCACAACGGCATGTTCTTCAGCACGTATGACAGAGACAATGACGGCTGGTTAACATCAGATCCCAGAAAACAGTGTTCT AAAGAAGACGGTGGTGGATGGTGGTATAATAGATGTCATGCAGCCAATCCAAACGGCAGATACTACTGGGGTGGACAGTACACCTGGGAC ATGGCAAAGCATGGCACAGATGATGGTGTAGTATGGATGAATTGGAAGGGGTCATGGTACTCAATGAGGAAGATGAGTATGAAGATCAGG CCCTTCTTCCCACAGCAATAGTCCCCAATACGTAGATTTTTGCTCTTCTGTATGTGACAACATTTTTGTACATTATGTTATTGGAATTTT CTTTCATACATTATATTCCTCTAAAACTCTCAAGCAGACGTGAGTGTGACTTTTTGAAAAAAGTATAGGATAAATTACATTAAAATAGCA CATGATTTTCTTTTGTTTTCTTCATTTCTCTTGCTCACCAAGAAGTAACAAAAGTATAGTTTTGACAGAGTTGGTGTTCATAATTTCAGT TCTAGTTGATTGCGAGAATTTTCAAATAAGGAAGAGGGGTCTTTTATCCTTGTCGTAGGAAAACCATGACGGAAAGGAAAAACTGATGTT TAAAAGTCCACTTTTAAAACTATATTTATTTATGTAGGATCTGTCAAAGAAAACTTCCAAAAAGATTTATTAATTAAACCAGACTCTGTT GCAATAA >5894_5894_1_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000353617_FGB_chr4_155486960_ENST00000302068_length(amino acids)=555AA_BP=102 MSSAVGQCEFVTAAVAPRPRAAEVRGGRGRRKRSGSRFRSVFARVEAIPESGGKDKPRLRSYRVTLRKESTMAQESPKNSAAEIPVTSNG EVDDSREHSFNRGFFSARGHRPLDKKREEAPSLRPAPPPISGGGYRARPAKAAATQKKVERKAPDAGGCLHADPDLGVLCPTGCQLQEAL LQQERPIRNSVDELNNNVEAVSQTSSSSFQYMYLLKDLWQKRQKQVKDNENVVNEYSSELEKHQLYIDETVNSNIPTNLRVLRSILENLR SKIQKLESDVSAQMEYCRTPCTVSCNIPVVSGKECEEIIRKGGETSEMYLIQPDSSVKPYRVYCDMNTENGGWTVIQNRQDGSVDFGRKW DPYKQGFGNVATNTDGKNYCGLPGEYWLGNDKISQLTRMGPTELLIEMEDWKGDKVKAHYGGFTVQNEANKYQISVNKYRGTAGNALMDG ASQLMGENRTMTIHNGMFFSTYDRDNDGWLTSDPRKQCSKEDGGGWWYNRCHAANPNGRYYWGGQYTWDMAKHGTDDGVVWMNWKGSWYS MRKMSMKIRPFFPQQ -------------------------------------------------------------- >5894_5894_2_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000356064_FGB_chr4_155486960_ENST00000302068_length(transcript)=2142nt_BP=344nt CTGGTTCTGGAGGCTGGGTTGAGAGGTCGCCGGTCCGACTGTCCTCGGCGGTTGGTCAGTGTGAATTTGTGACAGCTGCAGTTGCTCCCC GCCCCCGAGCAGCCGAGGTGCGTGGGGGAAGGGGAAGAAGGAAAAGGTCCGGGTCGCGTTTCCGCTCAGTTTTTGCCAGGGTTGAGGCGA TTCCAGAGAGCGGCGGAAAGGATAAGCCTCGACTTAGAAGCTACCGAGTTACCCTAAGAAAGGAGTCTACCATGGCTCAAGAATCTCCCA AAAATTCAGCAGCAGAAATTCCAGTGACTAGTAATGGAGAAGTTGATGACTCTCGTGAACATAGCTTTAATAGGGGTTTCTTCAGTGCCC GTGGTCATCGACCCCTTGACAAGAAGAGAGAAGAGGCTCCCAGCCTGAGGCCTGCCCCACCGCCCATCAGTGGAGGTGGCTATCGGGCTC GTCCAGCCAAAGCAGCTGCCACTCAAAAGAAAGTAGAAAGAAAAGCCCCTGATGCTGGAGGCTGTCTTCACGCTGACCCAGACCTGGGGG TGTTGTGTCCTACAGGATGTCAGTTGCAAGAGGCTTTGCTACAACAGGAAAGGCCAATCAGAAATAGTGTTGATGAGTTAAATAACAATG TGGAAGCTGTTTCCCAGACCTCCTCTTCTTCCTTTCAGTACATGTATTTGCTGAAAGACCTGTGGCAAAAGAGGCAGAAGCAAGTAAAAG ATAATGAAAATGTAGTCAATGAGTACTCCTCAGAACTGGAAAAGCACCAATTATATATAGATGAGACTGTGAATAGCAATATCCCAACTA ACCTTCGTGTGCTTCGTTCAATCCTGGAAAACCTGAGAAGCAAAATACAAAAGTTAGAATCTGATGTCTCAGCTCAAATGGAATATTGTC GCACCCCATGCACTGTCAGTTGCAATATTCCTGTGGTGTCTGGCAAAGAATGTGAGGAAATTATCAGGAAAGGAGGTGAAACATCTGAAA TGTATCTCATTCAACCTGACAGTTCTGTCAAACCGTATAGAGTATACTGTGACATGAATACAGAAAATGGAGGATGGACAGTGATTCAGA ACCGTCAAGACGGTAGTGTTGACTTTGGCAGGAAATGGGATCCATATAAACAGGGATTTGGAAATGTTGCAACCAACACAGATGGGAAGA ATTACTGTGGCCTACCAGGTGAATATTGGCTTGGAAATGATAAAATTAGCCAGCTTACCAGGATGGGACCCACAGAACTTTTGATAGAAA TGGAGGACTGGAAAGGAGACAAAGTAAAGGCTCACTATGGAGGATTCACTGTACAGAATGAAGCCAACAAATACCAGATCTCAGTGAACA AATACAGAGGAACAGCCGGTAATGCCCTCATGGATGGAGCATCTCAGCTGATGGGAGAAAACAGGACCATGACCATTCACAACGGCATGT TCTTCAGCACGTATGACAGAGACAATGACGGCTGGTTAACATCAGATCCCAGAAAACAGTGTTCTAAAGAAGACGGTGGTGGATGGTGGT ATAATAGATGTCATGCAGCCAATCCAAACGGCAGATACTACTGGGGTGGACAGTACACCTGGGACATGGCAAAGCATGGCACAGATGATG GTGTAGTATGGATGAATTGGAAGGGGTCATGGTACTCAATGAGGAAGATGAGTATGAAGATCAGGCCCTTCTTCCCACAGCAATAGTCCC CAATACGTAGATTTTTGCTCTTCTGTATGTGACAACATTTTTGTACATTATGTTATTGGAATTTTCTTTCATACATTATATTCCTCTAAA ACTCTCAAGCAGACGTGAGTGTGACTTTTTGAAAAAAGTATAGGATAAATTACATTAAAATAGCACATGATTTTCTTTTGTTTTCTTCAT TTCTCTTGCTCACCAAGAAGTAACAAAAGTATAGTTTTGACAGAGTTGGTGTTCATAATTTCAGTTCTAGTTGATTGCGAGAATTTTCAA ATAAGGAAGAGGGGTCTTTTATCCTTGTCGTAGGAAAACCATGACGGAAAGGAAAAACTGATGTTTAAAAGTCCACTTTTAAAACTATAT TTATTTATGTAGGATCTGTCAAAGAAAACTTCCAAAAAGATTTATTAATTAAACCAGACTCTGTTGCAATAA >5894_5894_2_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000356064_FGB_chr4_155486960_ENST00000302068_length(amino acids)=555AA_BP=102 MSSAVGQCEFVTAAVAPRPRAAEVRGGRGRRKRSGSRFRSVFARVEAIPESGGKDKPRLRSYRVTLRKESTMAQESPKNSAAEIPVTSNG EVDDSREHSFNRGFFSARGHRPLDKKREEAPSLRPAPPPISGGGYRARPAKAAATQKKVERKAPDAGGCLHADPDLGVLCPTGCQLQEAL LQQERPIRNSVDELNNNVEAVSQTSSSSFQYMYLLKDLWQKRQKQVKDNENVVNEYSSELEKHQLYIDETVNSNIPTNLRVLRSILENLR SKIQKLESDVSAQMEYCRTPCTVSCNIPVVSGKECEEIIRKGGETSEMYLIQPDSSVKPYRVYCDMNTENGGWTVIQNRQDGSVDFGRKW DPYKQGFGNVATNTDGKNYCGLPGEYWLGNDKISQLTRMGPTELLIEMEDWKGDKVKAHYGGFTVQNEANKYQISVNKYRGTAGNALMDG ASQLMGENRTMTIHNGMFFSTYDRDNDGWLTSDPRKQCSKEDGGGWWYNRCHAANPNGRYYWGGQYTWDMAKHGTDDGVVWMNWKGSWYS MRKMSMKIRPFFPQQ -------------------------------------------------------------- >5894_5894_3_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000405727_FGB_chr4_155486960_ENST00000302068_length(transcript)=2021nt_BP=223nt CGCTGCTCTTGGTTCTGGTTCTGGAGGCTGGGTTGAGAGGTCGCCGGTCCGACTGTCCTCGGCGGTTGGTCAGTGTGAATTTGTGACAGC TGCAGTTGCTCCCCGCCCCCGAGCAGCCGAGGAGTCTACCATGGCTCAAGAATCTCCCAAAAATTCAGCAGCAGAAATTCCAGTGACTAG TAATGGAGAAGTTGATGACTCTCGTGAACATAGCTTTAATAGGGGTTTCTTCAGTGCCCGTGGTCATCGACCCCTTGACAAGAAGAGAGA AGAGGCTCCCAGCCTGAGGCCTGCCCCACCGCCCATCAGTGGAGGTGGCTATCGGGCTCGTCCAGCCAAAGCAGCTGCCACTCAAAAGAA AGTAGAAAGAAAAGCCCCTGATGCTGGAGGCTGTCTTCACGCTGACCCAGACCTGGGGGTGTTGTGTCCTACAGGATGTCAGTTGCAAGA GGCTTTGCTACAACAGGAAAGGCCAATCAGAAATAGTGTTGATGAGTTAAATAACAATGTGGAAGCTGTTTCCCAGACCTCCTCTTCTTC CTTTCAGTACATGTATTTGCTGAAAGACCTGTGGCAAAAGAGGCAGAAGCAAGTAAAAGATAATGAAAATGTAGTCAATGAGTACTCCTC AGAACTGGAAAAGCACCAATTATATATAGATGAGACTGTGAATAGCAATATCCCAACTAACCTTCGTGTGCTTCGTTCAATCCTGGAAAA CCTGAGAAGCAAAATACAAAAGTTAGAATCTGATGTCTCAGCTCAAATGGAATATTGTCGCACCCCATGCACTGTCAGTTGCAATATTCC TGTGGTGTCTGGCAAAGAATGTGAGGAAATTATCAGGAAAGGAGGTGAAACATCTGAAATGTATCTCATTCAACCTGACAGTTCTGTCAA ACCGTATAGAGTATACTGTGACATGAATACAGAAAATGGAGGATGGACAGTGATTCAGAACCGTCAAGACGGTAGTGTTGACTTTGGCAG GAAATGGGATCCATATAAACAGGGATTTGGAAATGTTGCAACCAACACAGATGGGAAGAATTACTGTGGCCTACCAGGTGAATATTGGCT TGGAAATGATAAAATTAGCCAGCTTACCAGGATGGGACCCACAGAACTTTTGATAGAAATGGAGGACTGGAAAGGAGACAAAGTAAAGGC TCACTATGGAGGATTCACTGTACAGAATGAAGCCAACAAATACCAGATCTCAGTGAACAAATACAGAGGAACAGCCGGTAATGCCCTCAT GGATGGAGCATCTCAGCTGATGGGAGAAAACAGGACCATGACCATTCACAACGGCATGTTCTTCAGCACGTATGACAGAGACAATGACGG CTGGTTAACATCAGATCCCAGAAAACAGTGTTCTAAAGAAGACGGTGGTGGATGGTGGTATAATAGATGTCATGCAGCCAATCCAAACGG CAGATACTACTGGGGTGGACAGTACACCTGGGACATGGCAAAGCATGGCACAGATGATGGTGTAGTATGGATGAATTGGAAGGGGTCATG GTACTCAATGAGGAAGATGAGTATGAAGATCAGGCCCTTCTTCCCACAGCAATAGTCCCCAATACGTAGATTTTTGCTCTTCTGTATGTG ACAACATTTTTGTACATTATGTTATTGGAATTTTCTTTCATACATTATATTCCTCTAAAACTCTCAAGCAGACGTGAGTGTGACTTTTTG AAAAAAGTATAGGATAAATTACATTAAAATAGCACATGATTTTCTTTTGTTTTCTTCATTTCTCTTGCTCACCAAGAAGTAACAAAAGTA TAGTTTTGACAGAGTTGGTGTTCATAATTTCAGTTCTAGTTGATTGCGAGAATTTTCAAATAAGGAAGAGGGGTCTTTTATCCTTGTCGT AGGAAAACCATGACGGAAAGGAAAAACTGATGTTTAAAAGTCCACTTTTAAAACTATATTTATTTATGTAGGATCTGTCAAAGAAAACTT CCAAAAAGATTTATTAATTAAACCAGACTCTGTTGCAATAA >5894_5894_3_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000405727_FGB_chr4_155486960_ENST00000302068_length(amino acids)=510AA_BP=57 MSSAVGQCEFVTAAVAPRPRAAEESTMAQESPKNSAAEIPVTSNGEVDDSREHSFNRGFFSARGHRPLDKKREEAPSLRPAPPPISGGGY RARPAKAAATQKKVERKAPDAGGCLHADPDLGVLCPTGCQLQEALLQQERPIRNSVDELNNNVEAVSQTSSSSFQYMYLLKDLWQKRQKQ VKDNENVVNEYSSELEKHQLYIDETVNSNIPTNLRVLRSILENLRSKIQKLESDVSAQMEYCRTPCTVSCNIPVVSGKECEEIIRKGGET SEMYLIQPDSSVKPYRVYCDMNTENGGWTVIQNRQDGSVDFGRKWDPYKQGFGNVATNTDGKNYCGLPGEYWLGNDKISQLTRMGPTELL IEMEDWKGDKVKAHYGGFTVQNEANKYQISVNKYRGTAGNALMDGASQLMGENRTMTIHNGMFFSTYDRDNDGWLTSDPRKQCSKEDGGG WWYNRCHAANPNGRYYWGGQYTWDMAKHGTDDGVVWMNWKGSWYSMRKMSMKIRPFFPQQ -------------------------------------------------------------- >5894_5894_4_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000429148_FGB_chr4_155486960_ENST00000302068_length(transcript)=2053nt_BP=255nt AACGCTACTTCCGGTCTCCTCACTTCCGGCTTCGCTGCTCTTGGTTCTGGTTCTGGAGGCTGGGTTGAGAGGTCGCCGGTCCGACTGTCC TCGGCGGTTGGTCAGTGTGAATTTGTGACAGCTGCAGTTGCTCCCCGCCCCCGAGCAGCCGAGGAGTCTACCATGGCTCAAGAATCTCCC AAAAATTCAGCAGCAGAAATTCCAGTGACTAGTAATGGAGAAGTTGATGACTCTCGTGAACATAGCTTTAATAGGGGTTTCTTCAGTGCC CGTGGTCATCGACCCCTTGACAAGAAGAGAGAAGAGGCTCCCAGCCTGAGGCCTGCCCCACCGCCCATCAGTGGAGGTGGCTATCGGGCT CGTCCAGCCAAAGCAGCTGCCACTCAAAAGAAAGTAGAAAGAAAAGCCCCTGATGCTGGAGGCTGTCTTCACGCTGACCCAGACCTGGGG GTGTTGTGTCCTACAGGATGTCAGTTGCAAGAGGCTTTGCTACAACAGGAAAGGCCAATCAGAAATAGTGTTGATGAGTTAAATAACAAT GTGGAAGCTGTTTCCCAGACCTCCTCTTCTTCCTTTCAGTACATGTATTTGCTGAAAGACCTGTGGCAAAAGAGGCAGAAGCAAGTAAAA GATAATGAAAATGTAGTCAATGAGTACTCCTCAGAACTGGAAAAGCACCAATTATATATAGATGAGACTGTGAATAGCAATATCCCAACT AACCTTCGTGTGCTTCGTTCAATCCTGGAAAACCTGAGAAGCAAAATACAAAAGTTAGAATCTGATGTCTCAGCTCAAATGGAATATTGT CGCACCCCATGCACTGTCAGTTGCAATATTCCTGTGGTGTCTGGCAAAGAATGTGAGGAAATTATCAGGAAAGGAGGTGAAACATCTGAA ATGTATCTCATTCAACCTGACAGTTCTGTCAAACCGTATAGAGTATACTGTGACATGAATACAGAAAATGGAGGATGGACAGTGATTCAG AACCGTCAAGACGGTAGTGTTGACTTTGGCAGGAAATGGGATCCATATAAACAGGGATTTGGAAATGTTGCAACCAACACAGATGGGAAG AATTACTGTGGCCTACCAGGTGAATATTGGCTTGGAAATGATAAAATTAGCCAGCTTACCAGGATGGGACCCACAGAACTTTTGATAGAA ATGGAGGACTGGAAAGGAGACAAAGTAAAGGCTCACTATGGAGGATTCACTGTACAGAATGAAGCCAACAAATACCAGATCTCAGTGAAC AAATACAGAGGAACAGCCGGTAATGCCCTCATGGATGGAGCATCTCAGCTGATGGGAGAAAACAGGACCATGACCATTCACAACGGCATG TTCTTCAGCACGTATGACAGAGACAATGACGGCTGGTTAACATCAGATCCCAGAAAACAGTGTTCTAAAGAAGACGGTGGTGGATGGTGG TATAATAGATGTCATGCAGCCAATCCAAACGGCAGATACTACTGGGGTGGACAGTACACCTGGGACATGGCAAAGCATGGCACAGATGAT GGTGTAGTATGGATGAATTGGAAGGGGTCATGGTACTCAATGAGGAAGATGAGTATGAAGATCAGGCCCTTCTTCCCACAGCAATAGTCC CCAATACGTAGATTTTTGCTCTTCTGTATGTGACAACATTTTTGTACATTATGTTATTGGAATTTTCTTTCATACATTATATTCCTCTAA AACTCTCAAGCAGACGTGAGTGTGACTTTTTGAAAAAAGTATAGGATAAATTACATTAAAATAGCACATGATTTTCTTTTGTTTTCTTCA TTTCTCTTGCTCACCAAGAAGTAACAAAAGTATAGTTTTGACAGAGTTGGTGTTCATAATTTCAGTTCTAGTTGATTGCGAGAATTTTCA AATAAGGAAGAGGGGTCTTTTATCCTTGTCGTAGGAAAACCATGACGGAAAGGAAAAACTGATGTTTAAAAGTCCACTTTTAAAACTATA TTTATTTATGTAGGATCTGTCAAAGAAAACTTCCAAAAAGATTTATTAATTAAACCAGACTCTGTTGCAATAA >5894_5894_4_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000429148_FGB_chr4_155486960_ENST00000302068_length(amino acids)=510AA_BP=57 MSSAVGQCEFVTAAVAPRPRAAEESTMAQESPKNSAAEIPVTSNGEVDDSREHSFNRGFFSARGHRPLDKKREEAPSLRPAPPPISGGGY RARPAKAAATQKKVERKAPDAGGCLHADPDLGVLCPTGCQLQEALLQQERPIRNSVDELNNNVEAVSQTSSSSFQYMYLLKDLWQKRQKQ VKDNENVVNEYSSELEKHQLYIDETVNSNIPTNLRVLRSILENLRSKIQKLESDVSAQMEYCRTPCTVSCNIPVVSGKECEEIIRKGGET SEMYLIQPDSSVKPYRVYCDMNTENGGWTVIQNRQDGSVDFGRKWDPYKQGFGNVATNTDGKNYCGLPGEYWLGNDKISQLTRMGPTELL IEMEDWKGDKVKAHYGGFTVQNEANKYQISVNKYRGTAGNALMDGASQLMGENRTMTIHNGMFFSTYDRDNDGWLTSDPRKQCSKEDGGG WWYNRCHAANPNGRYYWGGQYTWDMAKHGTDDGVVWMNWKGSWYSMRKMSMKIRPFFPQQ -------------------------------------------------------------- >5894_5894_5_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000451320_FGB_chr4_155486960_ENST00000302068_length(transcript)=2055nt_BP=257nt ACAACGCTACTTCCGGTCTCCTCACTTCCGGCTTCGCTGCTCTTGGTTCTGGTTCTGGAGGCTGGGTTGAGAGGTCGCCGGTCCGACTGT CCTCGGCGGTTGGTCAGTGTGAATTTGTGACAGCTGCAGTTGCTCCCCGCCCCCGAGCAGCCGAGGAGTCTACCATGGCTCAAGAATCTC CCAAAAATTCAGCAGCAGAAATTCCAGTGACTAGTAATGGAGAAGTTGATGACTCTCGTGAACATAGCTTTAATAGGGGTTTCTTCAGTG CCCGTGGTCATCGACCCCTTGACAAGAAGAGAGAAGAGGCTCCCAGCCTGAGGCCTGCCCCACCGCCCATCAGTGGAGGTGGCTATCGGG CTCGTCCAGCCAAAGCAGCTGCCACTCAAAAGAAAGTAGAAAGAAAAGCCCCTGATGCTGGAGGCTGTCTTCACGCTGACCCAGACCTGG GGGTGTTGTGTCCTACAGGATGTCAGTTGCAAGAGGCTTTGCTACAACAGGAAAGGCCAATCAGAAATAGTGTTGATGAGTTAAATAACA ATGTGGAAGCTGTTTCCCAGACCTCCTCTTCTTCCTTTCAGTACATGTATTTGCTGAAAGACCTGTGGCAAAAGAGGCAGAAGCAAGTAA AAGATAATGAAAATGTAGTCAATGAGTACTCCTCAGAACTGGAAAAGCACCAATTATATATAGATGAGACTGTGAATAGCAATATCCCAA CTAACCTTCGTGTGCTTCGTTCAATCCTGGAAAACCTGAGAAGCAAAATACAAAAGTTAGAATCTGATGTCTCAGCTCAAATGGAATATT GTCGCACCCCATGCACTGTCAGTTGCAATATTCCTGTGGTGTCTGGCAAAGAATGTGAGGAAATTATCAGGAAAGGAGGTGAAACATCTG AAATGTATCTCATTCAACCTGACAGTTCTGTCAAACCGTATAGAGTATACTGTGACATGAATACAGAAAATGGAGGATGGACAGTGATTC AGAACCGTCAAGACGGTAGTGTTGACTTTGGCAGGAAATGGGATCCATATAAACAGGGATTTGGAAATGTTGCAACCAACACAGATGGGA AGAATTACTGTGGCCTACCAGGTGAATATTGGCTTGGAAATGATAAAATTAGCCAGCTTACCAGGATGGGACCCACAGAACTTTTGATAG AAATGGAGGACTGGAAAGGAGACAAAGTAAAGGCTCACTATGGAGGATTCACTGTACAGAATGAAGCCAACAAATACCAGATCTCAGTGA ACAAATACAGAGGAACAGCCGGTAATGCCCTCATGGATGGAGCATCTCAGCTGATGGGAGAAAACAGGACCATGACCATTCACAACGGCA TGTTCTTCAGCACGTATGACAGAGACAATGACGGCTGGTTAACATCAGATCCCAGAAAACAGTGTTCTAAAGAAGACGGTGGTGGATGGT GGTATAATAGATGTCATGCAGCCAATCCAAACGGCAGATACTACTGGGGTGGACAGTACACCTGGGACATGGCAAAGCATGGCACAGATG ATGGTGTAGTATGGATGAATTGGAAGGGGTCATGGTACTCAATGAGGAAGATGAGTATGAAGATCAGGCCCTTCTTCCCACAGCAATAGT CCCCAATACGTAGATTTTTGCTCTTCTGTATGTGACAACATTTTTGTACATTATGTTATTGGAATTTTCTTTCATACATTATATTCCTCT AAAACTCTCAAGCAGACGTGAGTGTGACTTTTTGAAAAAAGTATAGGATAAATTACATTAAAATAGCACATGATTTTCTTTTGTTTTCTT CATTTCTCTTGCTCACCAAGAAGTAACAAAAGTATAGTTTTGACAGAGTTGGTGTTCATAATTTCAGTTCTAGTTGATTGCGAGAATTTT CAAATAAGGAAGAGGGGTCTTTTATCCTTGTCGTAGGAAAACCATGACGGAAAGGAAAAACTGATGTTTAAAAGTCCACTTTTAAAACTA TATTTATTTATGTAGGATCTGTCAAAGAAAACTTCCAAAAAGATTTATTAATTAAACCAGACTCTGTTGCAATAA >5894_5894_5_ARFIP1-FGB_ARFIP1_chr4_153750878_ENST00000451320_FGB_chr4_155486960_ENST00000302068_length(amino acids)=510AA_BP=57 MSSAVGQCEFVTAAVAPRPRAAEESTMAQESPKNSAAEIPVTSNGEVDDSREHSFNRGFFSARGHRPLDKKREEAPSLRPAPPPISGGGY RARPAKAAATQKKVERKAPDAGGCLHADPDLGVLCPTGCQLQEALLQQERPIRNSVDELNNNVEAVSQTSSSSFQYMYLLKDLWQKRQKQ VKDNENVVNEYSSELEKHQLYIDETVNSNIPTNLRVLRSILENLRSKIQKLESDVSAQMEYCRTPCTVSCNIPVVSGKECEEIIRKGGET SEMYLIQPDSSVKPYRVYCDMNTENGGWTVIQNRQDGSVDFGRKWDPYKQGFGNVATNTDGKNYCGLPGEYWLGNDKISQLTRMGPTELL IEMEDWKGDKVKAHYGGFTVQNEANKYQISVNKYRGTAGNALMDGASQLMGENRTMTIHNGMFFSTYDRDNDGWLTSDPRKQCSKEDGGG WWYNRCHAANPNGRYYWGGQYTWDMAKHGTDDGVVWMNWKGSWYSMRKMSMKIRPFFPQQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ARFIP1-FGB |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ARFIP1-FGB |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Tgene | FGB | P02675 | DB11311 | Prothrombin | Cleavage | Biotech | Approved |
| Tgene | FGB | P02675 | DB11571 | Human thrombin | Activator | Biotech | Approved |
| Tgene | FGB | P02675 | DB11572 | Thrombin alfa | Activator | Biotech | Approved |
| Tgene | FGB | P02675 | DB11300 | Thrombin | Activator | Biotech | Approved|Investigational |
| Tgene | FGB | P02675 | DB13151 | Anti-inhibitor coagulant complex | Cleavage | Biotech | Approved|Investigational |
Top |
Related Diseases for ARFIP1-FGB |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C2584774 | Congenital hypofibrinogenemia | 7 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0272350 | Dysfibrinogenemia, Congenital | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0001787 | Osteoporosis, Age-Related | 1 | CTD_human | |
| Tgene | C0029456 | Osteoporosis | 1 | CTD_human | |
| Tgene | C0029459 | Osteoporosis, Senile | 1 | CTD_human | |
| Tgene | C0030567 | Parkinson Disease | 1 | CTD_human | |
| Tgene | C0751406 | Post-Traumatic Osteoporosis | 1 | CTD_human |
