
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GLRB-RPL13A (FusionGDB2 ID:HG2743TG23521) |
Fusion Gene Summary for GLRB-RPL13A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GLRB-RPL13A | Fusion gene ID: hg2743tg23521 | Hgene | Tgene | Gene symbol | GLRB | RPL13A | Gene ID | 2743 | 23521 |
| Gene name | glycine receptor beta | ribosomal protein L13a | |
| Synonyms | HKPX2 | L13A|TSTA1 | |
| Cytomap | ('GLRB')('RPL13A') 4q32.1 | 19q13.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | glycine receptor subunit betaglycine receptor 58 kDa subunitglycine receptor, beta subunit | 60S ribosomal protein L13a23 kDa highly basic proteinepididymis secretory sperm binding proteinlarge ribosomal subunit protein uL13tissue specific transplantation antigen 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P48167 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000512619, ENST00000541722, ENST00000264428, ENST00000509282, | ||
| Fusion gene scores | * DoF score | 4 X 4 X 3=48 | 15 X 16 X 9=2160 |
| # samples | 4 | 15 | |
| ** MAII score | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/2160*10)=-3.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GLRB [Title/Abstract] AND RPL13A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GLRB(158091782)-RPL13A(49994295), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | GLRB-RPL13A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GLRB-RPL13A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | GLRB | GO:0006811 | ion transport | 8717357 |
| Hgene | GLRB | GO:0007218 | neuropeptide signaling pathway | 8717357 |
| Tgene | RPL13A | GO:0017148 | negative regulation of translation | 14567916|23071094 |
| Tgene | RPL13A | GO:0071346 | cellular response to interferon-gamma | 15479637 |
| Tgene | RPL13A | GO:1901194 | negative regulation of formation of translation preinitiation complex | 17218275 |
 Fusion gene breakpoints across GLRB (5'-gene) Fusion gene breakpoints across GLRB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
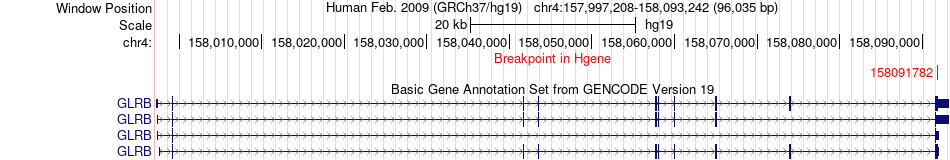 |
 Fusion gene breakpoints across RPL13A (3'-gene) Fusion gene breakpoints across RPL13A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
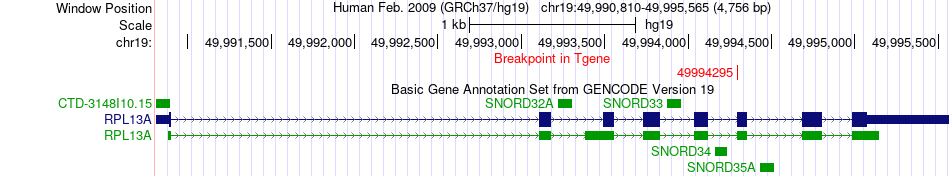 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-A4J9-01A | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
Top |
Fusion Gene ORF analysis for GLRB-RPL13A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000512619 | ENST00000391857 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| 3UTR-3CDS | ENST00000541722 | ENST00000391857 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| 3UTR-3UTR | ENST00000512619 | ENST00000477613 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| 3UTR-3UTR | ENST00000541722 | ENST00000477613 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| 5CDS-3UTR | ENST00000264428 | ENST00000477613 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| 5CDS-3UTR | ENST00000509282 | ENST00000477613 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| In-frame | ENST00000264428 | ENST00000391857 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
| In-frame | ENST00000509282 | ENST00000391857 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264428 | GLRB | chr4 | 158091782 | + | ENST00000391857 | RPL13A | chr19 | 49994295 | + | 3690 | 2927 | 129 | 1625 | 498 |
| ENST00000509282 | GLRB | chr4 | 158091782 | + | ENST00000391857 | RPL13A | chr19 | 49994295 | + | 2321 | 1558 | 23 | 1540 | 505 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264428 | ENST00000391857 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + | 0.000320575 | 0.99967945 |
| ENST00000509282 | ENST00000391857 | GLRB | chr4 | 158091782 | + | RPL13A | chr19 | 49994295 | + | 0.001497741 | 0.99850225 |
Top |
Fusion Genomic Features for GLRB-RPL13A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
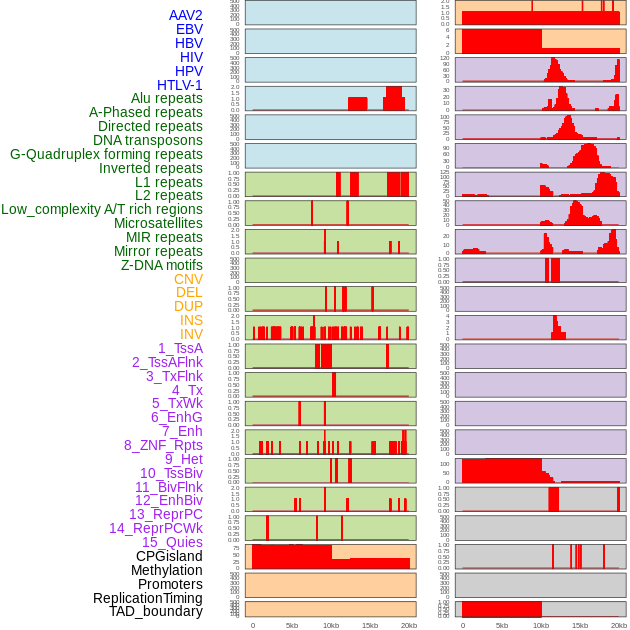 |
Top |
Fusion Protein Features for GLRB-RPL13A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:158091782/chr19:49994295) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GLRB | . |
| FUNCTION: Glycine receptors are ligand-gated chloride channels. GLRB does not form ligand-gated ion channels by itself, but is part of heteromeric ligand-gated chloride channels. Channel opening is triggered by extracellular glycine (PubMed:8717357, PubMed:15302677, PubMed:16144831, PubMed:22715885, PubMed:25445488, PubMed:11929858, PubMed:23238346). Heteropentameric channels composed of GLRB and GLRA1 are activated by lower glycine levels than homopentameric GLRA1 (PubMed:8717357). Plays an important role in the down-regulation of neuronal excitability (PubMed:11929858, PubMed:23238346). Contributes to the generation of inhibitory postsynaptic currents (PubMed:25445488). {ECO:0000269|PubMed:11929858, ECO:0000269|PubMed:15302677, ECO:0000269|PubMed:16144831, ECO:0000269|PubMed:22715885, ECO:0000269|PubMed:23238346, ECO:0000269|PubMed:25445488, ECO:0000269|PubMed:8717357}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 247_253 | 0 | 498.0 | Region | Strychnine-binding |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 247_253 | 0 | 498.0 | Region | Strychnine-binding |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 247_253 | 0 | 304.0 | Region | Strychnine-binding |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 23_268 | 0 | 498.0 | Topological domain | Extracellular |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 291_295 | 0 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 317_327 | 0 | 498.0 | Topological domain | Extracellular |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 349_475 | 0 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 23_268 | 0 | 498.0 | Topological domain | Extracellular |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 291_295 | 0 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 317_327 | 0 | 498.0 | Topological domain | Extracellular |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 349_475 | 0 | 498.0 | Topological domain | Cytoplasmic |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 23_268 | 0 | 304.0 | Topological domain | Extracellular |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 291_295 | 0 | 304.0 | Topological domain | Cytoplasmic |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 317_327 | 0 | 304.0 | Topological domain | Extracellular |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 349_475 | 0 | 304.0 | Topological domain | Cytoplasmic |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 269_290 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 296_316 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 328_348 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000264428 | + | 1 | 10 | 476_496 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 269_290 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 296_316 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 328_348 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000509282 | + | 1 | 10 | 476_496 | 0 | 498.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 269_290 | 0 | 304.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 296_316 | 0 | 304.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 328_348 | 0 | 304.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | GLRB | chr4:158091782 | chr19:49994295 | ENST00000541722 | + | 1 | 9 | 476_496 | 0 | 304.0 | Transmembrane | Helical%3B Name%3D4 |
Top |
Fusion Gene Sequence for GLRB-RPL13A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >33343_33343_1_GLRB-RPL13A_GLRB_chr4_158091782_ENST00000264428_RPL13A_chr19_49994295_ENST00000391857_length(transcript)=3690nt_BP=2927nt CCGCCCGCGGTCGCCAGGTTCCCCCGCGGCCCGCGCTGTCCCCGCTGCGGCCCCGACCGCGGCGCGCCACGGCGCAGGCTCCGCGGCCCG AGCAGGCGCGTCAGCCGAGCTCGGCGGTGGCGCGGGCGGCTGGGACGCAGCTGCCCCCGCTCGGCGCCCGCTGCACCCTTAGCAGCCACT GCCACCTGGGCCCGGAGCCTCCACGATCTCGCCCGGCGATTGTGGGCAGGGGCGCCTCCGGATCGATCTTCTGAAATTCAAGTTTTCAAG ATGAAGTTTTTATTGACAACTGCCTTTTTAATTTTAATTTCCTTGTGGGTGGAAGAAGCCTATTCTAAGGAAAAGTCTTCAAAGAAAGGG AAGGGGAAAAAGAAGCAGTATCTATGCCCATCTCAGCAGTCAGCAGAGGACCTTGCCCGAGTACCTGCCAACTCCACTAGCAATATCTTG AACAGGTTATTGGTCAGTTATGATCCCAGGATAAGACCAAACTTCAAAGGCATTCCTGTTGATGTAGTAGTCAACATTTTTATTAACAGT TTTGGATCCATTCAAGAAACAACAATGGACTATAGAGTTAACATCTTCCTGAGACAAAAATGGAATGACCCCAGGCTGAAGCTCCCCAGT GATTTTAGGGGTTCAGATGCACTGACAGTGGATCCAACAATGTACAAGTGTTTATGGAAACCTGATTTATTTTTTGCAAATGAAAAAAGT GCCAATTTTCATGATGTGACCCAGGAAAACATCCTCCTCTTTATTTTTCGTGATGGAGATGTCCTTGTCAGCATGAGGTTATCTATTACT CTTTCATGCCCTTTGGACTTGACATTGTTTCCCATGGATACACAACGTTGCAAGATGCAACTGGAGAGCTTTGGTTACACAACTGATGAT TTACGATTTATCTGGCAGTCAGGAGATCCTGTGCAATTAGAAAAAATTGCCTTGCCTCAATTTGATATCAAAAAGGAAGATATTGAATAT GGTAACTGTACAAAATACTATAAAGGCACGGGCTACTACACATGCGTGGAAGTCATCTTCACCCTGAGGAGGCAGGTCGGCTTTTACATG ATGGGGGTCTACGCCCCAACTCTGCTCATTGTTGTTCTCTCCTGGCTTTCCTTCTGGATCAACCCGGACGCGAGTGCTGCCAGAGTGCCC CTGGGTATCTTCTCAGTCCTCAGCTTGGCCTCTGAGTGCACAACCCTTGCCGCTGAGCTTCCCAAAGTTTCCTATGTGAAGGCTCTTGAT GTTTGGCTTATTGCTTGCCTTCTCTTTGGGTTTGCTTCCCTGGTGGAGTATGCAGTTGTCCAGGTGATGCTGAACAACCCCAAAAGGGTT GAAGCTGAAAAAGCCAGAATTGCTAAGGCTGAGCAAGCAGATGGAAAAGGTGGAAATGTGGCTAAAAAGAATACTGTGAATGGAACAGGG ACTCCTGTTCATATTAGCACTTTGCAGTTCCAACAGCAGCAAAGCGAATTGATCTTTATGCAAGAGCATTGTTTCCTTTCTGCTTCTTGT TCTTCAATGTTATATATTGGTCTATATATTTATGATAAATCTTTTCCATTTGTACAAAATAAAATTCCATTTCATTGTGACCTACTCCTT TCATAAATGCCAATCTGTGAGAACTTTTGAATTTTCATAGCAACATTGCATTTTGGATGCCATTTGATTGTAATAAAACTGTGGCACCTT AATTTTGAATGGCAGCATGATCATGTAATATCTGTGCTCTAATAACGATGTATATATGTATAGTGAACATATTGCTTAGTAACAAATGAA GGACAAGCATACTACATAATATAATCCATACAATTCTCTTCAGTTAGTGTAAACTGCAAATACTACAGATAATTCTGATAATAAAATGAT ATGCACGCTGAATCCTGCTATGGTCACCATTCTAATGTATGTAGTATTTCAAATTTCCTTCCTTGTAACTTTCAAAGAAAGCCATCTTAT TCTTGTAAAATTTTAGATGGTATTATCACAGATTTAAAAAGGTTGTATTACATATTGTTTAAACTTTGTAAGTAGAAATATATCTGTTAT AATTATACAGGCTCTGTGGAGAAATAAAGTTCAAAATATATTAATTTGTAAAATCAGCTCGTTTTAAAGTGTGCTTGTGTTGTCAAAAAT ATCAGATAGTAATACACAGTGAGCATTTTTAAACAAAGGGAAACCTATATTTATGTAACTGTATACTGAATTCTGACAAAATAAAAAAAG ATACCTTATTGACGAAATATTTAGGATAAACAAAATTCTATTTAATCCACCTTAAAACCTAAATGTATTTTCATGGATTTCATTTGTTGG TACATATTACACAAAACATTGTGCCTTAAAATGAGTCATACATCTTTTAAATTGGAATGCAGTAATAGATATGTGATTTTACATCATTTT TAAGAAACCAAGGGGAAGTAATAAGTTGAAAAAGAAATCCATAACTATTAAAAGATTTTAACTTTTTTATTTTATTAAAATGCTTGCATA TTTTAAGTAAAATTAAAAATGTTTACTGAATTTATTTTTTTATTTGAATATTTTGGGATTAGTTACAAAATATTTAGAGATTAAATAAAT AATAGCTTTGTTTGTGTTAGACTTCACTTCTACTTGTATTTTCTTTCTTGTTAAAAGTCTAGACAACTGAGATTTACTTACTTTGAACTT GTTCCAATCCAAAAGTAAGCACTAAGTCTCATTTTATGAGACCACCATTTCTTAATATCACATCCAGTGCACCTTTGTCTTTCTGCCATA TCTGAAATAAAACTATCAGTAATTCACATAGATAAACATAAGACTAAAGAAGTATATTTACATTATCTGGAGAGTTTTGTTGCAGCTACG AGTTTGTATGGCAAATTCAATAATAAAGTATTGTTTATGCAAATTGCAAAAAGCGGATGGTGGTTCCTGCTGCCCTCAAGGTCGTGCGTC TGAAGCCTACAAGAAAGTTTGCCTATCTGGGGCGCCTGGCTCACGAGGTTGGCTGGAAGTACCAGGCAGTGACAGCCACCCTGGAGGAGA AGAGGAAAGAGAAAGCCAAGATCCACTACCGGAAGAAGAAACAGCTCATGAGGCTACGGAAACAGGCCGAGAAGAACGTGGAGAAGAAAA TTGACAAATACACAGAGGTCCTCAAGACCCACGGACTCCTGGTCTGAGCCCAATAAAGACTGTTAATTCCTCATGCGTTGCCTGCCCTTC CTCCATTGTTGCCCTGGAATGTACGGGACCCAGGGGCAGCAGCAGTCCAGGTGCCACAGGCAGCCCTGGGACATAGGAAGCTGGGAGCAA GGAAAGGGTCTTAGTCACTGCCTCCCGAAGTTGCTTGAAAGCACTCGGAGAATTGTGCAGGTGTCATTTATCTATGACCAATAGGAAGAG CAACCAGTTACTATGAGTGAAAGGGAGCCAGAAGACTGATTGGAGGGCCCTATCTTGTGAGTGGGGCATCTGTTGGACTTTCCACCTGGT CATATACTCTGCAGCTGTTAGAATGTGCAAGCACTTGGGGACAGCATGAGCTTGCTGTTGTACACAGGGTATTTCTAGAAGCAGAAATAG ACTGGGAAGATGCACAACCAAGGGGTTACAGGCATCGCCCATGCTCCTCACCTGTATTTTGTAATCAGAAATAAATTGCTTTTAAAGAAA >33343_33343_1_GLRB-RPL13A_GLRB_chr4_158091782_ENST00000264428_RPL13A_chr19_49994295_ENST00000391857_length(amino acids)=498AA_BP= MGRSCPRSAPAAPLAATATWARSLHDLARRLWAGAPPDRSSEIQVFKMKFLLTTAFLILISLWVEEAYSKEKSSKKGKGKKKQYLCPSQQ SAEDLARVPANSTSNILNRLLVSYDPRIRPNFKGIPVDVVVNIFINSFGSIQETTMDYRVNIFLRQKWNDPRLKLPSDFRGSDALTVDPT MYKCLWKPDLFFANEKSANFHDVTQENILLFIFRDGDVLVSMRLSITLSCPLDLTLFPMDTQRCKMQLESFGYTTDDLRFIWQSGDPVQL EKIALPQFDIKKEDIEYGNCTKYYKGTGYYTCVEVIFTLRRQVGFYMMGVYAPTLLIVVLSWLSFWINPDASAARVPLGIFSVLSLASEC TTLAAELPKVSYVKALDVWLIACLLFGFASLVEYAVVQVMLNNPKRVEAEKARIAKAEQADGKGGNVAKKNTVNGTGTPVHISTLQFQQQ QSELIFMQEHCFLSASCSSMLYIGLYIYDKSFPFVQNKIPFHCDLLLS -------------------------------------------------------------- >33343_33343_2_GLRB-RPL13A_GLRB_chr4_158091782_ENST00000509282_RPL13A_chr19_49994295_ENST00000391857_length(transcript)=2321nt_BP=1558nt GGGGCGGCGGAGCGCGAGGAGCACTGCGGCGTGCCCGGGAGGACGCGCGCTGCCCTGCGCGGGAGAGGGGCGCGGGGCGCCTGGCAGCGG CAGCCGGGTCCCCGCTGCGGTGGCGCCCGCGGAGACGCTCCGGGTGGAGAGATGCAGAGGGCCGGGATCGATCTTCTGAAATTCAAGTTT TCAAGATGAAGTTTTTATTGACAACTGCCTTTTTAATTTTAATTTCCTTGTGGGTGGAAGAAGCCTATTCTAAGGAAAAGTCTTCAAAGA AAGGGAAGGGGAAAAAGAAGCAGTATCTATGCCCATCTCAGCAGTCAGCAGAGGACCTTGCCCGAGTACCTGCCAACTCCACTAGCAATA TCTTGAACAGGTTATTGGTCAGTTATGATCCCAGGATAAGACCAAACTTCAAAGGCATTCCTGTTGATGTAGTAGTCAACATTTTTATTA ACAGTTTTGGATCCATTCAAGAAACAACAATGGACTATAGAGTTAACATCTTCCTGAGACAAAAATGGAATGACCCCAGGCTGAAGCTCC CCAGTGATTTTAGGGGTTCAGATGCACTGACAGTGGATCCAACAATGTACAAGTGTTTATGGAAACCTGATTTATTTTTTGCAAATGAAA AAAGTGCCAATTTTCATGATGTGACCCAGGAAAACATCCTCCTCTTTATTTTTCGTGATGGAGATGTCCTTGTCAGCATGAGGTTATCTA TTACTCTTTCATGCCCTTTGGACTTGACATTGTTTCCCATGGATACACAACGTTGCAAGATGCAACTGGAGAGCTTTGGTTACACAACTG ATGATTTACGATTTATCTGGCAGTCAGGAGATCCTGTGCAATTAGAAAAAATTGCCTTGCCTCAATTTGATATCAAAAAGGAAGATATTG AATATGGTAACTGTACAAAATACTATAAAGGCACGGGCTACTACACATGCGTGGAAGTCATCTTCACCCTGAGGAGGCAGGTCGGCTTTT ACATGATGGGGGTCTACGCCCCAACTCTGCTCATTGTTGTTCTCTCCTGGCTTTCCTTCTGGATCAACCCGGACGCGAGTGCTGCCAGAG TGCCCCTGGGTATCTTCTCAGTCCTCAGCTTGGCCTCTGAGTGCACAACCCTTGCCGCTGAGCTTCCCAAAGTTTCCTATGTGAAGGCTC TTGATGTTTGGCTTATTGCTTGCCTTCTCTTTGGGTTTGCTTCCCTGGTGGAGTATGCAGTTGTCCAGGTGATGCTGAACAACCCCAAAA GGGTTGAAGCTGAAAAAGCCAGAATTGCTAAGGCTGAGCAAGCAGATGGAAAAGGTGGAAATGTGGCTAAAAAGAATACTGTGAATGGAA CAGGGACTCCTGTTCATATTAGCACTTTGCAGTTCCAACAGCAGCAAAGCGAATTGATCTTTATGCAAGAGCATTGTTTCCTTTCTGCTT CTTGTTCTTCAATGTTATATATTGGTCTATATATTTATGATAAATCTTTTCCATTTGTACAAAATAAAATTCCATTTCATTGTGACCTAC TCCTTTCATAAATGCCAATCTGTGAGAAAAAAAGCGGATGGTGGTTCCTGCTGCCCTCAAGGTCGTGCGTCTGAAGCCTACAAGAAAGTT TGCCTATCTGGGGCGCCTGGCTCACGAGGTTGGCTGGAAGTACCAGGCAGTGACAGCCACCCTGGAGGAGAAGAGGAAAGAGAAAGCCAA GATCCACTACCGGAAGAAGAAACAGCTCATGAGGCTACGGAAACAGGCCGAGAAGAACGTGGAGAAGAAAATTGACAAATACACAGAGGT CCTCAAGACCCACGGACTCCTGGTCTGAGCCCAATAAAGACTGTTAATTCCTCATGCGTTGCCTGCCCTTCCTCCATTGTTGCCCTGGAA TGTACGGGACCCAGGGGCAGCAGCAGTCCAGGTGCCACAGGCAGCCCTGGGACATAGGAAGCTGGGAGCAAGGAAAGGGTCTTAGTCACT GCCTCCCGAAGTTGCTTGAAAGCACTCGGAGAATTGTGCAGGTGTCATTTATCTATGACCAATAGGAAGAGCAACCAGTTACTATGAGTG AAAGGGAGCCAGAAGACTGATTGGAGGGCCCTATCTTGTGAGTGGGGCATCTGTTGGACTTTCCACCTGGTCATATACTCTGCAGCTGTT AGAATGTGCAAGCACTTGGGGACAGCATGAGCTTGCTGTTGTACACAGGGTATTTCTAGAAGCAGAAATAGACTGGGAAGATGCACAACC AAGGGGTTACAGGCATCGCCCATGCTCCTCACCTGTATTTTGTAATCAGAAATAAATTGCTTTTAAAGAAA >33343_33343_2_GLRB-RPL13A_GLRB_chr4_158091782_ENST00000509282_RPL13A_chr19_49994295_ENST00000391857_length(amino acids)=505AA_BP= MRRAREDARCPARERGAGRLAAAAGSPLRWRPRRRSGWRDAEGRDRSSEIQVFKMKFLLTTAFLILISLWVEEAYSKEKSSKKGKGKKKQ YLCPSQQSAEDLARVPANSTSNILNRLLVSYDPRIRPNFKGIPVDVVVNIFINSFGSIQETTMDYRVNIFLRQKWNDPRLKLPSDFRGSD ALTVDPTMYKCLWKPDLFFANEKSANFHDVTQENILLFIFRDGDVLVSMRLSITLSCPLDLTLFPMDTQRCKMQLESFGYTTDDLRFIWQ SGDPVQLEKIALPQFDIKKEDIEYGNCTKYYKGTGYYTCVEVIFTLRRQVGFYMMGVYAPTLLIVVLSWLSFWINPDASAARVPLGIFSV LSLASECTTLAAELPKVSYVKALDVWLIACLLFGFASLVEYAVVQVMLNNPKRVEAEKARIAKAEQADGKGGNVAKKNTVNGTGTPVHIS TLQFQQQQSELIFMQEHCFLSASCSSMLYIGLYIYDKSFPFVQNKIPFHCDLLLS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GLRB-RPL13A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GLRB-RPL13A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Hgene | GLRB | P48167 | DB00145 | Glycine | Ligand | Small molecule | Approved|Nutraceutical|Vet_approved |
| Hgene | GLRB | P48167 | DB00431 | Lindane | Antagonist | Small molecule | Approved|Withdrawn |
Top |
Related Diseases for GLRB-RPL13A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | GLRB | C3553291 | HYPEREKPLEXIA 2 | 3 | GENOMICS_ENGLAND;UNIPROT |
| Tgene | C0152013 | Adenocarcinoma of lung (disorder) | 1 | CTD_human |
