
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CD274-KDM4C (FusionGDB2 ID:HG29126TG23081) |
Fusion Gene Summary for CD274-KDM4C |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CD274-KDM4C | Fusion gene ID: hg29126tg23081 | Hgene | Tgene | Gene symbol | CD274 | KDM4C | Gene ID | 29126 | 23081 |
| Gene name | CD274 molecule | lysine demethylase 4C | |
| Synonyms | B7-H|B7H1|PD-L1|PDCD1L1|PDCD1LG1|PDL1|hPD-L1 | GASC1|JHDM3C|JMJD2C|TDRD14C | |
| Cytomap | ('CD274')('KDM4C') 9p24.1 | 9p24.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | programmed cell death 1 ligand 1B7 homolog 1CD274 antigenPDCD1 ligand 1 | lysine-specific demethylase 4CJmjC domain-containing histone demethylation protein 3Cgene amplified in squamous cell carcinoma 1 proteinjumonji domain-containing protein 2Clysine (K)-specific demethylase 4Ctudor domain containing 14C | |
| Modification date | 20200329 | 20200329 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000381573, ENST00000381577, ENST00000498261, | ||
| Fusion gene scores | * DoF score | 3 X 2 X 2=12 | 22 X 22 X 7=3388 |
| # samples | 3 | 26 | |
| ** MAII score | log2(3/12*10)=1.32192809488736 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(26/3388*10)=-3.70385034630374 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CD274 [Title/Abstract] AND KDM4C [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CD274(5465606)-KDM4C(6792971), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CD274-KDM4C seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CD274-KDM4C seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CD274-KDM4C seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CD274-KDM4C seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CD274 | GO:0006955 | immune response | 10581077 |
| Hgene | CD274 | GO:0007165 | signal transduction | 10581077 |
| Hgene | CD274 | GO:0007166 | cell surface receptor signaling pathway | 10581077 |
| Hgene | CD274 | GO:0031295 | T cell costimulation | 10581077 |
| Hgene | CD274 | GO:0034097 | response to cytokine | 17489864 |
| Hgene | CD274 | GO:1905399 | regulation of activated CD4-positive, alpha-beta T cell apoptotic process | 24187568 |
| Hgene | CD274 | GO:1905404 | positive regulation of activated CD8-positive, alpha-beta T cell apoptotic process | 24187568 |
| Hgene | CD274 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation | 24691993 |
| Hgene | CD274 | GO:2001181 | positive regulation of interleukin-10 secretion | 10581077 |
| Tgene | KDM4C | GO:0006357 | regulation of transcription by RNA polymerase II | 17277772 |
| Tgene | KDM4C | GO:0033169 | histone H3-K9 demethylation | 18066052|21914792 |
| Tgene | KDM4C | GO:0070544 | histone H3-K36 demethylation | 21914792 |
 Fusion gene breakpoints across CD274 (5'-gene) Fusion gene breakpoints across CD274 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
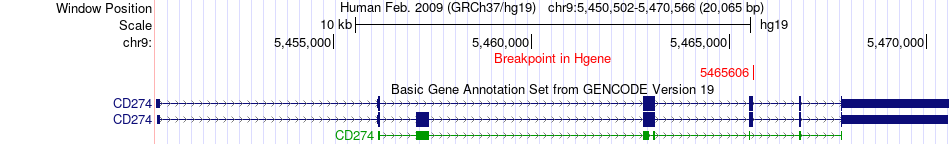 |
 Fusion gene breakpoints across KDM4C (3'-gene) Fusion gene breakpoints across KDM4C (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
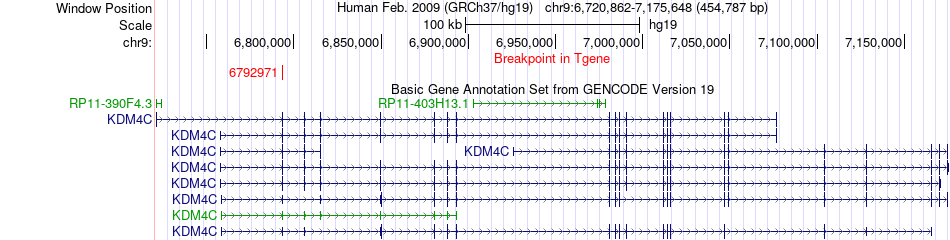 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-6706 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
Top |
Fusion Gene ORF analysis for CD274-KDM4C |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000381573 | ENST00000489243 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-3UTR | ENST00000381577 | ENST00000489243 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381573 | ENST00000381306 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381573 | ENST00000381309 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381573 | ENST00000401787 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381573 | ENST00000442236 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381573 | ENST00000536108 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381573 | ENST00000543771 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381577 | ENST00000381306 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381577 | ENST00000381309 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381577 | ENST00000401787 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381577 | ENST00000442236 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381577 | ENST00000536108 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-5UTR | ENST00000381577 | ENST00000543771 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-intron | ENST00000381573 | ENST00000428870 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| 5CDS-intron | ENST00000381577 | ENST00000428870 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| In-frame | ENST00000381573 | ENST00000535193 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| In-frame | ENST00000381577 | ENST00000535193 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-3CDS | ENST00000498261 | ENST00000535193 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-3UTR | ENST00000498261 | ENST00000489243 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-5UTR | ENST00000498261 | ENST00000381306 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-5UTR | ENST00000498261 | ENST00000381309 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-5UTR | ENST00000498261 | ENST00000401787 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-5UTR | ENST00000498261 | ENST00000442236 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-5UTR | ENST00000498261 | ENST00000536108 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-5UTR | ENST00000498261 | ENST00000543771 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
| intron-intron | ENST00000498261 | ENST00000428870 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000381573 | CD274 | chr9 | 5465606 | + | ENST00000535193 | KDM4C | chr9 | 6792971 | + | 3826 | 556 | 9 | 3014 | 1001 |
| ENST00000381577 | CD274 | chr9 | 5465606 | + | ENST00000535193 | KDM4C | chr9 | 6792971 | + | 4146 | 876 | 86 | 3334 | 1082 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000381573 | ENST00000535193 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + | 0.001786312 | 0.99821377 |
| ENST00000381577 | ENST00000535193 | CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + | 0.001817058 | 0.99818295 |
Top |
Fusion Genomic Features for CD274-KDM4C |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + | 2.80E-10 | 1 |
| CD274 | chr9 | 5465606 | + | KDM4C | chr9 | 6792971 | + | 2.80E-10 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
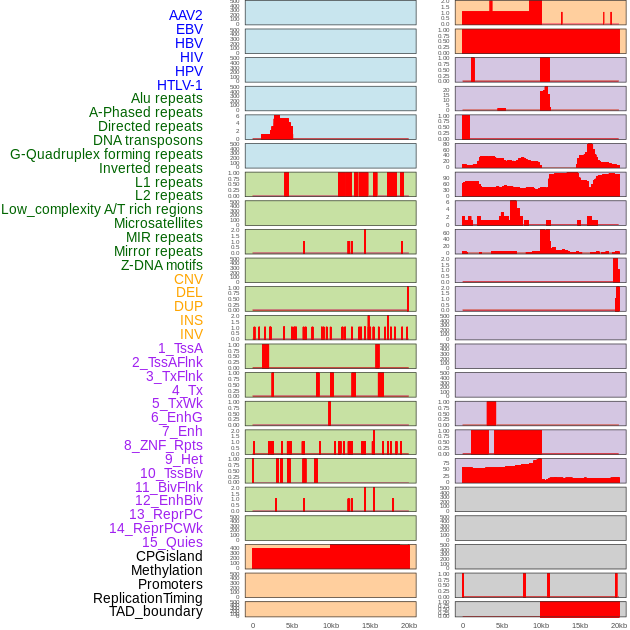 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
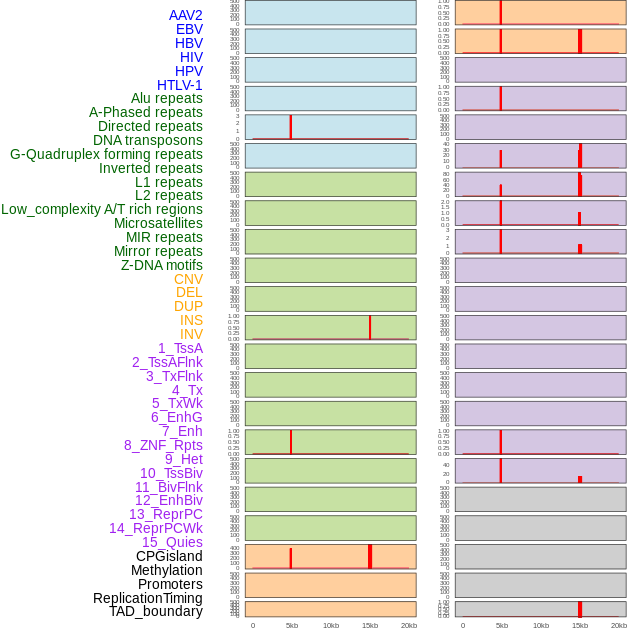 |
Top |
Fusion Protein Features for CD274-KDM4C |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:5465606/chr9:6792971) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381573 | + | 4 | 6 | 19_127 | 149 | 177.0 | Domain | Note=Ig-like V-type |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381577 | + | 5 | 7 | 133_225 | 263 | 291.0 | Domain | Note=Ig-like C2-type |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381577 | + | 5 | 7 | 19_127 | 263 | 291.0 | Domain | Note=Ig-like V-type |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381577 | + | 5 | 7 | 19_238 | 263 | 291.0 | Topological domain | Extracellular |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381577 | + | 5 | 7 | 239_259 | 263 | 291.0 | Transmembrane | Helical |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 144_310 | 0 | 1048.0 | Domain | JmjC | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 16_58 | 0 | 1048.0 | Domain | JmjN | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 877_934 | 0 | 1048.0 | Domain | Note=Tudor 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 935_991 | 0 | 1048.0 | Domain | Note=Tudor 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 144_310 | 0 | 1057.0 | Domain | JmjC | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 16_58 | 0 | 1057.0 | Domain | JmjN | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 877_934 | 0 | 1057.0 | Domain | Note=Tudor 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 935_991 | 0 | 1057.0 | Domain | Note=Tudor 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 144_310 | 16 | 836.0 | Domain | JmjC | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 16_58 | 16 | 836.0 | Domain | JmjN | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 877_934 | 16 | 836.0 | Domain | Note=Tudor 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 935_991 | 16 | 836.0 | Domain | Note=Tudor 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 144_310 | 0 | 814.0 | Domain | JmjC | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 16_58 | 0 | 814.0 | Domain | JmjN | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 877_934 | 0 | 814.0 | Domain | Note=Tudor 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 935_991 | 0 | 814.0 | Domain | Note=Tudor 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 689_747 | 0 | 1048.0 | Zinc finger | Note=PHD-type 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 752_785 | 0 | 1048.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381306 | 0 | 21 | 808_865 | 0 | 1048.0 | Zinc finger | PHD-type 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 689_747 | 0 | 1057.0 | Zinc finger | Note=PHD-type 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 752_785 | 0 | 1057.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000381309 | 0 | 22 | 808_865 | 0 | 1057.0 | Zinc finger | PHD-type 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 689_747 | 16 | 836.0 | Zinc finger | Note=PHD-type 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 752_785 | 16 | 836.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000535193 | 0 | 18 | 808_865 | 16 | 836.0 | Zinc finger | PHD-type 2 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 689_747 | 0 | 814.0 | Zinc finger | Note=PHD-type 1 | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 752_785 | 0 | 814.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KDM4C | chr9:5465606 | chr9:6792971 | ENST00000543771 | 0 | 18 | 808_865 | 0 | 814.0 | Zinc finger | PHD-type 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381573 | + | 4 | 6 | 133_225 | 149 | 177.0 | Domain | Note=Ig-like C2-type |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381573 | + | 4 | 6 | 19_238 | 149 | 177.0 | Topological domain | Extracellular |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381573 | + | 4 | 6 | 260_290 | 149 | 177.0 | Topological domain | Cytoplasmic |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381577 | + | 5 | 7 | 260_290 | 263 | 291.0 | Topological domain | Cytoplasmic |
| Hgene | CD274 | chr9:5465606 | chr9:6792971 | ENST00000381573 | + | 4 | 6 | 239_259 | 149 | 177.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CD274-KDM4C |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >14432_14432_1_CD274-KDM4C_CD274_chr9_5465606_ENST00000381573_KDM4C_chr9_6792971_ENST00000535193_length(transcript)=3826nt_BP=556nt GGCGCAACGCTGAGCAGCTGGCGCGTCCCGCGCGGCCCCAGTTCTGCGCAGCTTCCCGAGGCTCCGCACCAGCCGCGCTTCTGTCCGCCT GCAGGGCATTCCAGAAAGATGAGGATATTTGCTGTCTTTATATTCATGACCTACTGGCATTTGCTGAACGCCCCATACAACAAAATCAAC CAAAGAATTTTGGTTGTGGATCCAGTCACCTCTGAACATGAACTGACATGTCAGGCTGAGGGCTACCCCAAGGCCGAAGTCATCTGGACA AGCAGTGACCATCAAGTCCTGAGTGGTAAGACCACCACCACCAATTCCAAGAGAGAGGAGAAGCTTTTCAATGTGACCAGCACACTGAGA ATCAACACAACAACTAATGAGATTTTCTACTGCACTTTTAGGAGATTAGATCCTGAGGAAAACCATACAGCTGAATTGGTCATCCCAGAA CTACCTCTGGCACATCCTCCAAATGAAAGGACTCACTTGGTAATTCTGGGAGCCATCTTATTATGCCTTGGTGTAGCACTGACATTCATC TTCCGTTTAAGAAAAGACACTGCCCTAACCATCATGGAGGTGGCCGAGGTGGAAAGTCCTCTGAACCCCAGCTGTAAGATAATGACCTTC AGACCCTCCATGGAGGAGTTCCGGGAGTTCAACAAATACCTTGCATACATGGAGTCTAAAGGAGCCCATCGTGCGGGTCTTGCAAAGGTG ATTCCTCCTAAGGAGTGGAAGCCAAGACAGTGCTATGATGACATTGATAATTTGCTCATTCCAGCACCAATTCAGCAGATGGTCACAGGG CAGTCAGGACTGTTCACTCAGTACAACATCCAGAAAAAAGCGATGACTGTGAAGGAGTTCAGGCAGCTGGCCAACAGTGGCAAATATTGT ACTCCAAGATACTTGGATTACGAAGATTTGGAGCGCAAGTACTGGAAGAACTTAACTTTTGTGGCACCTATCTATGGTGCAGATATTAAT GGGAGCATATATGATGAGGGTGTGGATGAATGGAACATAGCTCGCCTCAATACAGTCTTGGATGTGGTTGAAGAAGAGTGTGGCATTTCT ATTGAGGGTGTAAATACCCCATATCTCTATTTTGGCATGTGGAAGACCACGTTTGCATGGCACACCGAAGACATGGACCTCTATAGCATT AATTATCTCCACTTTGGAGAGCCCAAGTCTTGGTATGCTATACCTCCGGAGCATGGAAAACGACTTGAAAGACTAGCTCAAGGTTTTTTC CCAAGCAGCTCCCAAGGGTGTGATGCATTTCTTCGCCACAAGATGACATTGATTTCTCCATCAGTATTGAAGAAATATGGTATTCCCTTT GACAAGATAACCCAGGAGGCTGGAGAATTCATGATCACTTTCCCATATGGCTACCATGCTGGTTTTAATCATGGTTTCAACTGTGCAGAA TCTACAAATTTTGCTACTGTCAGATGGATTGACTATGGAAAAGTTGCCAAATTGTGCACTTGCAGGAAAGACATGGTGAAGATTTCAATG GATATCTTTGTGAGGAAATTTCAGCCAGACAGATATCAGCTTTGGAAACAAGGAAAGGATATATACACCATTGATCACACGAAGCCTACT CCAGCATCCACCCCTGAAGTAAAAGCATGGCTGCAGAGGAGGAGGAAAGTAAGAAAAGCATCCCGAAGCTTCCAGTGTGCTAGGTCTACC TCTAAAAGGCCTAAGGCTGATGAGGAAGAGGAAGTGTCAGATGAAGTCGATGGGGCAGAGGTCCCTAACCCCGACTCAGTCACAGATGAC CTCAAGGTCAGTGAAAAGTCAGAAGCAGCAGTGAAGCTGAGGAACACAGAAGCATCTTCAGAAGAAGAGTCATCTGCTAGCAGGATGCAG GTGGAGCAGAATTTATCAGATCATATCAAACTCTCAGGAAACAGCTGCTTAAGTACATCTGTAACAGAAGACATAAAAACTGAGGATGAC AAAGCTTATGCATATAGAAGTGTACCTTCTATATCCAGTGAGGCTGATGATTCCATTCCATTGTCTAGTGGCTATGAGAAGCCCGAGAAA TCAGACCCATCCGAGCTTTCATGGCCAAAGTCACCTGAGTCATGCTCATCAGTGGCAGAGAGTAATGGTGTGTTAACAGAGGGAGAAGAG AGTGATGTGGAGAGCCATGGGAATGGCCTTGAACCTGGGGAAATCCCAGCGGTCCCCAGTGGAGAGAGAAATAGCTTCAAAGTCCCCAGT ATAGCAGAGGGAGAGAACAAAACCTCTAAGAGTTGGCGCCATCCACTTAGCAGGCCTCCAGCAAGATCTCCGATGACTCTTGTGAAGCAG CAGGCGCCAAGTGATGAAGAATTGCCTGAGGTTCTGTCCATTGAGGAGGAAGTGGAAGAAACAGAGTCTTGGGCGAAACCTCTCATCCAC CTTTGGCAGACGAAGTCCCCTAACTTCGCAGCTGAGCAAGAGTATAATGCAACAGTGGCCAGGATGAAGCCACACTGTGCCATCTGCACT CTGCTCATGCCGTACCACAAGCCAGATAGCAGCAATGAAGAAAATGATGCTAGATGGGAGACAAAATTAGATGAAGTCGTTACATCGGAG GGAAAGACTAAGCCCCTCATACCAGAGATGTGTTTTATTTATAGTGAAGAAAATATAGAATATTCTCCACCCAATGCCTTCCTTGAAGAG GATGGAACAAGTCTCCTTATTTCCTGTGCAAAGTGCTGCGTACGGGTTCATGCAAGTTGTTATGGTATTCCTTCTCATGAGATCTGTGAT GGATGGCTGTGTGCCCGGTGCAAAAGAAATGCGTGGACAGCAGAATGCTGTCTCTGCAATTTGAGAGGAGGTGCTCTTAAGCAAACGAAG AACAATAAGTGGGCCCATGTCATGTGCGCCGTTGCGGTCCCAGAAGTTCGATTCACTAATGTCCCAGAAAGGACACAAATAGATGTAGGC AGAATACCTTTACAGAGGTTAAAATTGGGAAGGCTGGGAATCTAGACACAGCAACAGTAACAACAACAACAATAACAATGAAGGCTCACT GTGAAAACATCAATTCATTAGGAAAGTTACATCCCCTCCGCCACTGTTTCAGATGCTGTTTCTCATTCTCCATGGGAGCAGCCAGCATCG TGTTTAGGATGGGGATACAATAGCCTGAGCTCCTGATTGAGTCAGACCCTGGACTTTCCCTTTTGTATCTGTCATTTTCCTCTGGATCCT CACACCTTTGTGTTTAATTTTTTTAAATTTCATTTGTTGACTTCCTATGTGGGGTAAAATGACTTCCTTTGATTAATGTACTTTCTGTAT CCTAGAGTTTCCCTTTCCCTTTGTGATTTAGTCCAACGTGTCCTACTACAGCCTCAAAAATTGTCATTGGAATCAAAAGCTTTCCAAGGT GCATCAGGTTAATGATTTGGATGCATCCTGAGGTTGTCTCCTGTAGCATGGAGGCATTATAATAGGAGTCTCAGAAGTTACACCATTTTT CTACAGCACATCACAGGCCCAGAGGCTTGAATCTAAGAGAATCCACTCTATGTCTGTCAGCCTGAATCCTTCCTTCTTGCTGCACAACAA AGCAACTGAACGTATTTTGAGTTTAGGTGAAAACTATCACTCATTCATCAAAGTTCTATCTGGGTGATATATGCTATCGAAACAGATGTG GACACTGGGCTGACAGATGTTGAGAACGGACCTGTTGCTGAAGCTCATGTTATCACACATGCTGTGGGAAGAGAGGTCATTGGCATTCAA GATGCAACACCTACAATATGGTGAATTAAAGATGCCATTTGCATGG >14432_14432_1_CD274-KDM4C_CD274_chr9_5465606_ENST00000381573_KDM4C_chr9_6792971_ENST00000535193_length(amino acids)=1001AA_BP=1 MSSWRVPRGPSSAQLPEAPHQPRFCPPAGHSRKMRIFAVFIFMTYWHLLNAPYNKINQRILVVDPVTSEHELTCQAEGYPKAEVIWTSSD HQVLSGKTTTTNSKREEKLFNVTSTLRINTTTNEIFYCTFRRLDPEENHTAELVIPELPLAHPPNERTHLVILGAILLCLGVALTFIFRL RKDTALTIMEVAEVESPLNPSCKIMTFRPSMEEFREFNKYLAYMESKGAHRAGLAKVIPPKEWKPRQCYDDIDNLLIPAPIQQMVTGQSG LFTQYNIQKKAMTVKEFRQLANSGKYCTPRYLDYEDLERKYWKNLTFVAPIYGADINGSIYDEGVDEWNIARLNTVLDVVEEECGISIEG VNTPYLYFGMWKTTFAWHTEDMDLYSINYLHFGEPKSWYAIPPEHGKRLERLAQGFFPSSSQGCDAFLRHKMTLISPSVLKKYGIPFDKI TQEAGEFMITFPYGYHAGFNHGFNCAESTNFATVRWIDYGKVAKLCTCRKDMVKISMDIFVRKFQPDRYQLWKQGKDIYTIDHTKPTPAS TPEVKAWLQRRRKVRKASRSFQCARSTSKRPKADEEEEVSDEVDGAEVPNPDSVTDDLKVSEKSEAAVKLRNTEASSEEESSASRMQVEQ NLSDHIKLSGNSCLSTSVTEDIKTEDDKAYAYRSVPSISSEADDSIPLSSGYEKPEKSDPSELSWPKSPESCSSVAESNGVLTEGEESDV ESHGNGLEPGEIPAVPSGERNSFKVPSIAEGENKTSKSWRHPLSRPPARSPMTLVKQQAPSDEELPEVLSIEEEVEETESWAKPLIHLWQ TKSPNFAAEQEYNATVARMKPHCAICTLLMPYHKPDSSNEENDARWETKLDEVVTSEGKTKPLIPEMCFIYSEENIEYSPPNAFLEEDGT SLLISCAKCCVRVHASCYGIPSHEICDGWLCARCKRNAWTAECCLCNLRGGALKQTKNNKWAHVMCAVAVPEVRFTNVPERTQIDVGRIP LQRLKLGRLGI -------------------------------------------------------------- >14432_14432_2_CD274-KDM4C_CD274_chr9_5465606_ENST00000381577_KDM4C_chr9_6792971_ENST00000535193_length(transcript)=4146nt_BP=876nt GCGTCCCGCGCGGCCCCAGTTCTGCGCAGCTTCCCGAGGCTCCGCACCAGCCGCGCTTCTGTCCGCCTGCAGGGCATTCCAGAAAGATGA GGATATTTGCTGTCTTTATATTCATGACCTACTGGCATTTGCTGAACGCATTTACTGTCACGGTTCCCAAGGACCTATATGTGGTAGAGT ATGGTAGCAATATGACAATTGAATGCAAATTCCCAGTAGAAAAACAATTAGACCTGGCTGCACTAATTGTCTATTGGGAAATGGAGGATA AGAACATTATTCAATTTGTGCATGGAGAGGAAGACCTGAAGGTTCAGCATAGTAGCTACAGACAGAGGGCCCGGCTGTTGAAGGACCAGC TCTCCCTGGGAAATGCTGCACTTCAGATCACAGATGTGAAATTGCAGGATGCAGGGGTGTACCGCTGCATGATCAGCTATGGTGGTGCCG ACTACAAGCGAATTACTGTGAAAGTCAATGCCCCATACAACAAAATCAACCAAAGAATTTTGGTTGTGGATCCAGTCACCTCTGAACATG AACTGACATGTCAGGCTGAGGGCTACCCCAAGGCCGAAGTCATCTGGACAAGCAGTGACCATCAAGTCCTGAGTGGTAAGACCACCACCA CCAATTCCAAGAGAGAGGAGAAGCTTTTCAATGTGACCAGCACACTGAGAATCAACACAACAACTAATGAGATTTTCTACTGCACTTTTA GGAGATTAGATCCTGAGGAAAACCATACAGCTGAATTGGTCATCCCAGAACTACCTCTGGCACATCCTCCAAATGAAAGGACTCACTTGG TAATTCTGGGAGCCATCTTATTATGCCTTGGTGTAGCACTGACATTCATCTTCCGTTTAAGAAAAGACACTGCCCTAACCATCATGGAGG TGGCCGAGGTGGAAAGTCCTCTGAACCCCAGCTGTAAGATAATGACCTTCAGACCCTCCATGGAGGAGTTCCGGGAGTTCAACAAATACC TTGCATACATGGAGTCTAAAGGAGCCCATCGTGCGGGTCTTGCAAAGGTGATTCCTCCTAAGGAGTGGAAGCCAAGACAGTGCTATGATG ACATTGATAATTTGCTCATTCCAGCACCAATTCAGCAGATGGTCACAGGGCAGTCAGGACTGTTCACTCAGTACAACATCCAGAAAAAAG CGATGACTGTGAAGGAGTTCAGGCAGCTGGCCAACAGTGGCAAATATTGTACTCCAAGATACTTGGATTACGAAGATTTGGAGCGCAAGT ACTGGAAGAACTTAACTTTTGTGGCACCTATCTATGGTGCAGATATTAATGGGAGCATATATGATGAGGGTGTGGATGAATGGAACATAG CTCGCCTCAATACAGTCTTGGATGTGGTTGAAGAAGAGTGTGGCATTTCTATTGAGGGTGTAAATACCCCATATCTCTATTTTGGCATGT GGAAGACCACGTTTGCATGGCACACCGAAGACATGGACCTCTATAGCATTAATTATCTCCACTTTGGAGAGCCCAAGTCTTGGTATGCTA TACCTCCGGAGCATGGAAAACGACTTGAAAGACTAGCTCAAGGTTTTTTCCCAAGCAGCTCCCAAGGGTGTGATGCATTTCTTCGCCACA AGATGACATTGATTTCTCCATCAGTATTGAAGAAATATGGTATTCCCTTTGACAAGATAACCCAGGAGGCTGGAGAATTCATGATCACTT TCCCATATGGCTACCATGCTGGTTTTAATCATGGTTTCAACTGTGCAGAATCTACAAATTTTGCTACTGTCAGATGGATTGACTATGGAA AAGTTGCCAAATTGTGCACTTGCAGGAAAGACATGGTGAAGATTTCAATGGATATCTTTGTGAGGAAATTTCAGCCAGACAGATATCAGC TTTGGAAACAAGGAAAGGATATATACACCATTGATCACACGAAGCCTACTCCAGCATCCACCCCTGAAGTAAAAGCATGGCTGCAGAGGA GGAGGAAAGTAAGAAAAGCATCCCGAAGCTTCCAGTGTGCTAGGTCTACCTCTAAAAGGCCTAAGGCTGATGAGGAAGAGGAAGTGTCAG ATGAAGTCGATGGGGCAGAGGTCCCTAACCCCGACTCAGTCACAGATGACCTCAAGGTCAGTGAAAAGTCAGAAGCAGCAGTGAAGCTGA GGAACACAGAAGCATCTTCAGAAGAAGAGTCATCTGCTAGCAGGATGCAGGTGGAGCAGAATTTATCAGATCATATCAAACTCTCAGGAA ACAGCTGCTTAAGTACATCTGTAACAGAAGACATAAAAACTGAGGATGACAAAGCTTATGCATATAGAAGTGTACCTTCTATATCCAGTG AGGCTGATGATTCCATTCCATTGTCTAGTGGCTATGAGAAGCCCGAGAAATCAGACCCATCCGAGCTTTCATGGCCAAAGTCACCTGAGT CATGCTCATCAGTGGCAGAGAGTAATGGTGTGTTAACAGAGGGAGAAGAGAGTGATGTGGAGAGCCATGGGAATGGCCTTGAACCTGGGG AAATCCCAGCGGTCCCCAGTGGAGAGAGAAATAGCTTCAAAGTCCCCAGTATAGCAGAGGGAGAGAACAAAACCTCTAAGAGTTGGCGCC ATCCACTTAGCAGGCCTCCAGCAAGATCTCCGATGACTCTTGTGAAGCAGCAGGCGCCAAGTGATGAAGAATTGCCTGAGGTTCTGTCCA TTGAGGAGGAAGTGGAAGAAACAGAGTCTTGGGCGAAACCTCTCATCCACCTTTGGCAGACGAAGTCCCCTAACTTCGCAGCTGAGCAAG AGTATAATGCAACAGTGGCCAGGATGAAGCCACACTGTGCCATCTGCACTCTGCTCATGCCGTACCACAAGCCAGATAGCAGCAATGAAG AAAATGATGCTAGATGGGAGACAAAATTAGATGAAGTCGTTACATCGGAGGGAAAGACTAAGCCCCTCATACCAGAGATGTGTTTTATTT ATAGTGAAGAAAATATAGAATATTCTCCACCCAATGCCTTCCTTGAAGAGGATGGAACAAGTCTCCTTATTTCCTGTGCAAAGTGCTGCG TACGGGTTCATGCAAGTTGTTATGGTATTCCTTCTCATGAGATCTGTGATGGATGGCTGTGTGCCCGGTGCAAAAGAAATGCGTGGACAG CAGAATGCTGTCTCTGCAATTTGAGAGGAGGTGCTCTTAAGCAAACGAAGAACAATAAGTGGGCCCATGTCATGTGCGCCGTTGCGGTCC CAGAAGTTCGATTCACTAATGTCCCAGAAAGGACACAAATAGATGTAGGCAGAATACCTTTACAGAGGTTAAAATTGGGAAGGCTGGGAA TCTAGACACAGCAACAGTAACAACAACAACAATAACAATGAAGGCTCACTGTGAAAACATCAATTCATTAGGAAAGTTACATCCCCTCCG CCACTGTTTCAGATGCTGTTTCTCATTCTCCATGGGAGCAGCCAGCATCGTGTTTAGGATGGGGATACAATAGCCTGAGCTCCTGATTGA GTCAGACCCTGGACTTTCCCTTTTGTATCTGTCATTTTCCTCTGGATCCTCACACCTTTGTGTTTAATTTTTTTAAATTTCATTTGTTGA CTTCCTATGTGGGGTAAAATGACTTCCTTTGATTAATGTACTTTCTGTATCCTAGAGTTTCCCTTTCCCTTTGTGATTTAGTCCAACGTG TCCTACTACAGCCTCAAAAATTGTCATTGGAATCAAAAGCTTTCCAAGGTGCATCAGGTTAATGATTTGGATGCATCCTGAGGTTGTCTC CTGTAGCATGGAGGCATTATAATAGGAGTCTCAGAAGTTACACCATTTTTCTACAGCACATCACAGGCCCAGAGGCTTGAATCTAAGAGA ATCCACTCTATGTCTGTCAGCCTGAATCCTTCCTTCTTGCTGCACAACAAAGCAACTGAACGTATTTTGAGTTTAGGTGAAAACTATCAC TCATTCATCAAAGTTCTATCTGGGTGATATATGCTATCGAAACAGATGTGGACACTGGGCTGACAGATGTTGAGAACGGACCTGTTGCTG AAGCTCATGTTATCACACATGCTGTGGGAAGAGAGGTCATTGGCATTCAAGATGCAACACCTACAATATGGTGAATTAAAGATGCCATTT GCATGG >14432_14432_2_CD274-KDM4C_CD274_chr9_5465606_ENST00000381577_KDM4C_chr9_6792971_ENST00000535193_length(amino acids)=1082AA_BP=0 MRIFAVFIFMTYWHLLNAFTVTVPKDLYVVEYGSNMTIECKFPVEKQLDLAALIVYWEMEDKNIIQFVHGEEDLKVQHSSYRQRARLLKD QLSLGNAALQITDVKLQDAGVYRCMISYGGADYKRITVKVNAPYNKINQRILVVDPVTSEHELTCQAEGYPKAEVIWTSSDHQVLSGKTT TTNSKREEKLFNVTSTLRINTTTNEIFYCTFRRLDPEENHTAELVIPELPLAHPPNERTHLVILGAILLCLGVALTFIFRLRKDTALTIM EVAEVESPLNPSCKIMTFRPSMEEFREFNKYLAYMESKGAHRAGLAKVIPPKEWKPRQCYDDIDNLLIPAPIQQMVTGQSGLFTQYNIQK KAMTVKEFRQLANSGKYCTPRYLDYEDLERKYWKNLTFVAPIYGADINGSIYDEGVDEWNIARLNTVLDVVEEECGISIEGVNTPYLYFG MWKTTFAWHTEDMDLYSINYLHFGEPKSWYAIPPEHGKRLERLAQGFFPSSSQGCDAFLRHKMTLISPSVLKKYGIPFDKITQEAGEFMI TFPYGYHAGFNHGFNCAESTNFATVRWIDYGKVAKLCTCRKDMVKISMDIFVRKFQPDRYQLWKQGKDIYTIDHTKPTPASTPEVKAWLQ RRRKVRKASRSFQCARSTSKRPKADEEEEVSDEVDGAEVPNPDSVTDDLKVSEKSEAAVKLRNTEASSEEESSASRMQVEQNLSDHIKLS GNSCLSTSVTEDIKTEDDKAYAYRSVPSISSEADDSIPLSSGYEKPEKSDPSELSWPKSPESCSSVAESNGVLTEGEESDVESHGNGLEP GEIPAVPSGERNSFKVPSIAEGENKTSKSWRHPLSRPPARSPMTLVKQQAPSDEELPEVLSIEEEVEETESWAKPLIHLWQTKSPNFAAE QEYNATVARMKPHCAICTLLMPYHKPDSSNEENDARWETKLDEVVTSEGKTKPLIPEMCFIYSEENIEYSPPNAFLEEDGTSLLISCAKC CVRVHASCYGIPSHEICDGWLCARCKRNAWTAECCLCNLRGGALKQTKNNKWAHVMCAVAVPEVRFTNVPERTQIDVGRIPLQRLKLGRL GI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CD274-KDM4C |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CD274-KDM4C |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CD274-KDM4C |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CD274 | C0020517 | Hypersensitivity | 2 | CTD_human |
| Hgene | CD274 | C1527304 | Allergic Reaction | 2 | CTD_human |
| Hgene | CD274 | C0013146 | Drug abuse | 1 | CTD_human |
| Hgene | CD274 | C0013170 | Drug habituation | 1 | CTD_human |
| Hgene | CD274 | C0013222 | Drug Use Disorders | 1 | CTD_human |
| Hgene | CD274 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Hgene | CD274 | C0024299 | Lymphoma | 1 | CTD_human |
| Hgene | CD274 | C0027666 | Neoplasms, Radiation-Induced | 1 | CTD_human |
| Hgene | CD274 | C0029231 | Organic Mental Disorders, Substance-Induced | 1 | CTD_human |
| Hgene | CD274 | C0038580 | Substance Dependence | 1 | CTD_human |
| Hgene | CD274 | C0038586 | Substance Use Disorders | 1 | CTD_human |
| Hgene | CD274 | C0235874 | Disease Exacerbation | 1 | CTD_human |
| Hgene | CD274 | C0236969 | Substance-Related Disorders | 1 | CTD_human |
| Hgene | CD274 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Hgene | CD274 | C0740858 | Substance abuse problem | 1 | CTD_human |
| Hgene | CD274 | C0751366 | Radiation-Induced Cancer | 1 | CTD_human |
| Hgene | CD274 | C0751552 | Malignant neoplasm of thymus | 1 | CTD_human |
| Hgene | CD274 | C1510472 | Drug Dependence | 1 | CTD_human |
| Hgene | CD274 | C2239176 | Liver carcinoma | 1 | CTD_human |
| Hgene | CD274 | C3714644 | Thymus Neoplasms | 1 | CTD_human |
| Hgene | CD274 | C4316881 | Prescription Drug Abuse | 1 | CTD_human |
| Tgene | C0004565 | Melanoma, B16 | 1 | CTD_human | |
| Tgene | C0009075 | Melanoma, Cloudman S91 | 1 | CTD_human | |
| Tgene | C0018598 | Melanoma, Harding-Passey | 1 | CTD_human | |
| Tgene | C0025149 | Medulloblastoma | 1 | CTD_human | |
| Tgene | C0025202 | melanoma | 1 | CTD_human | |
| Tgene | C0025205 | Melanoma, Experimental | 1 | CTD_human | |
| Tgene | C0079772 | T-Cell Lymphoma | 1 | CTD_human | |
| Tgene | C0205833 | Medullomyoblastoma | 1 | CTD_human | |
| Tgene | C0236663 | Alcohol withdrawal syndrome | 1 | PSYGENET | |
| Tgene | C0278510 | Childhood Medulloblastoma | 1 | CTD_human | |
| Tgene | C0278876 | Adult Medulloblastoma | 1 | CTD_human | |
| Tgene | C0751291 | Desmoplastic Medulloblastoma | 1 | CTD_human | |
| Tgene | C1275668 | Melanotic medulloblastoma | 1 | CTD_human |
