
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AGFG1-SND1 (FusionGDB2 ID:HG3267TG27044) |
Fusion Gene Summary for AGFG1-SND1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AGFG1-SND1 | Fusion gene ID: hg3267tg27044 | Hgene | Tgene | Gene symbol | AGFG1 | SND1 | Gene ID | 3267 | 27044 |
| Gene name | ArfGAP with FG repeats 1 | staphylococcal nuclease and tudor domain containing 1 | |
| Synonyms | HRB|RAB|RIP | TDRD11|Tudor-SN|p100 | |
| Cytomap | ('AGFG1')('SND1') 2q36.3 | 7q32.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | arf-GAP domain and FG repeat-containing protein 1HIV-1 Rev-binding proteinRab, Rev/Rex activation domain-binding proteinarf-GAP domain and FG repeats-containing protein 1hRIP, Rev interacting proteinnucleoporin-like protein RIPrev-interacting protei | staphylococcal nuclease domain-containing protein 1EBNA2 coactivator p100testis tissue sperm-binding protein Li 82Ptudor domain-containing protein 11 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q7KZF4 | |
| Ensembl transtripts involved in fusion gene | ENST00000310078, ENST00000373671, ENST00000409171, ENST00000409315, ENST00000409979, ENST00000486932, | ||
| Fusion gene scores | * DoF score | 13 X 9 X 8=936 | 29 X 24 X 11=7656 |
| # samples | 15 | 33 | |
| ** MAII score | log2(15/936*10)=-2.64154602908752 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(33/7656*10)=-4.53605290024021 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AGFG1 [Title/Abstract] AND SND1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AGFG1(228356356)-SND1(127484377), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | AGFG1-SND1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGFG1-SND1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGFG1-SND1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. AGFG1-SND1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | SND1 | GO:0010587 | miRNA catabolic process | 28546213 |
 Fusion gene breakpoints across AGFG1 (5'-gene) Fusion gene breakpoints across AGFG1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
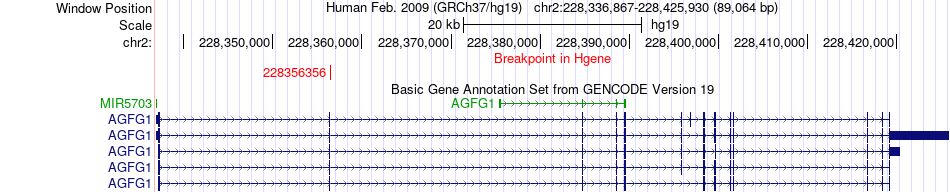 |
 Fusion gene breakpoints across SND1 (3'-gene) Fusion gene breakpoints across SND1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
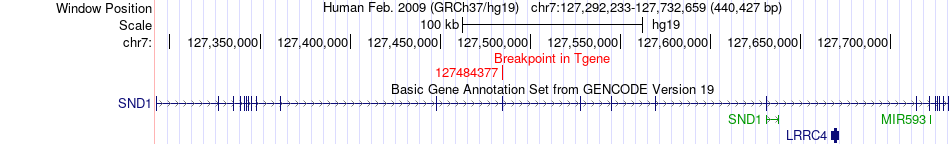 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-D7-8576-01A | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
Top |
Fusion Gene ORF analysis for AGFG1-SND1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000310078 | ENST00000467238 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| 5CDS-intron | ENST00000373671 | ENST00000467238 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| 5CDS-intron | ENST00000409171 | ENST00000467238 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| 5CDS-intron | ENST00000409315 | ENST00000467238 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| 5CDS-intron | ENST00000409979 | ENST00000467238 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| In-frame | ENST00000310078 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| In-frame | ENST00000373671 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| In-frame | ENST00000409171 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| In-frame | ENST00000409315 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| In-frame | ENST00000409979 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| intron-3CDS | ENST00000486932 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
| intron-intron | ENST00000486932 | ENST00000467238 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409979 | AGFG1 | chr2 | 228356356 | + | ENST00000354725 | SND1 | chr7 | 127484377 | + | 2571 | 531 | 45 | 2021 | 658 |
| ENST00000310078 | AGFG1 | chr2 | 228356356 | + | ENST00000354725 | SND1 | chr7 | 127484377 | + | 2561 | 521 | 35 | 2011 | 658 |
| ENST00000409315 | AGFG1 | chr2 | 228356356 | + | ENST00000354725 | SND1 | chr7 | 127484377 | + | 2354 | 314 | 23 | 1804 | 593 |
| ENST00000373671 | AGFG1 | chr2 | 228356356 | + | ENST00000354725 | SND1 | chr7 | 127484377 | + | 2351 | 311 | 20 | 1801 | 593 |
| ENST00000409171 | AGFG1 | chr2 | 228356356 | + | ENST00000354725 | SND1 | chr7 | 127484377 | + | 2351 | 311 | 20 | 1801 | 593 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409979 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + | 0.004163258 | 0.99583673 |
| ENST00000310078 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + | 0.004091817 | 0.99590814 |
| ENST00000409315 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + | 0.005074028 | 0.994926 |
| ENST00000373671 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + | 0.004929214 | 0.9950708 |
| ENST00000409171 | ENST00000354725 | AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484377 | + | 0.004929214 | 0.9950708 |
Top |
Fusion Genomic Features for AGFG1-SND1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484376 | + | 1.35E-05 | 0.9999865 |
| AGFG1 | chr2 | 228356356 | + | SND1 | chr7 | 127484376 | + | 1.35E-05 | 0.9999865 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
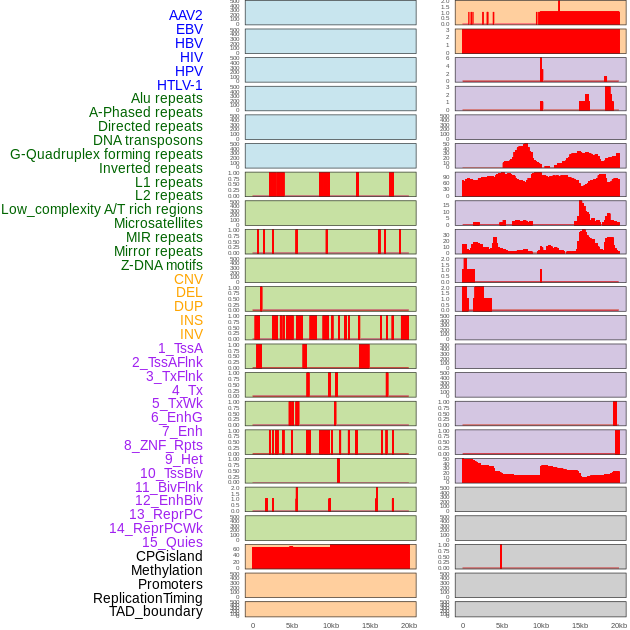 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
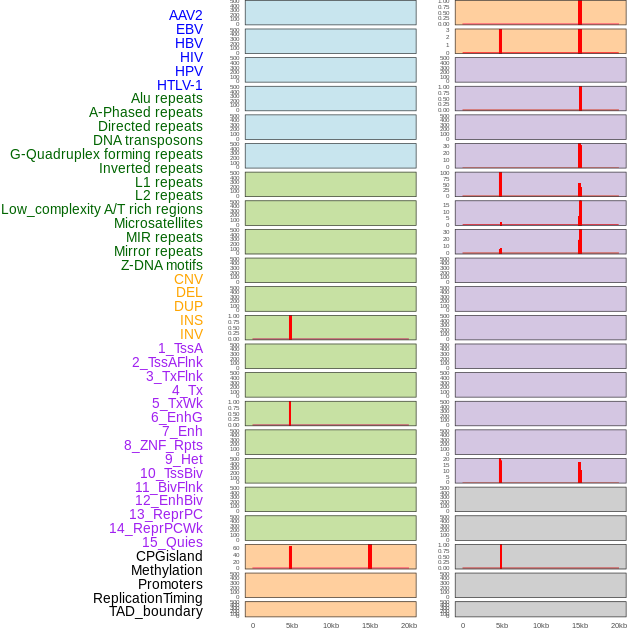 |
Top |
Fusion Protein Features for AGFG1-SND1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:228356356/chr7:127484377) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | SND1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Endonuclease that mediates miRNA decay of both protein-free and AGO2-loaded miRNAs (PubMed:28546213, PubMed:18453631). As part of its function in miRNA decay, regulates mRNAs involved in G1-to-S phase transition (PubMed:28546213). Functions as a bridging factor between STAT6 and the basal transcription factor (PubMed:12234934). Plays a role in PIM1 regulation of MYB activity (PubMed:9809063). Functions as a transcriptional coactivator for STAT5 (By similarity). {ECO:0000250|UniProtKB:Q78PY7, ECO:0000269|PubMed:12234934, ECO:0000269|PubMed:18453631, ECO:0000269|PubMed:28546213, ECO:0000269|PubMed:9809063}.; FUNCTION: (Microbial infection) Functions as a transcriptional coactivator for the Epstein-Barr virus nuclear antigen 2 (EBNA2). {ECO:0000269|PubMed:7651391}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGFG1 | chr2:228356356 | chr7:127484377 | ENST00000310078 | + | 2 | 13 | 29_52 | 87 | 563.0 | Zinc finger | C4-type |
| Hgene | AGFG1 | chr2:228356356 | chr7:127484377 | ENST00000373671 | + | 2 | 12 | 29_52 | 87 | 523.0 | Zinc finger | C4-type |
| Hgene | AGFG1 | chr2:228356356 | chr7:127484377 | ENST00000409171 | + | 2 | 13 | 29_52 | 87 | 561.0 | Zinc finger | C4-type |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 525_660 | 414 | 911.0 | Domain | TNase-like 4 | |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 729_787 | 414 | 911.0 | Domain | Tudor |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGFG1 | chr2:228356356 | chr7:127484377 | ENST00000310078 | + | 2 | 13 | 11_135 | 87 | 563.0 | Domain | Arf-GAP |
| Hgene | AGFG1 | chr2:228356356 | chr7:127484377 | ENST00000373671 | + | 2 | 12 | 11_135 | 87 | 523.0 | Domain | Arf-GAP |
| Hgene | AGFG1 | chr2:228356356 | chr7:127484377 | ENST00000409171 | + | 2 | 13 | 11_135 | 87 | 561.0 | Domain | Arf-GAP |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 18_166 | 414 | 911.0 | Domain | TNase-like 1 | |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 193_328 | 414 | 911.0 | Domain | TNase-like 2 | |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 341_496 | 414 | 911.0 | Domain | TNase-like 3 | |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 321_325 | 414 | 911.0 | Motif | Nuclear localization signal | |
| Tgene | SND1 | chr2:228356356 | chr7:127484377 | ENST00000354725 | 10 | 24 | 388_392 | 414 | 911.0 | Motif | Nuclear localization signal |
Top |
Fusion Gene Sequence for AGFG1-SND1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2873_2873_1_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000310078_SND1_chr7_127484377_ENST00000354725_length(transcript)=2561nt_BP=521nt AGGAGAAGTCGGGAAGGTGGCGGCGGCGGCGGCGGTTGTCCCGGCTGTGCCGGTTGGTGTGGCCCGTCAGCCCGCGTACCACAGCGCCCG GGCCGCGTCGAGCCCAGTACAGCCAAGCCGCTGCGGCCGGGTCCGGCGCGGGCGGCGCGCGCAGACGGAGGGCGGCGGCCGCGGCCAGGG CGGCCCGTGGGACCGCGGGCCCCCGGCGCAGCGCTGCCCGGCTCCCGGCCCTGCCGGCCTCCTCCCTTGGCGCCGCGGCCATGGCGGCCA GCGCGAAGCGGAAGCAGGAGGAGAAGCACCTGAAGATGCTGCGGGACATGACCGGCCTCCCGCACAACCGAAAGTGCTTCGACTGCGACC AGCGCGGCCCCACCTACGTTAACATGACGGTCGGCTCCTTCGTGTGTACCTCCTGCTCCGGCAGCCTGCGAGGATTAAATCCACCACACA GGGTGAAATCTATCTCCATGACAACATTCACACAACAGGAAATTGAATTCTTACAAAAACATGGAAATGAAGTCAATGTGACGGTGGACT ACATTAGACCAGCCAGCCCAGCCACAGAGACAGTGCCTGCCTTTTCAGAGCGTACCTGTGCCACTGTCACCATTGGAGGAATAAACATTG CTGAGGCTCTTGTCAGCAAAGGTCTAGCCACAGTGATCAGATACCGGCAGGATGATGACCAGAGATCATCACACTACGATGAACTGCTTG CTGCAGAGGCCAGAGCTATTAAGAATGGCAAAGGATTGCATAGCAAGAAGGAAGTGCCTATCCACCGTGTTGCAGATATATCTGGGGATA CCCAAAAAGCAAAGCAGTTCCTGCCTTTTCTTCAGCGGGCAGGTCGTTCTGAAGCTGTGGTGGAATACGTCTTCAGTGGTTCTCGTCTCA AACTCTATTTGCCAAAGGAAACTTGCCTTATCACCTTCTTGCTTGCAGGCATTGAATGCCCCAGAGGAGCCCGAAACCTCCCAGGCTTGG TGCAGGAAGGAGAGCCCTTCAGCGAGGAAGCTACACTTTTCACCAAGGAACTGGTGCTGCAGCGAGAGGTGGAGGTGGAGGTGGAGAGCA TGGACAAGGCCGGCAACTTTATCGGCTGGCTGCACATCGACGGTGCCAACCTGTCCGTCCTGCTGGTGGAGCACGCGCTCTCCAAGGTCC ACTTCACCGCCGAACGCAGCTCCTACTACAAGTCCCTGCTGTCTGCCGAGGAGGCCGCAAAGCAGAAGAAAGAGAAGGTCTGGGCCCACT ATGAGGAGCAGCCCGTGGAGGAGGTGATGCCAGTGCTGGAGGAGAAGGAGCGATCTGCTAGCTACAAGCCCGTGTTTGTGACTGAGATCA CTGATGACCTGCACTTCTACGTGCAGGATGTGGAGACCGGCACCCAGTTGGAGAAGCTGATGGAGAACATGCGCAATGACATTGCCAGTC ACCCCCCTGTAGAGGGCTCCTATGCCCCCCGCAGGGGAGAGTTCTGCATTGCCAAATTTGTAGATGGAGAATGGTACCGTGCCCGAGTAG AGAAAGTCGAGTCTCCTGCCAAAATACATGTCTTCTACATTGACTACGGCAACAGAGAGGTCCTGCCATCCACCCGCCTGGGTACCCTAT CACCTGCCTTCAGCACTCGGGTGCTGCCAGCTCAAGCCACGGAGTATGCCTTCGCCTTCATCCAGGTGCCCCAAGATGATGATGCCCGCA CGGACGCCGTGGACAGCGTAGTTCGGGATATCCAGAACACTCAGTGCCTGCTCAACGTGGAACACCTGAGTGCCGGCTGCCCCCATGTCA CCCTGCAGTTTGCAGATTCCAAGGGCGATGTGGGGCTGGGCTTGGTGAAGGAAGGGCTGGTCATGGTGGAGGTGCGCAAGGAGAAACAGT TCCAGAAAGTGATCACAGAATACCTGAATGCCCAAGAGTCAGCCAAGAGCGCCAGGCTGAACCTGTGGCGCTATGGAGACTTTCGAGCTG ATGATGCAGACGAATTTGGCTACAGCCGCTAAGGAGGGGATCGGGTTTGGCCCCCAGCCCCGCTCACGCCAGTCCCTCTTCCTCTGCCGG GAGGGTGTTTTCAACTCCAAACCCCAGAGAGGGGTTGTAGATTGGGTCCAGCTTTGCTTCAGTGTGTGGAAATGTCTCGTGGGGTGGCAT CGGGGCTGCGGGGTGGGGACCCCAAGGCTTTCTGGGGCAGACCCTTGTCCTCTGGGATGATGGGCACTGCTATCCACAGTCTCTGCCAGT TGGTTTTATTTGGAGGTTTGTGGGCTTTTTTTAAAAAAAAAAAGTCCTCAAATCAGGAAGAAACATCAAAGACTATGTCCTAGTGGAGGG AGTAATCCTAACACCCAGGCTGGCCGCCAGCTGGCACCTGCCTCTATCCCAGACTGCCCTCGTCCCAGCTCTCTGTCCAACTGTTGATTA TGTGATTTTTCTGATACGTCCATTCTCAAATGCCAGTGTGTTCACATCTTCGCTCTGGCCAGCCCATTCTGTATTTAAAGCTTTTGAGGC CCAATAAAATAGTACGTGCTGTCTGCAGCCCTTATTGATCA >2873_2873_1_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000310078_SND1_chr7_127484377_ENST00000354725_length(amino acids)=658AA_BP=162 MSRLCRLVWPVSPRTTAPGPRRAQYSQAAAAGSGAGGARRRRAAAAARAARGTAGPRRSAARLPALPASSLGAAAMAASAKRKQEEKHLK MLRDMTGLPHNRKCFDCDQRGPTYVNMTVGSFVCTSCSGSLRGLNPPHRVKSISMTTFTQQEIEFLQKHGNEVNVTVDYIRPASPATETV PAFSERTCATVTIGGINIAEALVSKGLATVIRYRQDDDQRSSHYDELLAAEARAIKNGKGLHSKKEVPIHRVADISGDTQKAKQFLPFLQ RAGRSEAVVEYVFSGSRLKLYLPKETCLITFLLAGIECPRGARNLPGLVQEGEPFSEEATLFTKELVLQREVEVEVESMDKAGNFIGWLH IDGANLSVLLVEHALSKVHFTAERSSYYKSLLSAEEAAKQKKEKVWAHYEEQPVEEVMPVLEEKERSASYKPVFVTEITDDLHFYVQDVE TGTQLEKLMENMRNDIASHPPVEGSYAPRRGEFCIAKFVDGEWYRARVEKVESPAKIHVFYIDYGNREVLPSTRLGTLSPAFSTRVLPAQ ATEYAFAFIQVPQDDDARTDAVDSVVRDIQNTQCLLNVEHLSAGCPHVTLQFADSKGDVGLGLVKEGLVMVEVRKEKQFQKVITEYLNAQ ESAKSARLNLWRYGDFRADDADEFGYSR -------------------------------------------------------------- >2873_2873_2_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000373671_SND1_chr7_127484377_ENST00000354725_length(transcript)=2351nt_BP=311nt GCGCTGCCCGGCTCCCGGCCCTGCCGGCCTCCTCCCTTGGCGCCGCGGCCATGGCGGCCAGCGCGAAGCGGAAGCAGGAGGAGAAGCACC TGAAGATGCTGCGGGACATGACCGGCCTCCCGCACAACCGAAAGTGCTTCGACTGCGACCAGCGCGGCCCCACCTACGTTAACATGACGG TCGGCTCCTTCGTGTGTACCTCCTGCTCCGGCAGCCTGCGAGGATTAAATCCACCACACAGGGTGAAATCTATCTCCATGACAACATTCA CACAACAGGAAATTGAATTCTTACAAAAACATGGAAATGAAGTCAATGTGACGGTGGACTACATTAGACCAGCCAGCCCAGCCACAGAGA CAGTGCCTGCCTTTTCAGAGCGTACCTGTGCCACTGTCACCATTGGAGGAATAAACATTGCTGAGGCTCTTGTCAGCAAAGGTCTAGCCA CAGTGATCAGATACCGGCAGGATGATGACCAGAGATCATCACACTACGATGAACTGCTTGCTGCAGAGGCCAGAGCTATTAAGAATGGCA AAGGATTGCATAGCAAGAAGGAAGTGCCTATCCACCGTGTTGCAGATATATCTGGGGATACCCAAAAAGCAAAGCAGTTCCTGCCTTTTC TTCAGCGGGCAGGTCGTTCTGAAGCTGTGGTGGAATACGTCTTCAGTGGTTCTCGTCTCAAACTCTATTTGCCAAAGGAAACTTGCCTTA TCACCTTCTTGCTTGCAGGCATTGAATGCCCCAGAGGAGCCCGAAACCTCCCAGGCTTGGTGCAGGAAGGAGAGCCCTTCAGCGAGGAAG CTACACTTTTCACCAAGGAACTGGTGCTGCAGCGAGAGGTGGAGGTGGAGGTGGAGAGCATGGACAAGGCCGGCAACTTTATCGGCTGGC TGCACATCGACGGTGCCAACCTGTCCGTCCTGCTGGTGGAGCACGCGCTCTCCAAGGTCCACTTCACCGCCGAACGCAGCTCCTACTACA AGTCCCTGCTGTCTGCCGAGGAGGCCGCAAAGCAGAAGAAAGAGAAGGTCTGGGCCCACTATGAGGAGCAGCCCGTGGAGGAGGTGATGC CAGTGCTGGAGGAGAAGGAGCGATCTGCTAGCTACAAGCCCGTGTTTGTGACTGAGATCACTGATGACCTGCACTTCTACGTGCAGGATG TGGAGACCGGCACCCAGTTGGAGAAGCTGATGGAGAACATGCGCAATGACATTGCCAGTCACCCCCCTGTAGAGGGCTCCTATGCCCCCC GCAGGGGAGAGTTCTGCATTGCCAAATTTGTAGATGGAGAATGGTACCGTGCCCGAGTAGAGAAAGTCGAGTCTCCTGCCAAAATACATG TCTTCTACATTGACTACGGCAACAGAGAGGTCCTGCCATCCACCCGCCTGGGTACCCTATCACCTGCCTTCAGCACTCGGGTGCTGCCAG CTCAAGCCACGGAGTATGCCTTCGCCTTCATCCAGGTGCCCCAAGATGATGATGCCCGCACGGACGCCGTGGACAGCGTAGTTCGGGATA TCCAGAACACTCAGTGCCTGCTCAACGTGGAACACCTGAGTGCCGGCTGCCCCCATGTCACCCTGCAGTTTGCAGATTCCAAGGGCGATG TGGGGCTGGGCTTGGTGAAGGAAGGGCTGGTCATGGTGGAGGTGCGCAAGGAGAAACAGTTCCAGAAAGTGATCACAGAATACCTGAATG CCCAAGAGTCAGCCAAGAGCGCCAGGCTGAACCTGTGGCGCTATGGAGACTTTCGAGCTGATGATGCAGACGAATTTGGCTACAGCCGCT AAGGAGGGGATCGGGTTTGGCCCCCAGCCCCGCTCACGCCAGTCCCTCTTCCTCTGCCGGGAGGGTGTTTTCAACTCCAAACCCCAGAGA GGGGTTGTAGATTGGGTCCAGCTTTGCTTCAGTGTGTGGAAATGTCTCGTGGGGTGGCATCGGGGCTGCGGGGTGGGGACCCCAAGGCTT TCTGGGGCAGACCCTTGTCCTCTGGGATGATGGGCACTGCTATCCACAGTCTCTGCCAGTTGGTTTTATTTGGAGGTTTGTGGGCTTTTT TTAAAAAAAAAAAGTCCTCAAATCAGGAAGAAACATCAAAGACTATGTCCTAGTGGAGGGAGTAATCCTAACACCCAGGCTGGCCGCCAG CTGGCACCTGCCTCTATCCCAGACTGCCCTCGTCCCAGCTCTCTGTCCAACTGTTGATTATGTGATTTTTCTGATACGTCCATTCTCAAA TGCCAGTGTGTTCACATCTTCGCTCTGGCCAGCCCATTCTGTATTTAAAGCTTTTGAGGCCCAATAAAATAGTACGTGCTGTCTGCAGCC CTTATTGATCA >2873_2873_2_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000373671_SND1_chr7_127484377_ENST00000354725_length(amino acids)=593AA_BP=97 MPASSLGAAAMAASAKRKQEEKHLKMLRDMTGLPHNRKCFDCDQRGPTYVNMTVGSFVCTSCSGSLRGLNPPHRVKSISMTTFTQQEIEF LQKHGNEVNVTVDYIRPASPATETVPAFSERTCATVTIGGINIAEALVSKGLATVIRYRQDDDQRSSHYDELLAAEARAIKNGKGLHSKK EVPIHRVADISGDTQKAKQFLPFLQRAGRSEAVVEYVFSGSRLKLYLPKETCLITFLLAGIECPRGARNLPGLVQEGEPFSEEATLFTKE LVLQREVEVEVESMDKAGNFIGWLHIDGANLSVLLVEHALSKVHFTAERSSYYKSLLSAEEAAKQKKEKVWAHYEEQPVEEVMPVLEEKE RSASYKPVFVTEITDDLHFYVQDVETGTQLEKLMENMRNDIASHPPVEGSYAPRRGEFCIAKFVDGEWYRARVEKVESPAKIHVFYIDYG NREVLPSTRLGTLSPAFSTRVLPAQATEYAFAFIQVPQDDDARTDAVDSVVRDIQNTQCLLNVEHLSAGCPHVTLQFADSKGDVGLGLVK EGLVMVEVRKEKQFQKVITEYLNAQESAKSARLNLWRYGDFRADDADEFGYSR -------------------------------------------------------------- >2873_2873_3_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000409171_SND1_chr7_127484377_ENST00000354725_length(transcript)=2351nt_BP=311nt GCGCTGCCCGGCTCCCGGCCCTGCCGGCCTCCTCCCTTGGCGCCGCGGCCATGGCGGCCAGCGCGAAGCGGAAGCAGGAGGAGAAGCACC TGAAGATGCTGCGGGACATGACCGGCCTCCCGCACAACCGAAAGTGCTTCGACTGCGACCAGCGCGGCCCCACCTACGTTAACATGACGG TCGGCTCCTTCGTGTGTACCTCCTGCTCCGGCAGCCTGCGAGGATTAAATCCACCACACAGGGTGAAATCTATCTCCATGACAACATTCA CACAACAGGAAATTGAATTCTTACAAAAACATGGAAATGAAGTCAATGTGACGGTGGACTACATTAGACCAGCCAGCCCAGCCACAGAGA CAGTGCCTGCCTTTTCAGAGCGTACCTGTGCCACTGTCACCATTGGAGGAATAAACATTGCTGAGGCTCTTGTCAGCAAAGGTCTAGCCA CAGTGATCAGATACCGGCAGGATGATGACCAGAGATCATCACACTACGATGAACTGCTTGCTGCAGAGGCCAGAGCTATTAAGAATGGCA AAGGATTGCATAGCAAGAAGGAAGTGCCTATCCACCGTGTTGCAGATATATCTGGGGATACCCAAAAAGCAAAGCAGTTCCTGCCTTTTC TTCAGCGGGCAGGTCGTTCTGAAGCTGTGGTGGAATACGTCTTCAGTGGTTCTCGTCTCAAACTCTATTTGCCAAAGGAAACTTGCCTTA TCACCTTCTTGCTTGCAGGCATTGAATGCCCCAGAGGAGCCCGAAACCTCCCAGGCTTGGTGCAGGAAGGAGAGCCCTTCAGCGAGGAAG CTACACTTTTCACCAAGGAACTGGTGCTGCAGCGAGAGGTGGAGGTGGAGGTGGAGAGCATGGACAAGGCCGGCAACTTTATCGGCTGGC TGCACATCGACGGTGCCAACCTGTCCGTCCTGCTGGTGGAGCACGCGCTCTCCAAGGTCCACTTCACCGCCGAACGCAGCTCCTACTACA AGTCCCTGCTGTCTGCCGAGGAGGCCGCAAAGCAGAAGAAAGAGAAGGTCTGGGCCCACTATGAGGAGCAGCCCGTGGAGGAGGTGATGC CAGTGCTGGAGGAGAAGGAGCGATCTGCTAGCTACAAGCCCGTGTTTGTGACTGAGATCACTGATGACCTGCACTTCTACGTGCAGGATG TGGAGACCGGCACCCAGTTGGAGAAGCTGATGGAGAACATGCGCAATGACATTGCCAGTCACCCCCCTGTAGAGGGCTCCTATGCCCCCC GCAGGGGAGAGTTCTGCATTGCCAAATTTGTAGATGGAGAATGGTACCGTGCCCGAGTAGAGAAAGTCGAGTCTCCTGCCAAAATACATG TCTTCTACATTGACTACGGCAACAGAGAGGTCCTGCCATCCACCCGCCTGGGTACCCTATCACCTGCCTTCAGCACTCGGGTGCTGCCAG CTCAAGCCACGGAGTATGCCTTCGCCTTCATCCAGGTGCCCCAAGATGATGATGCCCGCACGGACGCCGTGGACAGCGTAGTTCGGGATA TCCAGAACACTCAGTGCCTGCTCAACGTGGAACACCTGAGTGCCGGCTGCCCCCATGTCACCCTGCAGTTTGCAGATTCCAAGGGCGATG TGGGGCTGGGCTTGGTGAAGGAAGGGCTGGTCATGGTGGAGGTGCGCAAGGAGAAACAGTTCCAGAAAGTGATCACAGAATACCTGAATG CCCAAGAGTCAGCCAAGAGCGCCAGGCTGAACCTGTGGCGCTATGGAGACTTTCGAGCTGATGATGCAGACGAATTTGGCTACAGCCGCT AAGGAGGGGATCGGGTTTGGCCCCCAGCCCCGCTCACGCCAGTCCCTCTTCCTCTGCCGGGAGGGTGTTTTCAACTCCAAACCCCAGAGA GGGGTTGTAGATTGGGTCCAGCTTTGCTTCAGTGTGTGGAAATGTCTCGTGGGGTGGCATCGGGGCTGCGGGGTGGGGACCCCAAGGCTT TCTGGGGCAGACCCTTGTCCTCTGGGATGATGGGCACTGCTATCCACAGTCTCTGCCAGTTGGTTTTATTTGGAGGTTTGTGGGCTTTTT TTAAAAAAAAAAAGTCCTCAAATCAGGAAGAAACATCAAAGACTATGTCCTAGTGGAGGGAGTAATCCTAACACCCAGGCTGGCCGCCAG CTGGCACCTGCCTCTATCCCAGACTGCCCTCGTCCCAGCTCTCTGTCCAACTGTTGATTATGTGATTTTTCTGATACGTCCATTCTCAAA TGCCAGTGTGTTCACATCTTCGCTCTGGCCAGCCCATTCTGTATTTAAAGCTTTTGAGGCCCAATAAAATAGTACGTGCTGTCTGCAGCC CTTATTGATCA >2873_2873_3_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000409171_SND1_chr7_127484377_ENST00000354725_length(amino acids)=593AA_BP=97 MPASSLGAAAMAASAKRKQEEKHLKMLRDMTGLPHNRKCFDCDQRGPTYVNMTVGSFVCTSCSGSLRGLNPPHRVKSISMTTFTQQEIEF LQKHGNEVNVTVDYIRPASPATETVPAFSERTCATVTIGGINIAEALVSKGLATVIRYRQDDDQRSSHYDELLAAEARAIKNGKGLHSKK EVPIHRVADISGDTQKAKQFLPFLQRAGRSEAVVEYVFSGSRLKLYLPKETCLITFLLAGIECPRGARNLPGLVQEGEPFSEEATLFTKE LVLQREVEVEVESMDKAGNFIGWLHIDGANLSVLLVEHALSKVHFTAERSSYYKSLLSAEEAAKQKKEKVWAHYEEQPVEEVMPVLEEKE RSASYKPVFVTEITDDLHFYVQDVETGTQLEKLMENMRNDIASHPPVEGSYAPRRGEFCIAKFVDGEWYRARVEKVESPAKIHVFYIDYG NREVLPSTRLGTLSPAFSTRVLPAQATEYAFAFIQVPQDDDARTDAVDSVVRDIQNTQCLLNVEHLSAGCPHVTLQFADSKGDVGLGLVK EGLVMVEVRKEKQFQKVITEYLNAQESAKSARLNLWRYGDFRADDADEFGYSR -------------------------------------------------------------- >2873_2873_4_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000409315_SND1_chr7_127484377_ENST00000354725_length(transcript)=2354nt_BP=314nt GCAGCGCTGCCCGGCTCCCGGCCCTGCCGGCCTCCTCCCTTGGCGCCGCGGCCATGGCGGCCAGCGCGAAGCGGAAGCAGGAGGAGAAGC ACCTGAAGATGCTGCGGGACATGACCGGCCTCCCGCACAACCGAAAGTGCTTCGACTGCGACCAGCGCGGCCCCACCTACGTTAACATGA CGGTCGGCTCCTTCGTGTGTACCTCCTGCTCCGGCAGCCTGCGAGGATTAAATCCACCACACAGGGTGAAATCTATCTCCATGACAACAT TCACACAACAGGAAATTGAATTCTTACAAAAACATGGAAATGAAGTCAATGTGACGGTGGACTACATTAGACCAGCCAGCCCAGCCACAG AGACAGTGCCTGCCTTTTCAGAGCGTACCTGTGCCACTGTCACCATTGGAGGAATAAACATTGCTGAGGCTCTTGTCAGCAAAGGTCTAG CCACAGTGATCAGATACCGGCAGGATGATGACCAGAGATCATCACACTACGATGAACTGCTTGCTGCAGAGGCCAGAGCTATTAAGAATG GCAAAGGATTGCATAGCAAGAAGGAAGTGCCTATCCACCGTGTTGCAGATATATCTGGGGATACCCAAAAAGCAAAGCAGTTCCTGCCTT TTCTTCAGCGGGCAGGTCGTTCTGAAGCTGTGGTGGAATACGTCTTCAGTGGTTCTCGTCTCAAACTCTATTTGCCAAAGGAAACTTGCC TTATCACCTTCTTGCTTGCAGGCATTGAATGCCCCAGAGGAGCCCGAAACCTCCCAGGCTTGGTGCAGGAAGGAGAGCCCTTCAGCGAGG AAGCTACACTTTTCACCAAGGAACTGGTGCTGCAGCGAGAGGTGGAGGTGGAGGTGGAGAGCATGGACAAGGCCGGCAACTTTATCGGCT GGCTGCACATCGACGGTGCCAACCTGTCCGTCCTGCTGGTGGAGCACGCGCTCTCCAAGGTCCACTTCACCGCCGAACGCAGCTCCTACT ACAAGTCCCTGCTGTCTGCCGAGGAGGCCGCAAAGCAGAAGAAAGAGAAGGTCTGGGCCCACTATGAGGAGCAGCCCGTGGAGGAGGTGA TGCCAGTGCTGGAGGAGAAGGAGCGATCTGCTAGCTACAAGCCCGTGTTTGTGACTGAGATCACTGATGACCTGCACTTCTACGTGCAGG ATGTGGAGACCGGCACCCAGTTGGAGAAGCTGATGGAGAACATGCGCAATGACATTGCCAGTCACCCCCCTGTAGAGGGCTCCTATGCCC CCCGCAGGGGAGAGTTCTGCATTGCCAAATTTGTAGATGGAGAATGGTACCGTGCCCGAGTAGAGAAAGTCGAGTCTCCTGCCAAAATAC ATGTCTTCTACATTGACTACGGCAACAGAGAGGTCCTGCCATCCACCCGCCTGGGTACCCTATCACCTGCCTTCAGCACTCGGGTGCTGC CAGCTCAAGCCACGGAGTATGCCTTCGCCTTCATCCAGGTGCCCCAAGATGATGATGCCCGCACGGACGCCGTGGACAGCGTAGTTCGGG ATATCCAGAACACTCAGTGCCTGCTCAACGTGGAACACCTGAGTGCCGGCTGCCCCCATGTCACCCTGCAGTTTGCAGATTCCAAGGGCG ATGTGGGGCTGGGCTTGGTGAAGGAAGGGCTGGTCATGGTGGAGGTGCGCAAGGAGAAACAGTTCCAGAAAGTGATCACAGAATACCTGA ATGCCCAAGAGTCAGCCAAGAGCGCCAGGCTGAACCTGTGGCGCTATGGAGACTTTCGAGCTGATGATGCAGACGAATTTGGCTACAGCC GCTAAGGAGGGGATCGGGTTTGGCCCCCAGCCCCGCTCACGCCAGTCCCTCTTCCTCTGCCGGGAGGGTGTTTTCAACTCCAAACCCCAG AGAGGGGTTGTAGATTGGGTCCAGCTTTGCTTCAGTGTGTGGAAATGTCTCGTGGGGTGGCATCGGGGCTGCGGGGTGGGGACCCCAAGG CTTTCTGGGGCAGACCCTTGTCCTCTGGGATGATGGGCACTGCTATCCACAGTCTCTGCCAGTTGGTTTTATTTGGAGGTTTGTGGGCTT TTTTTAAAAAAAAAAAGTCCTCAAATCAGGAAGAAACATCAAAGACTATGTCCTAGTGGAGGGAGTAATCCTAACACCCAGGCTGGCCGC CAGCTGGCACCTGCCTCTATCCCAGACTGCCCTCGTCCCAGCTCTCTGTCCAACTGTTGATTATGTGATTTTTCTGATACGTCCATTCTC AAATGCCAGTGTGTTCACATCTTCGCTCTGGCCAGCCCATTCTGTATTTAAAGCTTTTGAGGCCCAATAAAATAGTACGTGCTGTCTGCA GCCCTTATTGATCA >2873_2873_4_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000409315_SND1_chr7_127484377_ENST00000354725_length(amino acids)=593AA_BP=97 MPASSLGAAAMAASAKRKQEEKHLKMLRDMTGLPHNRKCFDCDQRGPTYVNMTVGSFVCTSCSGSLRGLNPPHRVKSISMTTFTQQEIEF LQKHGNEVNVTVDYIRPASPATETVPAFSERTCATVTIGGINIAEALVSKGLATVIRYRQDDDQRSSHYDELLAAEARAIKNGKGLHSKK EVPIHRVADISGDTQKAKQFLPFLQRAGRSEAVVEYVFSGSRLKLYLPKETCLITFLLAGIECPRGARNLPGLVQEGEPFSEEATLFTKE LVLQREVEVEVESMDKAGNFIGWLHIDGANLSVLLVEHALSKVHFTAERSSYYKSLLSAEEAAKQKKEKVWAHYEEQPVEEVMPVLEEKE RSASYKPVFVTEITDDLHFYVQDVETGTQLEKLMENMRNDIASHPPVEGSYAPRRGEFCIAKFVDGEWYRARVEKVESPAKIHVFYIDYG NREVLPSTRLGTLSPAFSTRVLPAQATEYAFAFIQVPQDDDARTDAVDSVVRDIQNTQCLLNVEHLSAGCPHVTLQFADSKGDVGLGLVK EGLVMVEVRKEKQFQKVITEYLNAQESAKSARLNLWRYGDFRADDADEFGYSR -------------------------------------------------------------- >2873_2873_5_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000409979_SND1_chr7_127484377_ENST00000354725_length(transcript)=2571nt_BP=531nt TTTTCCCCTCAGGAGAAGTCGGGAAGGTGGCGGCGGCGGCGGCGGTTGTCCCGGCTGTGCCGGTTGGTGTGGCCCGTCAGCCCGCGTACC ACAGCGCCCGGGCCGCGTCGAGCCCAGTACAGCCAAGCCGCTGCGGCCGGGTCCGGCGCGGGCGGCGCGCGCAGACGGAGGGCGGCGGCC GCGGCCAGGGCGGCCCGTGGGACCGCGGGCCCCCGGCGCAGCGCTGCCCGGCTCCCGGCCCTGCCGGCCTCCTCCCTTGGCGCCGCGGCC ATGGCGGCCAGCGCGAAGCGGAAGCAGGAGGAGAAGCACCTGAAGATGCTGCGGGACATGACCGGCCTCCCGCACAACCGAAAGTGCTTC GACTGCGACCAGCGCGGCCCCACCTACGTTAACATGACGGTCGGCTCCTTCGTGTGTACCTCCTGCTCCGGCAGCCTGCGAGGATTAAAT CCACCACACAGGGTGAAATCTATCTCCATGACAACATTCACACAACAGGAAATTGAATTCTTACAAAAACATGGAAATGAAGTCAATGTG ACGGTGGACTACATTAGACCAGCCAGCCCAGCCACAGAGACAGTGCCTGCCTTTTCAGAGCGTACCTGTGCCACTGTCACCATTGGAGGA ATAAACATTGCTGAGGCTCTTGTCAGCAAAGGTCTAGCCACAGTGATCAGATACCGGCAGGATGATGACCAGAGATCATCACACTACGAT GAACTGCTTGCTGCAGAGGCCAGAGCTATTAAGAATGGCAAAGGATTGCATAGCAAGAAGGAAGTGCCTATCCACCGTGTTGCAGATATA TCTGGGGATACCCAAAAAGCAAAGCAGTTCCTGCCTTTTCTTCAGCGGGCAGGTCGTTCTGAAGCTGTGGTGGAATACGTCTTCAGTGGT TCTCGTCTCAAACTCTATTTGCCAAAGGAAACTTGCCTTATCACCTTCTTGCTTGCAGGCATTGAATGCCCCAGAGGAGCCCGAAACCTC CCAGGCTTGGTGCAGGAAGGAGAGCCCTTCAGCGAGGAAGCTACACTTTTCACCAAGGAACTGGTGCTGCAGCGAGAGGTGGAGGTGGAG GTGGAGAGCATGGACAAGGCCGGCAACTTTATCGGCTGGCTGCACATCGACGGTGCCAACCTGTCCGTCCTGCTGGTGGAGCACGCGCTC TCCAAGGTCCACTTCACCGCCGAACGCAGCTCCTACTACAAGTCCCTGCTGTCTGCCGAGGAGGCCGCAAAGCAGAAGAAAGAGAAGGTC TGGGCCCACTATGAGGAGCAGCCCGTGGAGGAGGTGATGCCAGTGCTGGAGGAGAAGGAGCGATCTGCTAGCTACAAGCCCGTGTTTGTG ACTGAGATCACTGATGACCTGCACTTCTACGTGCAGGATGTGGAGACCGGCACCCAGTTGGAGAAGCTGATGGAGAACATGCGCAATGAC ATTGCCAGTCACCCCCCTGTAGAGGGCTCCTATGCCCCCCGCAGGGGAGAGTTCTGCATTGCCAAATTTGTAGATGGAGAATGGTACCGT GCCCGAGTAGAGAAAGTCGAGTCTCCTGCCAAAATACATGTCTTCTACATTGACTACGGCAACAGAGAGGTCCTGCCATCCACCCGCCTG GGTACCCTATCACCTGCCTTCAGCACTCGGGTGCTGCCAGCTCAAGCCACGGAGTATGCCTTCGCCTTCATCCAGGTGCCCCAAGATGAT GATGCCCGCACGGACGCCGTGGACAGCGTAGTTCGGGATATCCAGAACACTCAGTGCCTGCTCAACGTGGAACACCTGAGTGCCGGCTGC CCCCATGTCACCCTGCAGTTTGCAGATTCCAAGGGCGATGTGGGGCTGGGCTTGGTGAAGGAAGGGCTGGTCATGGTGGAGGTGCGCAAG GAGAAACAGTTCCAGAAAGTGATCACAGAATACCTGAATGCCCAAGAGTCAGCCAAGAGCGCCAGGCTGAACCTGTGGCGCTATGGAGAC TTTCGAGCTGATGATGCAGACGAATTTGGCTACAGCCGCTAAGGAGGGGATCGGGTTTGGCCCCCAGCCCCGCTCACGCCAGTCCCTCTT CCTCTGCCGGGAGGGTGTTTTCAACTCCAAACCCCAGAGAGGGGTTGTAGATTGGGTCCAGCTTTGCTTCAGTGTGTGGAAATGTCTCGT GGGGTGGCATCGGGGCTGCGGGGTGGGGACCCCAAGGCTTTCTGGGGCAGACCCTTGTCCTCTGGGATGATGGGCACTGCTATCCACAGT CTCTGCCAGTTGGTTTTATTTGGAGGTTTGTGGGCTTTTTTTAAAAAAAAAAAGTCCTCAAATCAGGAAGAAACATCAAAGACTATGTCC TAGTGGAGGGAGTAATCCTAACACCCAGGCTGGCCGCCAGCTGGCACCTGCCTCTATCCCAGACTGCCCTCGTCCCAGCTCTCTGTCCAA CTGTTGATTATGTGATTTTTCTGATACGTCCATTCTCAAATGCCAGTGTGTTCACATCTTCGCTCTGGCCAGCCCATTCTGTATTTAAAG CTTTTGAGGCCCAATAAAATAGTACGTGCTGTCTGCAGCCCTTATTGATCA >2873_2873_5_AGFG1-SND1_AGFG1_chr2_228356356_ENST00000409979_SND1_chr7_127484377_ENST00000354725_length(amino acids)=658AA_BP=162 MSRLCRLVWPVSPRTTAPGPRRAQYSQAAAAGSGAGGARRRRAAAAARAARGTAGPRRSAARLPALPASSLGAAAMAASAKRKQEEKHLK MLRDMTGLPHNRKCFDCDQRGPTYVNMTVGSFVCTSCSGSLRGLNPPHRVKSISMTTFTQQEIEFLQKHGNEVNVTVDYIRPASPATETV PAFSERTCATVTIGGINIAEALVSKGLATVIRYRQDDDQRSSHYDELLAAEARAIKNGKGLHSKKEVPIHRVADISGDTQKAKQFLPFLQ RAGRSEAVVEYVFSGSRLKLYLPKETCLITFLLAGIECPRGARNLPGLVQEGEPFSEEATLFTKELVLQREVEVEVESMDKAGNFIGWLH IDGANLSVLLVEHALSKVHFTAERSSYYKSLLSAEEAAKQKKEKVWAHYEEQPVEEVMPVLEEKERSASYKPVFVTEITDDLHFYVQDVE TGTQLEKLMENMRNDIASHPPVEGSYAPRRGEFCIAKFVDGEWYRARVEKVESPAKIHVFYIDYGNREVLPSTRLGTLSPAFSTRVLPAQ ATEYAFAFIQVPQDDDARTDAVDSVVRDIQNTQCLLNVEHLSAGCPHVTLQFADSKGDVGLGLVKEGLVMVEVRKEKQFQKVITEYLNAQ ESAKSARLNLWRYGDFRADDADEFGYSR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AGFG1-SND1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AGFG1-SND1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AGFG1-SND1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | AGFG1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Tgene | C0004352 | Autistic Disorder | 1 | CTD_human | |
| Tgene | C0024121 | Lung Neoplasms | 1 | CTD_human | |
| Tgene | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
