
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:APLP1-DBH (FusionGDB2 ID:HG333TG1621) |
Fusion Gene Summary for APLP1-DBH |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: APLP1-DBH | Fusion gene ID: hg333tg1621 | Hgene | Tgene | Gene symbol | APLP1 | DBH | Gene ID | 333 | 1621 |
| Gene name | amyloid beta precursor like protein 1 | dopamine beta-hydroxylase | |
| Synonyms | APLP | DBM|ORTHYP1 | |
| Cytomap | ('APLP1')('DBH') 19q13.12 | 9q34.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | amyloid-like protein 1amyloid beta (A4) precursor-like protein 1amyloid precursor-like protein 1 | dopamine beta-hydroxylasedopamine beta-hydroxylase (dopamine beta-monooxygenase) | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P51693 | P09172 | |
| Ensembl transtripts involved in fusion gene | ENST00000589298, ENST00000221891, ENST00000537454, ENST00000586861, | ||
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 3 X 3 X 3=27 |
| # samples | 3 | 3 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: APLP1 [Title/Abstract] AND DBH [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | APLP1(36365771)-DBH(136523438), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | APLP1-DBH seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. APLP1-DBH seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. APLP1-DBH seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. APLP1-DBH seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | APLP1 | GO:0071874 | cellular response to norepinephrine stimulus | 16531006 |
| Hgene | APLP1 | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 16531006 |
| Tgene | DBH | GO:0042420 | dopamine catabolic process | 3443096|7961964 |
 Fusion gene breakpoints across APLP1 (5'-gene) Fusion gene breakpoints across APLP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
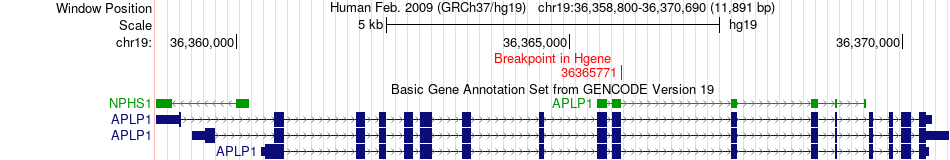 |
 Fusion gene breakpoints across DBH (3'-gene) Fusion gene breakpoints across DBH (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
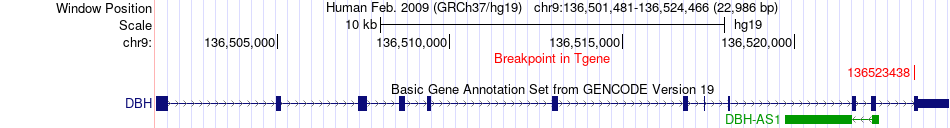 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PCPG | TCGA-SR-A6MT-01A | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + |
Top |
Fusion Gene ORF analysis for APLP1-DBH |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000589298 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + |
| In-frame | ENST00000221891 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + |
| In-frame | ENST00000537454 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + |
| In-frame | ENST00000586861 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000537454 | APLP1 | chr19 | 36365771 | + | ENST00000393056 | DBH | chr9 | 136523438 | + | 2601 | 1572 | 270 | 1703 | 477 |
| ENST00000221891 | APLP1 | chr19 | 36365771 | + | ENST00000393056 | DBH | chr9 | 136523438 | + | 2565 | 1536 | 192 | 1667 | 491 |
| ENST00000586861 | APLP1 | chr19 | 36365771 | + | ENST00000393056 | DBH | chr9 | 136523438 | + | 2420 | 1391 | 65 | 1522 | 485 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000537454 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + | 0.043739 | 0.956261 |
| ENST00000221891 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + | 0.08413941 | 0.9158606 |
| ENST00000586861 | ENST00000393056 | APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523438 | + | 0.08219274 | 0.9178073 |
Top |
Fusion Genomic Features for APLP1-DBH |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523437 | + | 5.03E-07 | 0.9999995 |
| APLP1 | chr19 | 36365771 | + | DBH | chr9 | 136523437 | + | 5.03E-07 | 0.9999995 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
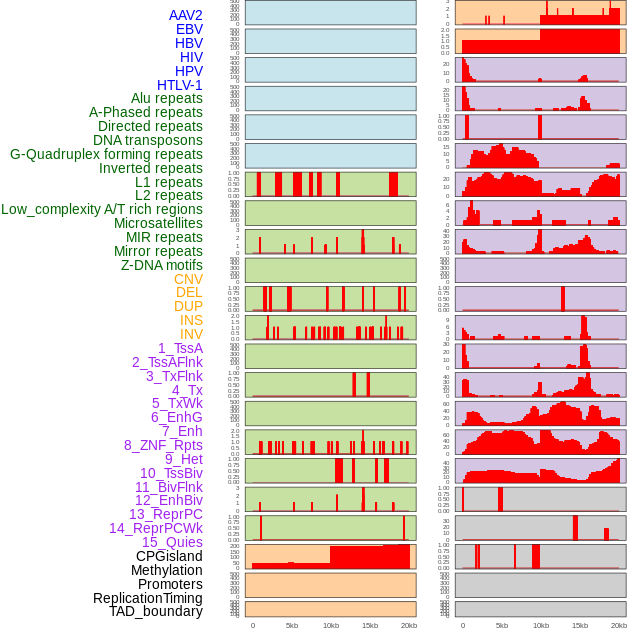 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
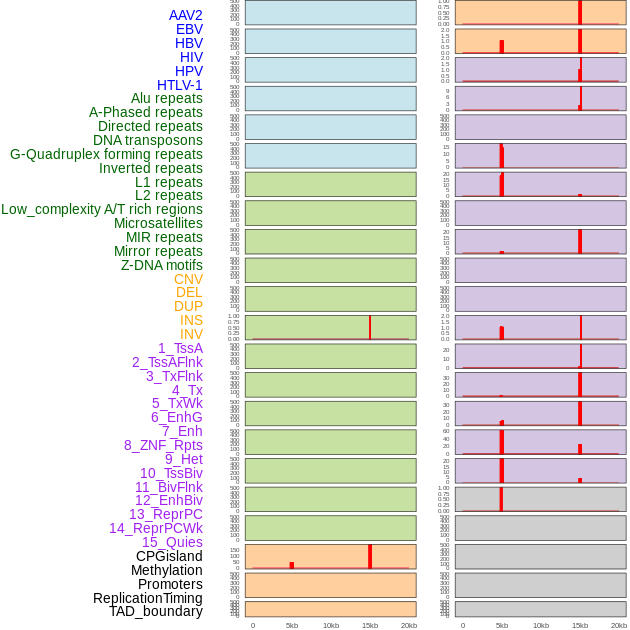 |
Top |
Fusion Protein Features for APLP1-DBH |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:36365771/chr9:136523438) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| APLP1 | DBH |
| FUNCTION: May play a role in postsynaptic function. The C-terminal gamma-secretase processed fragment, ALID1, activates transcription activation through APBB1 (Fe65) binding (By similarity). Couples to JIP signal transduction through C-terminal binding. May interact with cellular G-protein signaling pathways. Can regulate neurite outgrowth through binding to components of the extracellular matrix such as heparin and collagen I. {ECO:0000250}.; FUNCTION: The gamma-CTF peptide, C30, is a potent enhancer of neuronal apoptosis. {ECO:0000250}. | FUNCTION: Conversion of dopamine to noradrenaline. {ECO:0000269|PubMed:27148966, ECO:0000269|PubMed:3443096, ECO:0000269|PubMed:7961964, ECO:0000269|PubMed:8546710}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 241_247 | 448 | 652.0 | Compositional bias | Note=Poly-Glu |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 264_268 | 448 | 652.0 | Compositional bias | Note=Poly-Glu |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 50_212 | 448 | 652.0 | Domain | E1 |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 154_212 | 448 | 652.0 | Region | CuBD subdomain |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 158_178 | 448 | 652.0 | Region | Copper-binding |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 204_211 | 448 | 652.0 | Region | Note=Zinc-binding |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 285_305 | 448 | 652.0 | Region | Note=O-glycosylated at three sites |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 310_342 | 448 | 652.0 | Region | Heparin-binding |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 410_441 | 448 | 652.0 | Region | Heparin-binding |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 50_146 | 448 | 652.0 | Region | GFLD subdomain |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 293_484 | 448 | 652.0 | Domain | E2 |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 604_615 | 448 | 652.0 | Motif | Basolateral sorting signal |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 640_643 | 448 | 652.0 | Motif | Note=NPXY motif%3B contains endocytosis signal |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 442_459 | 448 | 652.0 | Region | Collagen-binding |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 39_580 | 448 | 652.0 | Topological domain | Extracellular |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 604_650 | 448 | 652.0 | Topological domain | Cytoplasmic |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 581_603 | 448 | 652.0 | Transmembrane | Helical |
| Tgene | DBH | chr19:36365771 | chr9:136523438 | ENST00000393056 | 10 | 12 | 57_173 | 574 | 618.0 | Domain | DOMON | |
| Tgene | DBH | chr19:36365771 | chr9:136523438 | ENST00000393056 | 10 | 12 | 1_16 | 574 | 618.0 | Topological domain | Cytoplasmic | |
| Tgene | DBH | chr19:36365771 | chr9:136523438 | ENST00000393056 | 10 | 12 | 38_617 | 574 | 618.0 | Topological domain | Intragranular | |
| Tgene | DBH | chr19:36365771 | chr9:136523438 | ENST00000393056 | 10 | 12 | 17_37 | 574 | 618.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for APLP1-DBH |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >5437_5437_1_APLP1-DBH_APLP1_chr19_36365771_ENST00000221891_DBH_chr9_136523438_ENST00000393056_length(transcript)=2565nt_BP=1536nt ACTCTGCGCGAAGCCGCCACGCTATTGTCCTGACCAGGAAGGCGGGGCCGGCGCGGGGCGGGGCTGGCGGCGCCGGCGCAGCCCGGGGGC GGCGGGAGGAGGAGGTGGCGGCGGTGGCGCTGGGAGCTCCTGTCACCGCTGGGGCCGGGCCGGGCGGGAGTGCAGGGGACGTGAGGGCGC AAGGGCCGGGACATGGGGCCCGCCAGCCCCGCTGCTCGCGGTCTAAGTCGCCGCCCGGGCCAGCCGCCGCTGCCGCTGCTGCTGCCACTA TTGCTGCTGCTTCTGCGCGCGCAGCCCGCCATCGGGAGCCTGGCCGGTGGGAGCCCCGGCGCGGCCGAGGCCCCGGGGTCGGCCCAGGTG GCTGGACTATGCGGGCGCCTAACCCTTCACCGGGACCTGCGCACCGGCCGCTGGGAACCAGACCCACAGCGCTCTCGACGCTGTCTCCGG GACCCGCAGCGCGTGCTGGAGTACTGCAGACAGATGTACCCGGAGCTGCAGATTGCACGTGTGGAGCAGGCTACGCAGGCCATCCCCATG GAGCGCTGGTGCGGGGGTTCCCGGAGCGGCAGCTGCGCCCACCCCCACCACCAGGTTGTGCCCTTCCGCTGCCTGCCTGGTGAATTTGTG AGTGAGGCCCTGCTGGTGCCTGAAGGCTGCCGGTTCTTGCACCAGGAGCGCATGGACCAATGTGAGAGTTCAACCCGGAGGCATCAGGAG GCACAGGAGGCCTGCAGCTCCCAGGGCCTCATCCTGCACGGCTCGGGCATGCTCTTACCCTGTGGCTCGGATCGGTTCCGTGGTGTGGAG TATGTGTGCTGTCCCCCTCCAGGGACCCCCGACCCATCTGGGACAGCAGTTGGTGACCCCTCCACCCGGTCCTGGCCCCCGGGGAGCAGA GTAGAGGGGGCTGAGGACGAGGAAGAGGAGGAATCCTTCCCACAGCCAGTAGATGATTACTTCGTGGAGCCTCCGCAGGCTGAAGAGGAA GAGGAAACGGTCCCACCCCCAAGCTCCCATACACTTGCAGTGGTCGGCAAAGTCACTCCCACCCCGAGGCCCACAGACGGTGTGGATATT TACTTTGGCATGCCTGGGGAAATCAGTGAGCACGAGGGGTTCCTGAGGGCCAAGATGGACCTGGAGGAGCGTAGGATGCGCCAGATTAAT GAGGTGATGCGTGAATGGGCCATGGCAGACAACCAGTCCAAGAACCTGCCTAAAGCCGACAGACAGGCCCTGAATGAGCACTTCCAGTCC ATTCTGCAGACTCTGGAGGAGCAGGTGTCTGGTGAGCGACAGCGCCTGGTGGAAACCCACGCCACCCGCGTCATCGCCCTTATCAACGAC CAGCGCCGGGCTGCCTTGGAGGGCTTCCTGGCAGCCCTGCAGGCAGATCCGCCTCAGGCGGAGCGTGTCCTGTTGGCCCTGCGGCGCTAC CTGCGTGCGGAGCAGAAGGAACAGAGGCACACGCTGCGCCACTACCAGCATGTGGCCGCCGTGGATCCCGAGAAGGCACAGCAGATGCGC TTCCAGGGTGAATGGAACCTGCAGCCCCTGCCCAAGGTCATCTCCACACTGGAAGAGCCCACCCCACAGTGCCCCACCAGCCAGGGCCGA AGCCCTGCTGGCCCCACCGTTGTCAGCATTGGTGGGGGCAAAGGCTGAGGGGGGACCTACTCCTCCCCCTCCTCCATGCTGTCCCTGTGG GCTCACACCGGCACTGTGCACTCTACTCTGCGACGATCCCCATGGAACAGCCCTGCACGCCCAGGATGAAGGGGCCAGACCACGCCCCTG CCTGAGACCACGGTCCAATCCAGCCTTCTTCCCCCAGGGTCCCCTGCATGGCTGAGAGGGTGTGGGTGCCCTGTTGACCTACCCTGGACC GAGTGGACCACGACCTCGTCCATTTAAACCCGGCTGACTCAGTGCAGGGACAGCCTGCACAGTGGTCCAGGGTCCAGCCCTCCGCCAGCC CTGTTCCGCCTCACTGGGTGTGGCCTGGCTTCTGGGACAGGCACCATGCTGGGCCGGGGTGTGGAATCACCGGGAACGCCCCCGCCCCCG CCCCGCTGCTCCCGGTGTGCAGCGGGTGCGGGTGCCGCTTAAACATTTCCCTGCTGAGTGGCTCGTGTTTCACAGTGGGCGGCTTCCCTG CGACGGAGGCAGGACCAGGCATTTAGCTAGTTAGAGACTCGCCTGGGAAATTGCTCCATTCCTGAGTAAACAGATATTTTCGCCCACCTA AAGGGAAGCCCTGACAACAACTATCACCAAAAGACGAGGCGGCAAAGATCCAGCGGGGCTTCTGGGCGCCGGTTCCACGTGGGGTGGAAT TATTAGCACCAGCTTGCTTCTCTGCCGGTGGGGCCAGCGCTGAACAGACCGGGGTGGAGTCAGGGCTGTGCTTTCCGCGTGGTTCTGCCA CTTAGGGAGTGTGCCTTGGGCGGGCCATTTCACATTCCTGACCCTCACTTTTCTCATCTGTAAAACCAGGCTGATGCCGTGCGGGCTAAT GAGCCAATAAAGCTCACACTTGGGCTGGCACCCACTGGAGGTGGG >5437_5437_1_APLP1-DBH_APLP1_chr19_36365771_ENST00000221891_DBH_chr9_136523438_ENST00000393056_length(amino acids)=491AA_BP=447 MGPASPAARGLSRRPGQPPLPLLLPLLLLLLRAQPAIGSLAGGSPGAAEAPGSAQVAGLCGRLTLHRDLRTGRWEPDPQRSRRCLRDPQR VLEYCRQMYPELQIARVEQATQAIPMERWCGGSRSGSCAHPHHQVVPFRCLPGEFVSEALLVPEGCRFLHQERMDQCESSTRRHQEAQEA CSSQGLILHGSGMLLPCGSDRFRGVEYVCCPPPGTPDPSGTAVGDPSTRSWPPGSRVEGAEDEEEEESFPQPVDDYFVEPPQAEEEEETV PPPSSHTLAVVGKVTPTPRPTDGVDIYFGMPGEISEHEGFLRAKMDLEERRMRQINEVMREWAMADNQSKNLPKADRQALNEHFQSILQT LEEQVSGERQRLVETHATRVIALINDQRRAALEGFLAALQADPPQAERVLLALRRYLRAEQKEQRHTLRHYQHVAAVDPEKAQQMRFQGE WNLQPLPKVISTLEEPTPQCPTSQGRSPAGPTVVSIGGGKG -------------------------------------------------------------- >5437_5437_2_APLP1-DBH_APLP1_chr19_36365771_ENST00000537454_DBH_chr9_136523438_ENST00000393056_length(transcript)=2601nt_BP=1572nt AATCCTGAGGTGCTGCATCCACAGTGGGCATGGTGCTGGTGCCTGCTTGGATGATCCTTAAAGAAAGGTCCTGGGGGCTTTGGTTCATGG ATCCTTGAGCTAGGAGTTAAAGGTCCAGGCCCCTGGGACCCTTGGGAAGCAGAGCAAGAAGAGTGAACTCCTGGGTCTGAAGGAGAATGG GCTGGGGGCTTGGTCTCTGGTCCTGAGAGAGAAGGTGCCCAGACTTCTGGATCTGAAAGAGGAAGGGACTAGGTCTCAACTGCTGCCTTC TTGACTGGGGACATTTTGGAGGCCTGTATTCCTGAGCCCTCAACAGAGGAATGTACTAGGGGATGGGGGTCTCTGATGCTTGCATCCTTG GAAAAGGACAAAACTGCCCCGGGGTCGGCCCAGGTGGCTGGACTATGCGGGCGCCTAACCCTTCACCGGGACCTGCGCACCGGCCGCTGG GAACCAGACCCACAGCGCTCTCGACGCTGTCTCCGGGACCCGCAGCGCGTGCTGGAGTACTGCAGACAGATGTACCCGGAGCTGCAGATT GCACGTGTGGAGCAGGCTACGCAGGCCATCCCCATGGAGCGCTGGTGCGGGGGTTCCCGGAGCGGCAGCTGCGCCCACCCCCACCACCAG GTTGTGCCCTTCCGCTGCCTGCCTGGTGAATTTGTGAGTGAGGCCCTGCTGGTGCCTGAAGGCTGCCGGTTCTTGCACCAGGAGCGCATG GACCAATGTGAGAGTTCAACCCGGAGGCATCAGGAGGCACAGGAGGCCTGCAGCTCCCAGGGCCTCATCCTGCACGGCTCGGGCATGCTC TTACCCTGTGGCTCGGATCGGTTCCGTGGTGTGGAGTATGTGTGCTGTCCCCCTCCAGGGACCCCCGACCCATCTGGGACAGCAGTTGGT GACCCCTCCACCCGGTCCTGGCCCCCGGGGAGCAGAGTAGAGGGGGCTGAGGACGAGGAAGAGGAGGAATCCTTCCCACAGCCAGTAGAT GATTACTTCGTGGAGCCTCCGCAGGCTGAAGAGGAAGAGGAAACGGTCCCACCCCCAAGCTCCCATACACTTGCAGTGGTCGGCAAAGTC ACTCCCACCCCGAGGCCCACAGACGGTGTGGATATTTACTTTGGCATGCCTGGGGAAATCAGTGAGCACGAGGGGTTCCTGAGGGCCAAG ATGGACCTGGAGGAGCGTAGGATGCGCCAGATTAATGAGGTGATGCGTGAATGGGCCATGGCAGACAACCAGTCCAAGAACCTGCCTAAA GCCGACAGACAGGCCCTGAATGAGCACTTCCAGTCCATTCTGCAGACTCTGGAGGAGCAGGTGTCTGGTGAGCGACAGCGCCTGGTGGAA ACCCACGCCACCCGCGTCATCGCCCTTATCAACGACCAGCGCCGGGCTGCCTTGGAGGGCTTCCTGGCAGCCCTGCAGGCAGATCCGCCT CAGGCGGAGCGTGTCCTGTTGGCCCTGCGGCGCTACCTGCGTGCGGAGCAGAAGGAACAGAGGCACACGCTGCGCCACTACCAGCATGTG GCCGCCGTGGATCCCGAGAAGGCACAGCAGATGCGCTTCCAGGGTGAATGGAACCTGCAGCCCCTGCCCAAGGTCATCTCCACACTGGAA GAGCCCACCCCACAGTGCCCCACCAGCCAGGGCCGAAGCCCTGCTGGCCCCACCGTTGTCAGCATTGGTGGGGGCAAAGGCTGAGGGGGG ACCTACTCCTCCCCCTCCTCCATGCTGTCCCTGTGGGCTCACACCGGCACTGTGCACTCTACTCTGCGACGATCCCCATGGAACAGCCCT GCACGCCCAGGATGAAGGGGCCAGACCACGCCCCTGCCTGAGACCACGGTCCAATCCAGCCTTCTTCCCCCAGGGTCCCCTGCATGGCTG AGAGGGTGTGGGTGCCCTGTTGACCTACCCTGGACCGAGTGGACCACGACCTCGTCCATTTAAACCCGGCTGACTCAGTGCAGGGACAGC CTGCACAGTGGTCCAGGGTCCAGCCCTCCGCCAGCCCTGTTCCGCCTCACTGGGTGTGGCCTGGCTTCTGGGACAGGCACCATGCTGGGC CGGGGTGTGGAATCACCGGGAACGCCCCCGCCCCCGCCCCGCTGCTCCCGGTGTGCAGCGGGTGCGGGTGCCGCTTAAACATTTCCCTGC TGAGTGGCTCGTGTTTCACAGTGGGCGGCTTCCCTGCGACGGAGGCAGGACCAGGCATTTAGCTAGTTAGAGACTCGCCTGGGAAATTGC TCCATTCCTGAGTAAACAGATATTTTCGCCCACCTAAAGGGAAGCCCTGACAACAACTATCACCAAAAGACGAGGCGGCAAAGATCCAGC GGGGCTTCTGGGCGCCGGTTCCACGTGGGGTGGAATTATTAGCACCAGCTTGCTTCTCTGCCGGTGGGGCCAGCGCTGAACAGACCGGGG TGGAGTCAGGGCTGTGCTTTCCGCGTGGTTCTGCCACTTAGGGAGTGTGCCTTGGGCGGGCCATTTCACATTCCTGACCCTCACTTTTCT CATCTGTAAAACCAGGCTGATGCCGTGCGGGCTAATGAGCCAATAAAGCTCACACTTGGGCTGGCACCCACTGGAGGTGGG >5437_5437_2_APLP1-DBH_APLP1_chr19_36365771_ENST00000537454_DBH_chr9_136523438_ENST00000393056_length(amino acids)=477AA_BP=433 MTGDILEACIPEPSTEECTRGWGSLMLASLEKDKTAPGSAQVAGLCGRLTLHRDLRTGRWEPDPQRSRRCLRDPQRVLEYCRQMYPELQI ARVEQATQAIPMERWCGGSRSGSCAHPHHQVVPFRCLPGEFVSEALLVPEGCRFLHQERMDQCESSTRRHQEAQEACSSQGLILHGSGML LPCGSDRFRGVEYVCCPPPGTPDPSGTAVGDPSTRSWPPGSRVEGAEDEEEEESFPQPVDDYFVEPPQAEEEEETVPPPSSHTLAVVGKV TPTPRPTDGVDIYFGMPGEISEHEGFLRAKMDLEERRMRQINEVMREWAMADNQSKNLPKADRQALNEHFQSILQTLEEQVSGERQRLVE THATRVIALINDQRRAALEGFLAALQADPPQAERVLLALRRYLRAEQKEQRHTLRHYQHVAAVDPEKAQQMRFQGEWNLQPLPKVISTLE EPTPQCPTSQGRSPAGPTVVSIGGGKG -------------------------------------------------------------- >5437_5437_3_APLP1-DBH_APLP1_chr19_36365771_ENST00000586861_DBH_chr9_136523438_ENST00000393056_length(transcript)=2420nt_BP=1391nt ATAGCAACGGGAGCGCTGGGGCGCCCCCGCCCTACGGGAGCTGTTTCCCAGGGAACGGTGCCTCCATGGAGGCGGTGTGCGGTGCTTGGG GGAGGGGGCTGGTGCTGGGGGTCTCGGTCCTAGGGAGCAAAGAACCAGGGGACCCTCATGCCAACGCCCCCCGAGCCCTCACTGTCCTTT CCACTTCCATCCAGGCCCCGGGGTCGGCCCAGGTGGCTGGACTATGCGGGCGCCTAACCCTTCACCGGGACCTGCGCACCGGCCGCTGGG AACCAGACCCACAGCGCTCTCGACGCTGTCTCCGGGACCCGCAGCGCGTGCTGGAGTACTGCAGACAGATGTACCCGGAGCTGCAGATTG CACGTGTGGAGCAGGCTACGCAGGCCATCCCCATGGAGCGCTGGTGCGGGGGTTCCCGGAGCGGCAGCTGCGCCCACCCCCACCACCAGG TTGTGCCCTTCCGCTGCCTGCCTGGTGAATTTGTGAGTGAGGCCCTGCTGGTGCCTGAAGGCTGCCGGTTCTTGCACCAGGAGCGCATGG ACCAATGTGAGAGTTCAACCCGGAGGCATCAGGAGGCACAGGAGGCCTGCAGCTCCCAGGGCCTCATCCTGCACGGCTCGGGCATGCTCT TACCCTGTGGCTCGGATCGGTTCCGTGGTGTGGAGTATGTGTGCTGTCCCCCTCCAGGGACCCCCGACCCATCTGGGACAGCAGTTGGTG ACCCCTCCACCCGGTCCTGGCCCCCGGGGAGCAGAGTAGAGGGGGCTGAGGACGAGGAAGAGGAGGAATCCTTCCCACAGCCAGTAGATG ATTACTTCGTGGAGCCTCCGCAGGCTGAAGAGGAAGAGGAAACGGTCCCACCCCCAAGCTCCCATACACTTGCAGTGGTCGGCAAAGTCA CTCCCACCCCGAGGCCCACAGACGGTGTGGATATTTACTTTGGCATGCCTGGGGAAATCAGTGAGCACGAGGGGTTCCTGAGGGCCAAGA TGGACCTGGAGGAGCGTAGGATGCGCCAGATTAATGAGGTGATGCGTGAATGGGCCATGGCAGACAACCAGTCCAAGAACCTGCCTAAAG CCGACAGACAGGCCCTGAATGAGCACTTCCAGTCCATTCTGCAGACTCTGGAGGAGCAGGTGTCTGGTGAGCGACAGCGCCTGGTGGAAA CCCACGCCACCCGCGTCATCGCCCTTATCAACGACCAGCGCCGGGCTGCCTTGGAGGGCTTCCTGGCAGCCCTGCAGGCAGATCCGCCTC AGGCGGAGCGTGTCCTGTTGGCCCTGCGGCGCTACCTGCGTGCGGAGCAGAAGGAACAGAGGCACACGCTGCGCCACTACCAGCATGTGG CCGCCGTGGATCCCGAGAAGGCACAGCAGATGCGCTTCCAGGGTGAATGGAACCTGCAGCCCCTGCCCAAGGTCATCTCCACACTGGAAG AGCCCACCCCACAGTGCCCCACCAGCCAGGGCCGAAGCCCTGCTGGCCCCACCGTTGTCAGCATTGGTGGGGGCAAAGGCTGAGGGGGGA CCTACTCCTCCCCCTCCTCCATGCTGTCCCTGTGGGCTCACACCGGCACTGTGCACTCTACTCTGCGACGATCCCCATGGAACAGCCCTG CACGCCCAGGATGAAGGGGCCAGACCACGCCCCTGCCTGAGACCACGGTCCAATCCAGCCTTCTTCCCCCAGGGTCCCCTGCATGGCTGA GAGGGTGTGGGTGCCCTGTTGACCTACCCTGGACCGAGTGGACCACGACCTCGTCCATTTAAACCCGGCTGACTCAGTGCAGGGACAGCC TGCACAGTGGTCCAGGGTCCAGCCCTCCGCCAGCCCTGTTCCGCCTCACTGGGTGTGGCCTGGCTTCTGGGACAGGCACCATGCTGGGCC GGGGTGTGGAATCACCGGGAACGCCCCCGCCCCCGCCCCGCTGCTCCCGGTGTGCAGCGGGTGCGGGTGCCGCTTAAACATTTCCCTGCT GAGTGGCTCGTGTTTCACAGTGGGCGGCTTCCCTGCGACGGAGGCAGGACCAGGCATTTAGCTAGTTAGAGACTCGCCTGGGAAATTGCT CCATTCCTGAGTAAACAGATATTTTCGCCCACCTAAAGGGAAGCCCTGACAACAACTATCACCAAAAGACGAGGCGGCAAAGATCCAGCG GGGCTTCTGGGCGCCGGTTCCACGTGGGGTGGAATTATTAGCACCAGCTTGCTTCTCTGCCGGTGGGGCCAGCGCTGAACAGACCGGGGT GGAGTCAGGGCTGTGCTTTCCGCGTGGTTCTGCCACTTAGGGAGTGTGCCTTGGGCGGGCCATTTCACATTCCTGACCCTCACTTTTCTC ATCTGTAAAACCAGGCTGATGCCGTGCGGGCTAATGAGCCAATAAAGCTCACACTTGGGCTGGCACCCACTGGAGGTGGG >5437_5437_3_APLP1-DBH_APLP1_chr19_36365771_ENST00000586861_DBH_chr9_136523438_ENST00000393056_length(amino acids)=485AA_BP=441 MEAVCGAWGRGLVLGVSVLGSKEPGDPHANAPRALTVLSTSIQAPGSAQVAGLCGRLTLHRDLRTGRWEPDPQRSRRCLRDPQRVLEYCR QMYPELQIARVEQATQAIPMERWCGGSRSGSCAHPHHQVVPFRCLPGEFVSEALLVPEGCRFLHQERMDQCESSTRRHQEAQEACSSQGL ILHGSGMLLPCGSDRFRGVEYVCCPPPGTPDPSGTAVGDPSTRSWPPGSRVEGAEDEEEEESFPQPVDDYFVEPPQAEEEEETVPPPSSH TLAVVGKVTPTPRPTDGVDIYFGMPGEISEHEGFLRAKMDLEERRMRQINEVMREWAMADNQSKNLPKADRQALNEHFQSILQTLEEQVS GERQRLVETHATRVIALINDQRRAALEGFLAALQADPPQAERVLLALRRYLRAEQKEQRHTLRHYQHVAAVDPEKAQQMRFQGEWNLQPL PKVISTLEEPTPQCPTSQGRSPAGPTVVSIGGGKG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for APLP1-DBH |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 632_649 | 448.0 | 652.0 | DAB1 |
| Hgene | APLP1 | chr19:36365771 | chr9:136523438 | ENST00000221891 | + | 10 | 17 | 636_650 | 448.0 | 652.0 | DAB2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for APLP1-DBH |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Hgene | APLP1 | P51693 | DB14548 | Zinc sulfate, unspecified form | Ligand | Small molecule | Approved|Experimental |
| Hgene | APLP1 | P51693 | DB01593 | Zinc | Cofactor | Small molecule | Approved|Investigational |
| Hgene | APLP1 | P51693 | DB09130 | Copper | Cofactor | Small molecule | Approved|Investigational |
| Hgene | APLP1 | P51693 | DB14487 | Zinc acetate | Small molecule | Approved|Investigational | |
| Hgene | APLP1 | P51693 | DB14533 | Zinc chloride | Ligand | Small molecule | Approved|Investigational |
| Tgene | DBH | P09172 | DB00822 | Disulfiram | Inhibitor | Small molecule | Approved |
| Tgene | DBH | P09172 | DB00988 | Dopamine | Ligand | Small molecule | Approved |
| Tgene | DBH | P09172 | DB00126 | Ascorbic acid | Cofactor | Small molecule | Approved|Nutraceutical |
Top |
Related Diseases for APLP1-DBH |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C0001973 | Alcoholic Intoxication, Chronic | 5 | PSYGENET | |
| Tgene | C0005586 | Bipolar Disorder | 5 | PSYGENET | |
| Tgene | C0011570 | Mental Depression | 5 | PSYGENET | |
| Tgene | C0011581 | Depressive disorder | 5 | PSYGENET | |
| Tgene | C0600427 | Cocaine Dependence | 5 | PSYGENET | |
| Tgene | C0525045 | Mood Disorders | 4 | PSYGENET | |
| Tgene | C4746777 | ORTHOSTATIC HYPOTENSION 1 | 4 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0041696 | Unipolar Depression | 3 | PSYGENET | |
| Tgene | C1269683 | Major Depressive Disorder | 3 | PSYGENET | |
| Tgene | C1306067 | Drug-induced paranoid state | 3 | PSYGENET | |
| Tgene | C1456784 | Paranoia | 3 | CTD_human | |
| Tgene | C0027819 | Neuroblastoma | 1 | CTD_human | |
| Tgene | C0031511 | Pheochromocytoma | 1 | CTD_human | |
| Tgene | C1257877 | Pheochromocytoma, Extra-Adrenal | 1 | CTD_human | |
| Tgene | C2239176 | Liver carcinoma | 1 | CTD_human |
