
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ANKRD42-PICALM (FusionGDB2 ID:HG338699TG8301) |
Fusion Gene Summary for ANKRD42-PICALM |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ANKRD42-PICALM | Fusion gene ID: hg338699tg8301 | Hgene | Tgene | Gene symbol | ANKRD42 | PICALM | Gene ID | 338699 | 8301 |
| Gene name | ankyrin repeat domain 42 | phosphatidylinositol binding clathrin assembly protein | |
| Synonyms | PPP1R79|SARP | CALM|CLTH|LAP | |
| Cytomap | ('ANKRD42')('PICALM') 11q14.1 | 11q14.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ankyrin repeat domain-containing protein 42protein phosphatase 1, regulatory subunit 79several ankyrin repeat protein | phosphatidylinositol-binding clathrin assembly proteinclathrin assembly lymphoid myeloid leukemia protein | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q8N9B4 | Q13492 | |
| Ensembl transtripts involved in fusion gene | ENST00000260047, ENST00000393389, ENST00000526731, ENST00000531895, ENST00000533342, ENST00000528722, ENST00000393392, ENST00000528190, | ||
| Fusion gene scores | * DoF score | 6 X 6 X 3=108 | 35 X 26 X 12=10920 |
| # samples | 6 | 44 | |
| ** MAII score | log2(6/108*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(44/10920*10)=-4.63332552228256 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ANKRD42 [Title/Abstract] AND PICALM [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ANKRD42(82909684)-PICALM(85670103), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ANKRD42-PICALM seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ANKRD42-PICALM seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ANKRD42-PICALM seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. ANKRD42-PICALM seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | PICALM | GO:0006897 | endocytosis | 22118466 |
| Tgene | PICALM | GO:0006898 | receptor-mediated endocytosis | 10436022 |
| Tgene | PICALM | GO:0032880 | regulation of protein localization | 10436022 |
| Tgene | PICALM | GO:0045893 | positive regulation of transcription, DNA-templated | 11425879 |
| Tgene | PICALM | GO:0048261 | negative regulation of receptor-mediated endocytosis | 10436022 |
| Tgene | PICALM | GO:1905224 | clathrin-coated pit assembly | 16262731 |
 Fusion gene breakpoints across ANKRD42 (5'-gene) Fusion gene breakpoints across ANKRD42 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
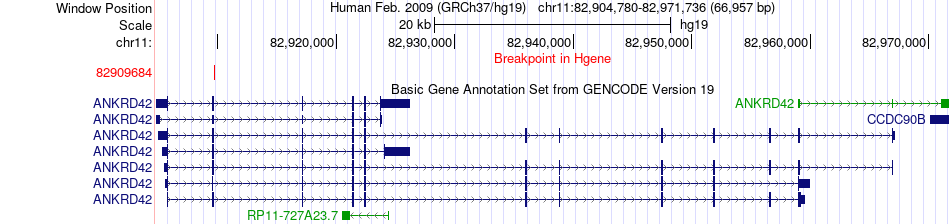 |
 Fusion gene breakpoints across PICALM (3'-gene) Fusion gene breakpoints across PICALM (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
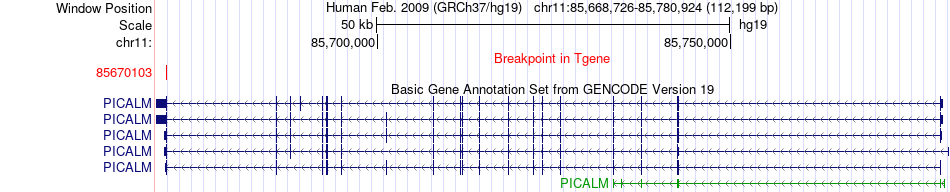 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CV-7422-01A | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
Top |
Fusion Gene ORF analysis for ANKRD42-PICALM |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000260047 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5CDS-intron | ENST00000393389 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5CDS-intron | ENST00000526731 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5CDS-intron | ENST00000531895 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5CDS-intron | ENST00000533342 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5UTR-3CDS | ENST00000528722 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5UTR-3CDS | ENST00000528722 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5UTR-3CDS | ENST00000528722 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5UTR-3CDS | ENST00000528722 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5UTR-3CDS | ENST00000528722 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| 5UTR-intron | ENST00000528722 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000260047 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000260047 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000260047 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000393389 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000393389 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000393389 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000526731 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000526731 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000526731 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000531895 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000531895 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000531895 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000533342 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000533342 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| Frame-shift | ENST00000533342 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000260047 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000260047 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000393389 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000393389 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000526731 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000526731 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000531895 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000531895 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000533342 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| In-frame | ENST00000533342 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000393392 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000393392 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000393392 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000393392 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000393392 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000528190 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000528190 | ENST00000393346 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000528190 | ENST00000526033 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000528190 | ENST00000528398 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-3CDS | ENST00000528190 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-intron | ENST00000393392 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
| intron-intron | ENST00000528190 | ENST00000528411 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000393389 | ANKRD42 | chr11 | 82909684 | + | ENST00000532317 | PICALM | chr11 | 85670103 | - | 2531 | 1154 | 32 | 1168 | 378 |
| ENST00000393389 | ANKRD42 | chr11 | 82909684 | + | ENST00000356360 | PICALM | chr11 | 85670103 | - | 1228 | 1154 | 32 | 1168 | 378 |
| ENST00000260047 | ANKRD42 | chr11 | 82909684 | + | ENST00000532317 | PICALM | chr11 | 85670103 | - | 2394 | 1017 | 150 | 1031 | 293 |
| ENST00000260047 | ANKRD42 | chr11 | 82909684 | + | ENST00000356360 | PICALM | chr11 | 85670103 | - | 1091 | 1017 | 150 | 1031 | 293 |
| ENST00000526731 | ANKRD42 | chr11 | 82909684 | + | ENST00000532317 | PICALM | chr11 | 85670103 | - | 2021 | 644 | 576 | 205 | 123 |
| ENST00000526731 | ANKRD42 | chr11 | 82909684 | + | ENST00000356360 | PICALM | chr11 | 85670103 | - | 718 | 644 | 576 | 205 | 123 |
| ENST00000531895 | ANKRD42 | chr11 | 82909684 | + | ENST00000532317 | PICALM | chr11 | 85670103 | - | 1872 | 495 | 427 | 56 | 123 |
| ENST00000531895 | ANKRD42 | chr11 | 82909684 | + | ENST00000356360 | PICALM | chr11 | 85670103 | - | 569 | 495 | 427 | 56 | 123 |
| ENST00000533342 | ANKRD42 | chr11 | 82909684 | + | ENST00000532317 | PICALM | chr11 | 85670103 | - | 1625 | 248 | 26 | 262 | 78 |
| ENST00000533342 | ANKRD42 | chr11 | 82909684 | + | ENST00000356360 | PICALM | chr11 | 85670103 | - | 322 | 248 | 26 | 262 | 78 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000393389 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.7582044 | 0.24179566 |
| ENST00000393389 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.43496865 | 0.56503135 |
| ENST00000260047 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.68972266 | 0.31027734 |
| ENST00000260047 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.38373676 | 0.6162633 |
| ENST00000526731 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.7190709 | 0.28092906 |
| ENST00000526731 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.43616393 | 0.5638361 |
| ENST00000531895 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.31233755 | 0.6876624 |
| ENST00000531895 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.1829078 | 0.8170922 |
| ENST00000533342 | ENST00000532317 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.30648735 | 0.6935127 |
| ENST00000533342 | ENST00000356360 | ANKRD42 | chr11 | 82909684 | + | PICALM | chr11 | 85670103 | - | 0.15115194 | 0.848848 |
Top |
Fusion Genomic Features for ANKRD42-PICALM |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
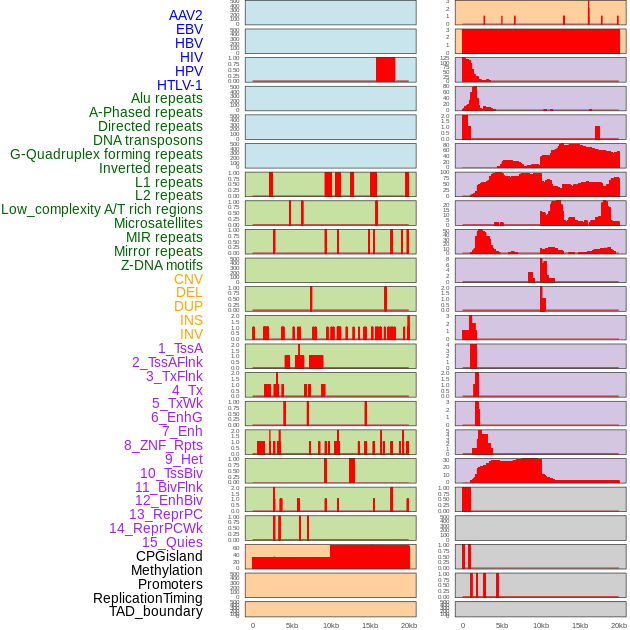 |
Top |
Fusion Protein Features for ANKRD42-PICALM |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:82909684/chr11:85670103) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ANKRD42 | PICALM |
| FUNCTION: Cytoplasmic adapter protein that plays a critical role in clathrin-mediated endocytosis which is important in processes such as internalization of cell receptors, synaptic transmission or removal of apoptotic cells. Recruits AP-2 and attaches clathrin triskelions to the cytoplasmic side of plasma membrane leading to clathrin-coated vesicles (CCVs) assembly (PubMed:10436022, PubMed:16262731, PubMed:27574975). Furthermore, regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature (PubMed:25898166). In addition to binding to clathrin, mediates the endocytosis of small R-SNARES (Soluble NSF Attachment Protein REceptors) between plasma membranes and endosomes including VAMP2, VAMP3, VAMP4, VAMP7 or VAMP8 (PubMed:22118466, PubMed:21808019, PubMed:23741335). In turn, PICALM-dependent SNARE endocytosis is required for the formation and maturation of autophagic precursors (PubMed:25241929). Modulates thereby autophagy and the turnover of autophagy substrates such as MAPT/TAU or amyloid precursor protein cleaved C-terminal fragment (APP-CTF) (PubMed:25241929, PubMed:24067654). {ECO:0000269|PubMed:10436022, ECO:0000269|PubMed:16262731, ECO:0000269|PubMed:21808019, ECO:0000269|PubMed:22118466, ECO:0000269|PubMed:23741335, ECO:0000269|PubMed:24067654, ECO:0000269|PubMed:25241929, ECO:0000269|PubMed:25898166, ECO:0000269|PubMed:27574975}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 25_60 | 74 | 199.0 | Repeat | Note=ANK 1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 130_159 | 74 | 199.0 | Repeat | Note=ANK 4 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 163_192 | 74 | 199.0 | Repeat | Note=ANK 5 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 200_232 | 74 | 199.0 | Repeat | Note=ANK 6 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 235_265 | 74 | 199.0 | Repeat | Note=ANK 7 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 269_298 | 74 | 199.0 | Repeat | Note=ANK 8 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 302_332 | 74 | 199.0 | Repeat | Note=ANK 9 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 64_93 | 74 | 199.0 | Repeat | Note=ANK 2 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393389 | + | 2 | 6 | 97_126 | 74 | 199.0 | Repeat | Note=ANK 3 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 130_159 | 0 | 390.0 | Repeat | Note=ANK 4 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 163_192 | 0 | 390.0 | Repeat | Note=ANK 5 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 200_232 | 0 | 390.0 | Repeat | Note=ANK 6 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 235_265 | 0 | 390.0 | Repeat | Note=ANK 7 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 25_60 | 0 | 390.0 | Repeat | Note=ANK 1 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 269_298 | 0 | 390.0 | Repeat | Note=ANK 8 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 302_332 | 0 | 390.0 | Repeat | Note=ANK 9 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 64_93 | 0 | 390.0 | Repeat | Note=ANK 2 |
| Hgene | ANKRD42 | chr11:82909684 | chr11:85670103 | ENST00000393392 | + | 1 | 10 | 97_126 | 0 | 390.0 | Repeat | Note=ANK 3 |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000356360 | 17 | 19 | 14_145 | 628 | 633.0 | Domain | ENTH | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000393346 | 18 | 20 | 14_145 | 648 | 653.0 | Domain | ENTH | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000526033 | 18 | 20 | 14_145 | 641 | 646.0 | Domain | ENTH | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000528398 | 17 | 19 | 14_145 | 547 | 552.0 | Domain | ENTH | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000532317 | 18 | 20 | 14_145 | 606 | 611.0 | Domain | ENTH |
Top |
Fusion Gene Sequence for ANKRD42-PICALM |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >4731_4731_1_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000260047_PICALM_chr11_85670103_ENST00000356360_length(transcript)=1091nt_BP=1017nt TGGAGGATAAACGGTCTACACGGCCATTCCGGCGCCGAGTCTAGGGAAAGAGTTAGCGACGACGGGGAAAGAAAATGTGAAGAGAGCGAC CGCCGCTCCAGGGTCGCTGCAGGAAGCCTAAGTGCAGACGCCGGCTTCTCCCGCAGTGACTTGAGAAGGGTCAGTGAAAACCTCGGCCAC TGCCGCAGCGTCTCTAGGGAGAGAGTTAGGGGAGATAGTGGCCACAGTCACAGCTGCTCTTGGGAGAGAGTTAGGGGAGACAGCACCTTC TGCAGCAGCGACGTGAATTTTAGTGAAGTTGGAGGCCACCAAACTACCGACTCCAGGGGAACAGCCAGAGAAGACCGAGGCCTCCGCCTC AGTGGTCCTTGGGAGGGAGTCAGTGACATTCGGGACCCGCGAACTAGTGACTTCGGGGATAGAGTCAGTGACGATCGCAGTCGCCGCTTC AGTGGCTCCTGGGAGGGAGGGAGTGTCGAAGGCGGCCACAGCGTTGGTAGTTCTTGGGAGGAAGTAAGTGGAGACCGCGGCTACGCAGCC AGCGACTCCTCTGGTGTGAGCGGCAGTGAAGACGCCAGCTACCGCTTCAGTGGCTTTTGGGAGAGAGAAAGTGAAGACGAAGGTTTCCGC TGCAGCTTCTGGGAGAGAGCAAGAGAGGACCTTGGGCCCCGTCCTAGTGACGACGGAGAAGAGGGCCGCTGCCGCTGCAGTGGCTCGTGG GTGAGAGCAAGTGAAGACCGCCGCAGCATCAGGGGCCTGGACTCAACTCCTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCC AATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCAGGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGA GCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCATTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACAT TGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAACTTGATGGAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCA GCAAAATCCTT >4731_4731_1_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000260047_PICALM_chr11_85670103_ENST00000356360_length(amino acids)=293AA_BP= MRRVSENLGHCRSVSRERVRGDSGHSHSCSWERVRGDSTFCSSDVNFSEVGGHQTTDSRGTAREDRGLRLSGPWEGVSDIRDPRTSDFGD RVSDDRSRRFSGSWEGGSVEGGHSVGSSWEEVSGDRGYAASDSSGVSGSEDASYRFSGFWERESEDEGFRCSFWERAREDLGPRPSDDGE EGRCRCSGSWVRASEDRRSIRGLDSTPPQSRRCCAMPGVANSGPSTSSRETANPCSRKKVHFGSIHDAVRAGDVKQLSEIVVRGASINEL DVLHKFTPLHWAAHSGSLEIQFM -------------------------------------------------------------- >4731_4731_2_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000260047_PICALM_chr11_85670103_ENST00000532317_length(transcript)=2394nt_BP=1017nt TGGAGGATAAACGGTCTACACGGCCATTCCGGCGCCGAGTCTAGGGAAAGAGTTAGCGACGACGGGGAAAGAAAATGTGAAGAGAGCGAC CGCCGCTCCAGGGTCGCTGCAGGAAGCCTAAGTGCAGACGCCGGCTTCTCCCGCAGTGACTTGAGAAGGGTCAGTGAAAACCTCGGCCAC TGCCGCAGCGTCTCTAGGGAGAGAGTTAGGGGAGATAGTGGCCACAGTCACAGCTGCTCTTGGGAGAGAGTTAGGGGAGACAGCACCTTC TGCAGCAGCGACGTGAATTTTAGTGAAGTTGGAGGCCACCAAACTACCGACTCCAGGGGAACAGCCAGAGAAGACCGAGGCCTCCGCCTC AGTGGTCCTTGGGAGGGAGTCAGTGACATTCGGGACCCGCGAACTAGTGACTTCGGGGATAGAGTCAGTGACGATCGCAGTCGCCGCTTC AGTGGCTCCTGGGAGGGAGGGAGTGTCGAAGGCGGCCACAGCGTTGGTAGTTCTTGGGAGGAAGTAAGTGGAGACCGCGGCTACGCAGCC AGCGACTCCTCTGGTGTGAGCGGCAGTGAAGACGCCAGCTACCGCTTCAGTGGCTTTTGGGAGAGAGAAAGTGAAGACGAAGGTTTCCGC TGCAGCTTCTGGGAGAGAGCAAGAGAGGACCTTGGGCCCCGTCCTAGTGACGACGGAGAAGAGGGCCGCTGCCGCTGCAGTGGCTCGTGG GTGAGAGCAAGTGAAGACCGCCGCAGCATCAGGGGCCTGGACTCAACTCCTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCC AATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCAGGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGA GCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCATTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACAT TGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAACTTGATGGAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCA GCAAAATCCTTACTTCCAGCAAAATCCAAACTGCTGTCTCTTAAATCTCTTAAACTCTCTTCTTCCATTAGAATGCTACAAGTAACTCAG TGAAGGCCCATGAAGGAAATTGGGACTAGTTTATAGGAGAACGTATCAATACAGTTTATAAAGCCAAGAATTGCTATGATTTAAGACTAA GATCTGTCTTTTTGGTGACTAACCCTTCAATTCTTTCAACTCCTGTTAATACCCATAATCAGTAACCTATCAAGAAAAGCCCTTATTTGG AAAGTGTGAAATTTGTATTTGGAAAAGCTGCCTGGAGAGAAGAACTGTGTCCTTTACTGTATTTCAACAGGACTCTTTTGGGGGATCAAA ATTAAAATTCCTAATTATGCATTATCTTTCTTTTCTCCAGTCCTCACAAATACAGAAACAATAACTGAAATTAACTTTTCTTTTTTTAAA AAAAATTATATTCAGTTTGCAGTAGACATTCCTTAAGTATTTGTATTTATTTATGATTATCAATTTTACATAACATTAATATTGTATCAG ACCTCCTTATGAAAATGAGTATGGATGTGCACAGTATGTTTGATTTTTATCCACAAGAATGAATCTGATTCAGAATGCTTTTCTCAGCTG ACATACAGAGCACTAAATATTTTAAGGCAAGTCCATAGGTCTGAATCTCTTAAGAATTCTCGGCCTCTGTGGGATTTAGGGAAGCATTAT AAATGCATTAATCCTTATAGTCAATTCTGTGCCTAGGATTTTGCCAGGGAACAGTTCACTGACTAGGAAAAGCACTACATTTTAAATTCA GCATTAGTGCATTGGGAAGGATCTTTACTGCTTTGTGCTTGGCATGTCATTATTTTCCATTTGACATTAGGGCCTTTCCAAAATGAATGT GAGGAATTGCTTTCACTTCAAGACTTTCCTTCTTTTCACTAAAACTCTAGAAGGTGTTACAAGGGGGAGGGAAGGGGGGCAAAGTCCTTG AACATTTTCTTTGGCTCGTGCCATGTTATGATCATATACCTTTTAAATAAGGGGAAATAGTATCTTTAAAGTTAATGTCTAGCCAAGAGT TTAGTAAACGAAGAATTAAACTGCACTGTTGATCGGTGCTTTGTGTAAATACATCTTTAACATTTGGGTGGAGAGGGGCCTTAAGAAGGA CAGTTCATTGTAGGAAAGCAATTCTGTACATGAGTTTAAGCATTCTTGTTGCATTGTCTCTGCAGATTCTATTTTTGTTTACAATATTAA AATGTATGTTAGCAAAATGGGTGGATTTTCAAATAAAATGCAGCTTCCACAAAA >4731_4731_2_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000260047_PICALM_chr11_85670103_ENST00000532317_length(amino acids)=293AA_BP= MRRVSENLGHCRSVSRERVRGDSGHSHSCSWERVRGDSTFCSSDVNFSEVGGHQTTDSRGTAREDRGLRLSGPWEGVSDIRDPRTSDFGD RVSDDRSRRFSGSWEGGSVEGGHSVGSSWEEVSGDRGYAASDSSGVSGSEDASYRFSGFWERESEDEGFRCSFWERAREDLGPRPSDDGE EGRCRCSGSWVRASEDRRSIRGLDSTPPQSRRCCAMPGVANSGPSTSSRETANPCSRKKVHFGSIHDAVRAGDVKQLSEIVVRGASINEL DVLHKFTPLHWAAHSGSLEIQFM -------------------------------------------------------------- >4731_4731_3_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000393389_PICALM_chr11_85670103_ENST00000356360_length(transcript)=1228nt_BP=1154nt GCCTCTCCAGCCAAGTGGCTGGAGTCGGGAGGCTGGAAAGAGACTCCGAGAAAGTACCAGCGGAAGGCGGCCGCCGCTACGGCGATTCGC AGGGAGTAGCAGACGAAGACGGTGGCCGCCGCACTAGCCACCACGTGTGGAGGATAAACGGTCTACACGGCCATTCCGGCGCCGAGTCTA GGGAAAGAGTTAGCGACGACGGGGAAAGAAAATGTGAAGAGAGCGACCGCCGCTCCAGGGTCGCTGCAGGAAGCCTAAGTGCAGACGCCG GCTTCTCCCGCAGTGACTTGAGAAGGGTCAGTGAAAACCTCGGCCACTGCCGCAGCGTCTCTAGGGAGAGAGTTAGGGGAGATAGTGGCC ACAGTCACAGCTGCTCTTGGGAGAGAGTTAGGGGAGACAGCACCTTCTGCAGCAGCGACGTGAATTTTAGTGAAGTTGGAGGCCACCAAA CTACCGACTCCAGGGGAACAGCCAGAGAAGACCGAGGCCTCCGCCTCAGTGGTCCTTGGGAGGGAGTCAGTGACATTCGGGACCCGCGAA CTAGTGACTTCGGGGATAGAGTCAGTGACGATCGCAGTCGCCGCTTCAGTGGCTCCTGGGAGGGAGGGAGTGTCGAAGGCGGCCACAGCG TTGGTAGTTCTTGGGAGGAAGTAAGTGGAGACCGCGGCTACGCAGCCAGCGACTCCTCTGGTGTGAGCGGCAGTGAAGACGCCAGCTACC GCTTCAGTGGCTTTTGGGAGAGAGAAAGTGAAGACGAAGGTTTCCGCTGCAGCTTCTGGGAGAGAGCAAGAGAGGACCTTGGGCCCCGTC CTAGTGACGACGGAGAAGAGGGCCGCTGCCGCTGCAGTGGCTCGTGGGTGAGAGCAAGTGAAGACCGCCGCAGCATCAGGGGCCTGGACT CAACTCCTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCCAATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCT GTTCCAGGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGAGCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAG CCAGCATTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACATTGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAAC TTGATGGAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCAGCAAAATCCTT >4731_4731_3_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000393389_PICALM_chr11_85670103_ENST00000356360_length(amino acids)=378AA_BP= MERDSEKVPAEGGRRYGDSQGVADEDGGRRTSHHVWRINGLHGHSGAESRERVSDDGERKCEESDRRSRVAAGSLSADAGFSRSDLRRVS ENLGHCRSVSRERVRGDSGHSHSCSWERVRGDSTFCSSDVNFSEVGGHQTTDSRGTAREDRGLRLSGPWEGVSDIRDPRTSDFGDRVSDD RSRRFSGSWEGGSVEGGHSVGSSWEEVSGDRGYAASDSSGVSGSEDASYRFSGFWERESEDEGFRCSFWERAREDLGPRPSDDGEEGRCR CSGSWVRASEDRRSIRGLDSTPPQSRRCCAMPGVANSGPSTSSRETANPCSRKKVHFGSIHDAVRAGDVKQLSEIVVRGASINELDVLHK FTPLHWAAHSGSLEIQFM -------------------------------------------------------------- >4731_4731_4_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000393389_PICALM_chr11_85670103_ENST00000532317_length(transcript)=2531nt_BP=1154nt GCCTCTCCAGCCAAGTGGCTGGAGTCGGGAGGCTGGAAAGAGACTCCGAGAAAGTACCAGCGGAAGGCGGCCGCCGCTACGGCGATTCGC AGGGAGTAGCAGACGAAGACGGTGGCCGCCGCACTAGCCACCACGTGTGGAGGATAAACGGTCTACACGGCCATTCCGGCGCCGAGTCTA GGGAAAGAGTTAGCGACGACGGGGAAAGAAAATGTGAAGAGAGCGACCGCCGCTCCAGGGTCGCTGCAGGAAGCCTAAGTGCAGACGCCG GCTTCTCCCGCAGTGACTTGAGAAGGGTCAGTGAAAACCTCGGCCACTGCCGCAGCGTCTCTAGGGAGAGAGTTAGGGGAGATAGTGGCC ACAGTCACAGCTGCTCTTGGGAGAGAGTTAGGGGAGACAGCACCTTCTGCAGCAGCGACGTGAATTTTAGTGAAGTTGGAGGCCACCAAA CTACCGACTCCAGGGGAACAGCCAGAGAAGACCGAGGCCTCCGCCTCAGTGGTCCTTGGGAGGGAGTCAGTGACATTCGGGACCCGCGAA CTAGTGACTTCGGGGATAGAGTCAGTGACGATCGCAGTCGCCGCTTCAGTGGCTCCTGGGAGGGAGGGAGTGTCGAAGGCGGCCACAGCG TTGGTAGTTCTTGGGAGGAAGTAAGTGGAGACCGCGGCTACGCAGCCAGCGACTCCTCTGGTGTGAGCGGCAGTGAAGACGCCAGCTACC GCTTCAGTGGCTTTTGGGAGAGAGAAAGTGAAGACGAAGGTTTCCGCTGCAGCTTCTGGGAGAGAGCAAGAGAGGACCTTGGGCCCCGTC CTAGTGACGACGGAGAAGAGGGCCGCTGCCGCTGCAGTGGCTCGTGGGTGAGAGCAAGTGAAGACCGCCGCAGCATCAGGGGCCTGGACT CAACTCCTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCCAATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCT GTTCCAGGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGAGCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAG CCAGCATTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACATTGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAAC TTGATGGAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCAGCAAAATCCTTACTTCCAGCAAAATCCAAACTGCTGTCTCTTA AATCTCTTAAACTCTCTTCTTCCATTAGAATGCTACAAGTAACTCAGTGAAGGCCCATGAAGGAAATTGGGACTAGTTTATAGGAGAACG TATCAATACAGTTTATAAAGCCAAGAATTGCTATGATTTAAGACTAAGATCTGTCTTTTTGGTGACTAACCCTTCAATTCTTTCAACTCC TGTTAATACCCATAATCAGTAACCTATCAAGAAAAGCCCTTATTTGGAAAGTGTGAAATTTGTATTTGGAAAAGCTGCCTGGAGAGAAGA ACTGTGTCCTTTACTGTATTTCAACAGGACTCTTTTGGGGGATCAAAATTAAAATTCCTAATTATGCATTATCTTTCTTTTCTCCAGTCC TCACAAATACAGAAACAATAACTGAAATTAACTTTTCTTTTTTTAAAAAAAATTATATTCAGTTTGCAGTAGACATTCCTTAAGTATTTG TATTTATTTATGATTATCAATTTTACATAACATTAATATTGTATCAGACCTCCTTATGAAAATGAGTATGGATGTGCACAGTATGTTTGA TTTTTATCCACAAGAATGAATCTGATTCAGAATGCTTTTCTCAGCTGACATACAGAGCACTAAATATTTTAAGGCAAGTCCATAGGTCTG AATCTCTTAAGAATTCTCGGCCTCTGTGGGATTTAGGGAAGCATTATAAATGCATTAATCCTTATAGTCAATTCTGTGCCTAGGATTTTG CCAGGGAACAGTTCACTGACTAGGAAAAGCACTACATTTTAAATTCAGCATTAGTGCATTGGGAAGGATCTTTACTGCTTTGTGCTTGGC ATGTCATTATTTTCCATTTGACATTAGGGCCTTTCCAAAATGAATGTGAGGAATTGCTTTCACTTCAAGACTTTCCTTCTTTTCACTAAA ACTCTAGAAGGTGTTACAAGGGGGAGGGAAGGGGGGCAAAGTCCTTGAACATTTTCTTTGGCTCGTGCCATGTTATGATCATATACCTTT TAAATAAGGGGAAATAGTATCTTTAAAGTTAATGTCTAGCCAAGAGTTTAGTAAACGAAGAATTAAACTGCACTGTTGATCGGTGCTTTG TGTAAATACATCTTTAACATTTGGGTGGAGAGGGGCCTTAAGAAGGACAGTTCATTGTAGGAAAGCAATTCTGTACATGAGTTTAAGCAT TCTTGTTGCATTGTCTCTGCAGATTCTATTTTTGTTTACAATATTAAAATGTATGTTAGCAAAATGGGTGGATTTTCAAATAAAATGCAG CTTCCACAAAA >4731_4731_4_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000393389_PICALM_chr11_85670103_ENST00000532317_length(amino acids)=378AA_BP= MERDSEKVPAEGGRRYGDSQGVADEDGGRRTSHHVWRINGLHGHSGAESRERVSDDGERKCEESDRRSRVAAGSLSADAGFSRSDLRRVS ENLGHCRSVSRERVRGDSGHSHSCSWERVRGDSTFCSSDVNFSEVGGHQTTDSRGTAREDRGLRLSGPWEGVSDIRDPRTSDFGDRVSDD RSRRFSGSWEGGSVEGGHSVGSSWEEVSGDRGYAASDSSGVSGSEDASYRFSGFWERESEDEGFRCSFWERAREDLGPRPSDDGEEGRCR CSGSWVRASEDRRSIRGLDSTPPQSRRCCAMPGVANSGPSTSSRETANPCSRKKVHFGSIHDAVRAGDVKQLSEIVVRGASINELDVLHK FTPLHWAAHSGSLEIQFM -------------------------------------------------------------- >4731_4731_5_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000526731_PICALM_chr11_85670103_ENST00000356360_length(transcript)=718nt_BP=644nt AGGGAGTCAGTGACATTCGGGACCCGCGAACTAGTGACTTCGGGGATAGAGTCAGTGACGATCGCAGTCGCCGCTTCAGTGGCTCCTGGG AGGGAGGGAGTGTCGAAGGCGGCCACAGCGTTGGTAGTTCTTGGGAGGAAGTAAGTGGAGACCGCGGCTACGCAGCCAGCGACTCCTCTG GTGTGAGCGGCAGTGAAGACGCCAGCTACCGCTTCAGTGGCTTTTGGGAGAGAGAAAGTGAAGACGAAGGTTTCCGCTGCAGCTTCTGGG AGAGAGCAAGAGAGGACCTTGGGCCCCGTCCTAGTGACGACGGAGAAGAGGGCCGCTGCCGCTGCAGTGGCTCGTGGGTGAGAGCAAGTG AAGACCGCCGCAGCATCAGGGGCCTGGACTCAACTCCTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCCAATTCAGGCCCCT CCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCAGGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGAGCTGGAGATGTAA AGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCATTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACATTGGGCAGCACATT CTGGAAGTTTGGAGATACAGTTTATGTAACTTGATGGAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCAGCAAAATCCTT >4731_4731_5_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000526731_PICALM_chr11_85670103_ENST00000356360_length(amino acids)=123AA_BP= MLAPRTTISESCFTSPARTASCMLPKCTFFLEQGFAVSLEEVEGPELATPGMAQHLRLWGGVESRPLMLRRSSLALTHEPLQRQRPSSPS SLGRGPRSSLALSQKLQRKPSSSLSLSQKPLKR -------------------------------------------------------------- >4731_4731_6_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000526731_PICALM_chr11_85670103_ENST00000532317_length(transcript)=2021nt_BP=644nt AGGGAGTCAGTGACATTCGGGACCCGCGAACTAGTGACTTCGGGGATAGAGTCAGTGACGATCGCAGTCGCCGCTTCAGTGGCTCCTGGG AGGGAGGGAGTGTCGAAGGCGGCCACAGCGTTGGTAGTTCTTGGGAGGAAGTAAGTGGAGACCGCGGCTACGCAGCCAGCGACTCCTCTG GTGTGAGCGGCAGTGAAGACGCCAGCTACCGCTTCAGTGGCTTTTGGGAGAGAGAAAGTGAAGACGAAGGTTTCCGCTGCAGCTTCTGGG AGAGAGCAAGAGAGGACCTTGGGCCCCGTCCTAGTGACGACGGAGAAGAGGGCCGCTGCCGCTGCAGTGGCTCGTGGGTGAGAGCAAGTG AAGACCGCCGCAGCATCAGGGGCCTGGACTCAACTCCTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCCAATTCAGGCCCCT CCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCAGGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGAGCTGGAGATGTAA AGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCATTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACATTGGGCAGCACATT CTGGAAGTTTGGAGATACAGTTTATGTAACTTGATGGAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCAGCAAAATCCTTAC TTCCAGCAAAATCCAAACTGCTGTCTCTTAAATCTCTTAAACTCTCTTCTTCCATTAGAATGCTACAAGTAACTCAGTGAAGGCCCATGA AGGAAATTGGGACTAGTTTATAGGAGAACGTATCAATACAGTTTATAAAGCCAAGAATTGCTATGATTTAAGACTAAGATCTGTCTTTTT GGTGACTAACCCTTCAATTCTTTCAACTCCTGTTAATACCCATAATCAGTAACCTATCAAGAAAAGCCCTTATTTGGAAAGTGTGAAATT TGTATTTGGAAAAGCTGCCTGGAGAGAAGAACTGTGTCCTTTACTGTATTTCAACAGGACTCTTTTGGGGGATCAAAATTAAAATTCCTA ATTATGCATTATCTTTCTTTTCTCCAGTCCTCACAAATACAGAAACAATAACTGAAATTAACTTTTCTTTTTTTAAAAAAAATTATATTC AGTTTGCAGTAGACATTCCTTAAGTATTTGTATTTATTTATGATTATCAATTTTACATAACATTAATATTGTATCAGACCTCCTTATGAA AATGAGTATGGATGTGCACAGTATGTTTGATTTTTATCCACAAGAATGAATCTGATTCAGAATGCTTTTCTCAGCTGACATACAGAGCAC TAAATATTTTAAGGCAAGTCCATAGGTCTGAATCTCTTAAGAATTCTCGGCCTCTGTGGGATTTAGGGAAGCATTATAAATGCATTAATC CTTATAGTCAATTCTGTGCCTAGGATTTTGCCAGGGAACAGTTCACTGACTAGGAAAAGCACTACATTTTAAATTCAGCATTAGTGCATT GGGAAGGATCTTTACTGCTTTGTGCTTGGCATGTCATTATTTTCCATTTGACATTAGGGCCTTTCCAAAATGAATGTGAGGAATTGCTTT CACTTCAAGACTTTCCTTCTTTTCACTAAAACTCTAGAAGGTGTTACAAGGGGGAGGGAAGGGGGGCAAAGTCCTTGAACATTTTCTTTG GCTCGTGCCATGTTATGATCATATACCTTTTAAATAAGGGGAAATAGTATCTTTAAAGTTAATGTCTAGCCAAGAGTTTAGTAAACGAAG AATTAAACTGCACTGTTGATCGGTGCTTTGTGTAAATACATCTTTAACATTTGGGTGGAGAGGGGCCTTAAGAAGGACAGTTCATTGTAG GAAAGCAATTCTGTACATGAGTTTAAGCATTCTTGTTGCATTGTCTCTGCAGATTCTATTTTTGTTTACAATATTAAAATGTATGTTAGC AAAATGGGTGGATTTTCAAATAAAATGCAGCTTCCACAAAA >4731_4731_6_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000526731_PICALM_chr11_85670103_ENST00000532317_length(amino acids)=123AA_BP= MLAPRTTISESCFTSPARTASCMLPKCTFFLEQGFAVSLEEVEGPELATPGMAQHLRLWGGVESRPLMLRRSSLALTHEPLQRQRPSSPS SLGRGPRSSLALSQKLQRKPSSSLSLSQKPLKR -------------------------------------------------------------- >4731_4731_7_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000531895_PICALM_chr11_85670103_ENST00000356360_length(transcript)=569nt_BP=495nt GACCGCGGCTACGCAGCCAGCGACTCCTCTGGTGTGAGCGGCAGTGAAGACGCCAGCTACCGCTTCAGTGGCTTTTGGGAGAGAGAAAGT GAAGACGAAGGTTTCCGCTGCAGCTTCTGGGAGAGAGCAAGAGAGGACCTTGGGCCCCGTCCTAGTGACGACGGAGAAGAGGGCCGCTGC CGCTGCAGTGGCTCGTGGGTGAGAGCAAGTGAAGACCGCCGCAGCATCAGGGGCCTGGACTCAACTCCTCCCCAGAGTCGGAGGTGTTGC GCCATGCCCGGGGTGGCCAATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCAGGAAGAAGGTGCATTTTGGCAGC ATACATGATGCAGTACGAGCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCATTAATGAACTTGATGTTCTCCAT AAGTTTACCCCTTTACATTGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAACTTGATGGAAGAAAATGGAATTACTCCAAA AAGACAAGTGCTCAAGCAGCAAAATCCTT >4731_4731_7_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000531895_PICALM_chr11_85670103_ENST00000356360_length(amino acids)=123AA_BP= MLAPRTTISESCFTSPARTASCMLPKCTFFLEQGFAVSLEEVEGPELATPGMAQHLRLWGGVESRPLMLRRSSLALTHEPLQRQRPSSPS SLGRGPRSSLALSQKLQRKPSSSLSLSQKPLKR -------------------------------------------------------------- >4731_4731_8_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000531895_PICALM_chr11_85670103_ENST00000532317_length(transcript)=1872nt_BP=495nt GACCGCGGCTACGCAGCCAGCGACTCCTCTGGTGTGAGCGGCAGTGAAGACGCCAGCTACCGCTTCAGTGGCTTTTGGGAGAGAGAAAGT GAAGACGAAGGTTTCCGCTGCAGCTTCTGGGAGAGAGCAAGAGAGGACCTTGGGCCCCGTCCTAGTGACGACGGAGAAGAGGGCCGCTGC CGCTGCAGTGGCTCGTGGGTGAGAGCAAGTGAAGACCGCCGCAGCATCAGGGGCCTGGACTCAACTCCTCCCCAGAGTCGGAGGTGTTGC GCCATGCCCGGGGTGGCCAATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCAGGAAGAAGGTGCATTTTGGCAGC ATACATGATGCAGTACGAGCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCATTAATGAACTTGATGTTCTCCAT AAGTTTACCCCTTTACATTGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAACTTGATGGAAGAAAATGGAATTACTCCAAA AAGACAAGTGCTCAAGCAGCAAAATCCTTACTTCCAGCAAAATCCAAACTGCTGTCTCTTAAATCTCTTAAACTCTCTTCTTCCATTAGA ATGCTACAAGTAACTCAGTGAAGGCCCATGAAGGAAATTGGGACTAGTTTATAGGAGAACGTATCAATACAGTTTATAAAGCCAAGAATT GCTATGATTTAAGACTAAGATCTGTCTTTTTGGTGACTAACCCTTCAATTCTTTCAACTCCTGTTAATACCCATAATCAGTAACCTATCA AGAAAAGCCCTTATTTGGAAAGTGTGAAATTTGTATTTGGAAAAGCTGCCTGGAGAGAAGAACTGTGTCCTTTACTGTATTTCAACAGGA CTCTTTTGGGGGATCAAAATTAAAATTCCTAATTATGCATTATCTTTCTTTTCTCCAGTCCTCACAAATACAGAAACAATAACTGAAATT AACTTTTCTTTTTTTAAAAAAAATTATATTCAGTTTGCAGTAGACATTCCTTAAGTATTTGTATTTATTTATGATTATCAATTTTACATA ACATTAATATTGTATCAGACCTCCTTATGAAAATGAGTATGGATGTGCACAGTATGTTTGATTTTTATCCACAAGAATGAATCTGATTCA GAATGCTTTTCTCAGCTGACATACAGAGCACTAAATATTTTAAGGCAAGTCCATAGGTCTGAATCTCTTAAGAATTCTCGGCCTCTGTGG GATTTAGGGAAGCATTATAAATGCATTAATCCTTATAGTCAATTCTGTGCCTAGGATTTTGCCAGGGAACAGTTCACTGACTAGGAAAAG CACTACATTTTAAATTCAGCATTAGTGCATTGGGAAGGATCTTTACTGCTTTGTGCTTGGCATGTCATTATTTTCCATTTGACATTAGGG CCTTTCCAAAATGAATGTGAGGAATTGCTTTCACTTCAAGACTTTCCTTCTTTTCACTAAAACTCTAGAAGGTGTTACAAGGGGGAGGGA AGGGGGGCAAAGTCCTTGAACATTTTCTTTGGCTCGTGCCATGTTATGATCATATACCTTTTAAATAAGGGGAAATAGTATCTTTAAAGT TAATGTCTAGCCAAGAGTTTAGTAAACGAAGAATTAAACTGCACTGTTGATCGGTGCTTTGTGTAAATACATCTTTAACATTTGGGTGGA GAGGGGCCTTAAGAAGGACAGTTCATTGTAGGAAAGCAATTCTGTACATGAGTTTAAGCATTCTTGTTGCATTGTCTCTGCAGATTCTAT TTTTGTTTACAATATTAAAATGTATGTTAGCAAAATGGGTGGATTTTCAAATAAAATGCAGCTTCCACAAAA >4731_4731_8_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000531895_PICALM_chr11_85670103_ENST00000532317_length(amino acids)=123AA_BP= MLAPRTTISESCFTSPARTASCMLPKCTFFLEQGFAVSLEEVEGPELATPGMAQHLRLWGGVESRPLMLRRSSLALTHEPLQRQRPSSPS SLGRGPRSSLALSQKLQRKPSSSLSLSQKPLKR -------------------------------------------------------------- >4731_4731_9_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000533342_PICALM_chr11_85670103_ENST00000356360_length(transcript)=322nt_BP=248nt CTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCCAATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCA GGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGAGCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCA TTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACATTGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAACTTGATG GAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCAGCAAAATCCTT >4731_4731_9_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000533342_PICALM_chr11_85670103_ENST00000356360_length(amino acids)=78AA_BP= MPGVANSGPSTSSRETANPCSRKKVHFGSIHDAVRAGDVKQLSEIVVRGASINELDVLHKFTPLHWAAHSGSLEIQFM -------------------------------------------------------------- >4731_4731_10_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000533342_PICALM_chr11_85670103_ENST00000532317_length(transcript)=1625nt_BP=248nt CTCCCCAGAGTCGGAGGTGTTGCGCCATGCCCGGGGTGGCCAATTCAGGCCCCTCCACTTCCTCTAGGGAGACTGCAAACCCCTGTTCCA GGAAGAAGGTGCATTTTGGCAGCATACATGATGCAGTACGAGCTGGAGATGTAAAGCAGCTTTCAGAAATAGTGGTACGTGGAGCCAGCA TTAATGAACTTGATGTTCTCCATAAGTTTACCCCTTTACATTGGGCAGCACATTCTGGAAGTTTGGAGATACAGTTTATGTAACTTGATG GAAGAAAATGGAATTACTCCAAAAAGACAAGTGCTCAAGCAGCAAAATCCTTACTTCCAGCAAAATCCAAACTGCTGTCTCTTAAATCTC TTAAACTCTCTTCTTCCATTAGAATGCTACAAGTAACTCAGTGAAGGCCCATGAAGGAAATTGGGACTAGTTTATAGGAGAACGTATCAA TACAGTTTATAAAGCCAAGAATTGCTATGATTTAAGACTAAGATCTGTCTTTTTGGTGACTAACCCTTCAATTCTTTCAACTCCTGTTAA TACCCATAATCAGTAACCTATCAAGAAAAGCCCTTATTTGGAAAGTGTGAAATTTGTATTTGGAAAAGCTGCCTGGAGAGAAGAACTGTG TCCTTTACTGTATTTCAACAGGACTCTTTTGGGGGATCAAAATTAAAATTCCTAATTATGCATTATCTTTCTTTTCTCCAGTCCTCACAA ATACAGAAACAATAACTGAAATTAACTTTTCTTTTTTTAAAAAAAATTATATTCAGTTTGCAGTAGACATTCCTTAAGTATTTGTATTTA TTTATGATTATCAATTTTACATAACATTAATATTGTATCAGACCTCCTTATGAAAATGAGTATGGATGTGCACAGTATGTTTGATTTTTA TCCACAAGAATGAATCTGATTCAGAATGCTTTTCTCAGCTGACATACAGAGCACTAAATATTTTAAGGCAAGTCCATAGGTCTGAATCTC TTAAGAATTCTCGGCCTCTGTGGGATTTAGGGAAGCATTATAAATGCATTAATCCTTATAGTCAATTCTGTGCCTAGGATTTTGCCAGGG AACAGTTCACTGACTAGGAAAAGCACTACATTTTAAATTCAGCATTAGTGCATTGGGAAGGATCTTTACTGCTTTGTGCTTGGCATGTCA TTATTTTCCATTTGACATTAGGGCCTTTCCAAAATGAATGTGAGGAATTGCTTTCACTTCAAGACTTTCCTTCTTTTCACTAAAACTCTA GAAGGTGTTACAAGGGGGAGGGAAGGGGGGCAAAGTCCTTGAACATTTTCTTTGGCTCGTGCCATGTTATGATCATATACCTTTTAAATA AGGGGAAATAGTATCTTTAAAGTTAATGTCTAGCCAAGAGTTTAGTAAACGAAGAATTAAACTGCACTGTTGATCGGTGCTTTGTGTAAA TACATCTTTAACATTTGGGTGGAGAGGGGCCTTAAGAAGGACAGTTCATTGTAGGAAAGCAATTCTGTACATGAGTTTAAGCATTCTTGT TGCATTGTCTCTGCAGATTCTATTTTTGTTTACAATATTAAAATGTATGTTAGCAAAATGGGTGGATTTTCAAATAAAATGCAGCTTCCA CAAAA >4731_4731_10_ANKRD42-PICALM_ANKRD42_chr11_82909684_ENST00000533342_PICALM_chr11_85670103_ENST00000532317_length(amino acids)=78AA_BP= MPGVANSGPSTSSRETANPCSRKKVHFGSIHDAVRAGDVKQLSEIVVRGASINELDVLHKFTPLHWAAHSGSLEIQFM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ANKRD42-PICALM |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000356360 | 17 | 19 | 221_294 | 628.0 | 633.0 | PIMREG | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000393346 | 18 | 20 | 221_294 | 648.0 | 653.0 | PIMREG | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000526033 | 18 | 20 | 221_294 | 641.0 | 646.0 | PIMREG | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000528398 | 17 | 19 | 221_294 | 547.0 | 552.0 | PIMREG | |
| Tgene | PICALM | chr11:82909684 | chr11:85670103 | ENST00000532317 | 18 | 20 | 221_294 | 606.0 | 611.0 | PIMREG |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ANKRD42-PICALM |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ANKRD42-PICALM |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C0002395 | Alzheimer's Disease | 2 | CTD_human | |
| Tgene | C0011265 | Presenile dementia | 2 | CTD_human | |
| Tgene | C0276496 | Familial Alzheimer Disease (FAD) | 2 | CTD_human | |
| Tgene | C0494463 | Alzheimer Disease, Late Onset | 2 | CTD_human | |
| Tgene | C0546126 | Acute Confusional Senile Dementia | 2 | CTD_human | |
| Tgene | C0750900 | Alzheimer's Disease, Focal Onset | 2 | CTD_human | |
| Tgene | C0750901 | Alzheimer Disease, Early Onset | 2 | CTD_human | |
| Tgene | C0234985 | Mental deterioration | 1 | CTD_human | |
| Tgene | C0338656 | Impaired cognition | 1 | CTD_human | |
| Tgene | C1270972 | Mild cognitive disorder | 1 | CTD_human |
