
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AGRN-PIEZO1 (FusionGDB2 ID:HG375790TG9780) |
Fusion Gene Summary for AGRN-PIEZO1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AGRN-PIEZO1 | Fusion gene ID: hg375790tg9780 | Hgene | Tgene | Gene symbol | AGRN | PIEZO1 | Gene ID | 375790 | 9780 |
| Gene name | agrin | piezo type mechanosensitive ion channel component 1 | |
| Synonyms | AGRIN|CMS8|CMSPPD | DHS|FAM38A|LMPH3|LMPHM6|Mib | |
| Cytomap | ('AGRN')('PIEZO1') 1p36.33 | 16q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | agrinagrin proteoglycan | piezo-type mechanosensitive ion channel component 1family with sequence similarity 38, member Amembrane protein induced by beta-amyloid treatment | |
| Modification date | 20200315 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000477585, ENST00000379370, | ||
| Fusion gene scores | * DoF score | 14 X 7 X 10=980 | 6 X 4 X 2=48 |
| # samples | 18 | 6 | |
| ** MAII score | log2(18/980*10)=-2.4447848426729 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/48*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: AGRN [Title/Abstract] AND PIEZO1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AGRN(970704)-PIEZO1(88815887), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | AGRN-PIEZO1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGRN-PIEZO1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGRN-PIEZO1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. AGRN-PIEZO1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AGRN | GO:0043113 | receptor clustering | 15340048 |
 Fusion gene breakpoints across AGRN (5'-gene) Fusion gene breakpoints across AGRN (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
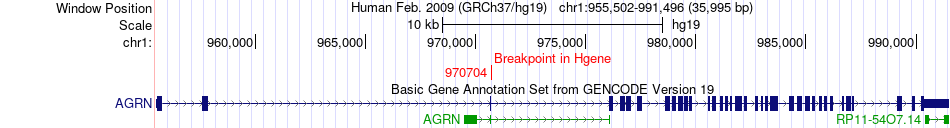 |
 Fusion gene breakpoints across PIEZO1 (3'-gene) Fusion gene breakpoints across PIEZO1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
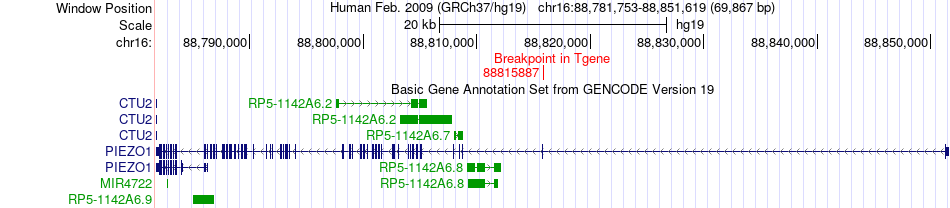 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-IN-7808-01A | AGRN | chr1 | 970704 | + | PIEZO1 | chr16 | 88815887 | - |
Top |
Fusion Gene ORF analysis for AGRN-PIEZO1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000477585 | ENST00000301015 | AGRN | chr1 | 970704 | + | PIEZO1 | chr16 | 88815887 | - |
| 3UTR-intron | ENST00000477585 | ENST00000327397 | AGRN | chr1 | 970704 | + | PIEZO1 | chr16 | 88815887 | - |
| 5CDS-intron | ENST00000379370 | ENST00000327397 | AGRN | chr1 | 970704 | + | PIEZO1 | chr16 | 88815887 | - |
| In-frame | ENST00000379370 | ENST00000301015 | AGRN | chr1 | 970704 | + | PIEZO1 | chr16 | 88815887 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379370 | AGRN | chr1 | 970704 | + | ENST00000301015 | PIEZO1 | chr16 | 88815887 | - | 8322 | 561 | 50 | 8062 | 2670 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379370 | ENST00000301015 | AGRN | chr1 | 970704 | + | PIEZO1 | chr16 | 88815887 | - | 0.002609934 | 0.9973901 |
Top |
Fusion Genomic Features for AGRN-PIEZO1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
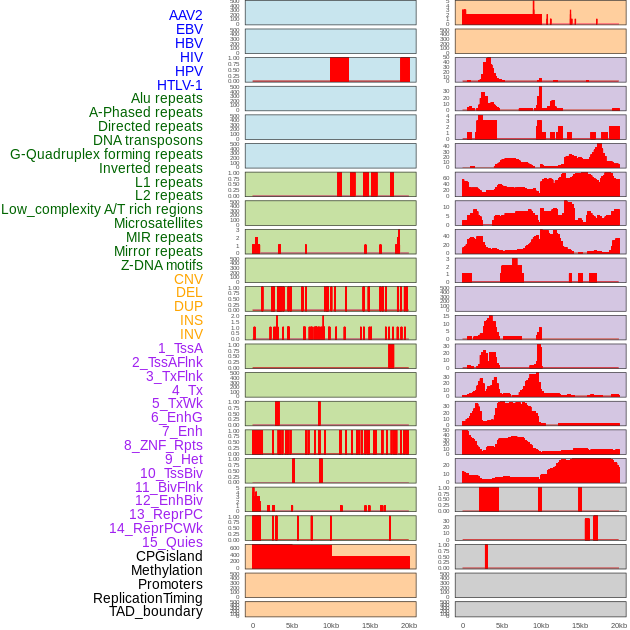 |
Top |
Fusion Protein Features for AGRN-PIEZO1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:970704/chr16:88815887) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 30_157 | 170 | 2046.0 | Domain | NtA |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1339_1368 | 21 | 2522.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2198_2431 | 21 | 2522.0 | Topological domain | Extracellular | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1043_1063 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1160_1180 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1184_1204 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 118_138 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1218_1240 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1247_1264 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1277_1297 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1678_1698 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1700_1720 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1734_1754 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 194_214 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 1962_1982 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2003_2023 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2032_2052 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2061_2081 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2100_2122 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2129_2149 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2177_2197 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 218_238 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 2432_2452 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 248_268 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 27_47 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 302_322 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 428_448 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 460_480 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 514_534 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 579_599 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 602_622 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 629_649 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 64_84 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 681_701 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 823_843 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 852_872 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 926_946 | 21 | 2522.0 | Transmembrane | Helical | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 987_1007 | 21 | 2522.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1941_2009 | 170 | 2046.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1058_1097 | 170 | 2046.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1254_1324 | 170 | 2046.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 671_677 | 170 | 2046.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 974_1099 | 170 | 2046.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1130_1252 | 170 | 2046.0 | Domain | SEA |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1329_1367 | 170 | 2046.0 | Domain | EGF-like 1 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1372_1548 | 170 | 2046.0 | Domain | Laminin G-like 1 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1549_1586 | 170 | 2046.0 | Domain | EGF-like 2 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1588_1625 | 170 | 2046.0 | Domain | EGF-like 3 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1635_1822 | 170 | 2046.0 | Domain | Laminin G-like 2 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1818_1857 | 170 | 2046.0 | Domain | EGF-like 4 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 1868_2065 | 170 | 2046.0 | Domain | Laminin G-like 3 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 191_244 | 170 | 2046.0 | Domain | Kazal-like 1 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 264_319 | 170 | 2046.0 | Domain | Kazal-like 2 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 337_391 | 170 | 2046.0 | Domain | Kazal-like 3 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 408_463 | 170 | 2046.0 | Domain | Kazal-like 4 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 484_536 | 170 | 2046.0 | Domain | Kazal-like 5 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 540_601 | 170 | 2046.0 | Domain | Kazal-like 6 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 607_666 | 170 | 2046.0 | Domain | Kazal-like 7 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 699_752 | 170 | 2046.0 | Domain | Kazal-like 8 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 793_846 | 170 | 2046.0 | Domain | Laminin EGF-like 1 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 847_893 | 170 | 2046.0 | Domain | Laminin EGF-like 2 |
| Hgene | AGRN | chr1:970704 | chr16:88815887 | ENST00000379370 | + | 3 | 36 | 917_971 | 170 | 2046.0 | Domain | Kazal-like 9 |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 6_91 | 21 | 2522.0 | Compositional bias | Note=Leu-rich | |
| Tgene | PIEZO1 | chr1:970704 | chr16:88815887 | ENST00000301015 | 0 | 51 | 5_25 | 21 | 2522.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for AGRN-PIEZO1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >3004_3004_1_AGRN-PIEZO1_AGRN_chr1_970704_ENST00000379370_PIEZO1_chr16_88815887_ENST00000301015_length(transcript)=8322nt_BP=561nt CCCGTCCCCGGCGCGGCCCGCGCGCTCCTCCGCCGCCTCTCGCCTGCGCCATGGCCGGCCGGTCCCACCCGGGCCCGCTGCGGCCGCTGC TGCCGCTCCTTGTGGTGGCCGCGTGCGTCCTGCCCGGAGCCGGCGGGACATGCCCGGAGCGCGCGCTGGAGCGGCGCGAGGAGGAGGCGA ACGTGGTGCTCACCGGGACGGTGGAGGAGATCCTCAACGTGGACCCGGTGCAGCACACGTACTCCTGCAAGGTTCGGGTCTGGCGGTACT TGAAGGGCAAAGACCTGGTGGCCCGGGAGAGCCTGCTGGACGGCGGCAACAAGGTGGTGATCAGCGGCTTTGGAGACCCCCTCATCTGTG ACAACCAGGTGTCCACTGGGGACACCAGGATCTTCTTTGTGAACCCTGCACCCCCATACCTGTGGCCAGCCCACAAGAACGAGCTGATGC TCAACTCCAGCCTCATGCGGATCACCCTGCGGAACCTGGAGGAGGTGGAGTTCTGTGTGGAAGATAAACCCGGGACCCACTTCACTCCAG TGCCTCCGACGCCTCCTGATGCCTGCCTGCTCCGCTTCAGCGGACTCTCGCTGGTCTACCTGCTCTTCCTGCTGCTGCTGCCCTGGTTCC CCGGCCCCACCCGATGCGGCCTCCAAGGTCACACAGGCCGCCTCCTGCGGGCATTGCTGGGCCTCAGCCTGCTCTTCCTGGTGGCCCATC TCGCCCTCCAGATCTGCCTGCATATTGTGCCCCGCCTGGACCAGCTCCTGGGACCCAGCTGCAGCCGCTGGGAGACCCTCTCGCGACACA TAGGGGTCACAAGGCTGGACCTGAAGGACATCCCCAACGCCATCCGGCTGGTGGCCCCTGACCTGGGCATCTTGGTGGTCTCCTCTGTCT GCCTCGGCATCTGCGGGCGCCTTGCAAGGAACACCCGGCAGAGCCCACATCCACGGGAGCTGGATGATGATGAGAGGGATGTGGATGCCA GCCCGACGGCAGGGCTGCAGGAAGCAGCAACGCTGGCCCCTACACGGAGGTCACGGCTGGCCGCTCGTTTCCGAGTCACGGCCCACTGGC TGCTGGTGGCGGCTGGGCGGGTCCTGGCCGTAACACTGCTTGCACTGGCAGGCATCGCCCACCCCTCGGCCCTCTCCAGTGTCTACCTGC TGCTCTTCCTGGCCCTCTGCACCTGGTGGGCCTGCCACTTTCCCATCAGCACTCGGGGCTTCAGCAGACTCTGCGTCGCGGTGGGGTGCT TCGGCGCCGGCCATCTCATCTGCCTCTACTGCTACCAGATGCCCTTGGCACAGGCTCTGCTCCCGCCTGCCGGCATCTGGGCTAGGGTGC TGGGTCTCAAGGACTTCGTGGGTCCCACCAACTGCTCCAGCCCCCACGCGCTGGTCCTCAACACCGGCCTGGACTGGCCTGTGTATGCCA GCCCCGGCGTCCTCCTGCTGCTGTGCTACGCCACGGCCTCTCTGCGCAAGCTCCGCGCGTACCGCCCCTCCGGCCAGAGGAAGGAGGCGG CAAAGGGGTATGAGGCTCGGGAGCTGGAGCTAGCAGAGCTGGACCAGTGGCCCCAGGAACGGGAGTCTGACCAGCACGTGGTGCCCACAG CACCCGACACCGAGGCTGATAACTGCATCGTGCACGAGCTGACCGGCCAGAGCTCCGTCCTGCGGCGGCCTGTGCGGCCCAAGCGGGCTG AGCCCAGGGAGGCGTCTCCGCTCCACAGCCTGGGCCACCTCATCATGGACCAGAGCTATGTGTGCGCGCTCATTGCCATGATGGTATGGA GCATCACCTACCACAGCTGGCTGACCTTCGTACTGCTGCTCTGGGCCTGCCTCATCTGGACGGTGCGCAGCCGCCACCAACTGGCCATGC TGTGCTCGCCCTGCATCCTGCTGTATGGGATGACGCTGTGCTGCCTACGCTACGTGTGGGCCATGGACCTGCGCCCTGAGCTGCCCACCA CCCTGGGCCCCGTCAGCCTGCGCCAGCTGGGGCTGGAGCACACCCGCTACCCCTGTCTGGACCTTGGTGCCATGTTGCTCTACACCCTGA CCTTCTGGCTCCTGCTGCGCCAGTTTGTGAAAGAGAAGCTGCTGAAGTGGGCAGAGTCTCCAGCTGCGCTGACGGAGGTCACCGTGGCAG ACACAGAGCCCACGCGGACGCAGACGCTGTTGCAGAGCCTGGGGGAGCTGGTGAAGGGCGTGTACGCCAAGTACTGGATCTATGTGTGTG CTGGCATGTTCATCGTGGTCAGCTTCGCCGGCCGCCTCGTGGTCTACAAGATTGTCTACATGTTCCTCTTCCTGCTCTGCCTCACCCTCT TCCAGGTCTACTACAGCCTGTGGCGGAAGCTGCTCAAGGCCTTCTGGTGGCTCGTGGTGGCCTACACCATGCTGGTCCTCATCGCCGTCT ACACCTTCCAGTTCCAGGACTTCCCTGCCTACTGGCGCAACCTCACTGGCTTCACCGACGAGCAGCTGGGGGACCTGGGCCTGGAGCAGT TCAGCGTGTCCGAGCTCTTCTCCAGCATCCTGGTGCCCGGCTTCTTCCTCCTGGCCTGCATCCTGCAGCTGCACTACTTCCACAGGCCCT TCATGCAGCTCACCGACATGGAGCACGTGTCCCTGCCTGGCACGCGCCTCCCGCGCTGGGCTCACAGGCAGGATGCAGTGAGTGGGACCC CACTGCTGCGGGAGGAGCAGCAGGAGCATCAGCAGCAGCAGCAGGAGGAGGAGGAGGAGGAGGAGGACTCCAGGGACGAGGGGCTGGGCG TGGCCACTCCCCACCAGGCCACGCAGGTGCCTGAAGGGGCAGCCAAGTGGGGCCTGGTGGCTGAGCGGCTGCTGGAGCTGGCAGCCGGCT TCTCGGACGTCCTCTCACGCGTGCAGGTGTTCCTGCGGCGGCTGCTGGAGCTTCACGTTTTCAAGCTGGTGGCCCTGTACACCGTCTGGG TGGCCCTGAAGGAGGTGTCGGTGATGAACCTGCTGCTGGTGGTGCTGTGGGCCTTCGCCCTGCCCTACCCACGCTTCCGGCCCATGGCCT CCTGCCTGTCCACCGTGTGGACCTGCGTCATCATCGTGTGTAAGATGCTGTACCAGCTCAAGGTTGTCAACCCCCAGGAGTATTCCAGCA ACTGCACCGAGCCCTTCCCCAACAGCACCAACTTGCTGCCCACGGAGATCAGCCAGTCCCTGCTGTACCGGGGGCCCGTGGACCCTGCCA ACTGGTTTGGGGTGCGGAAAGGGTTCCCCAACCTGGGCTACATCCAGAACCACCTGCAAGTGCTGCTGCTGCTGGTATTCGAGGCCATCG TGTACCGGCGCCAGGAGCACTACCGCCGGCAGCACCAGCTGGCCCCGCTGCCTGCCCAGGCCGTGTTTGCCAGCGGCACCCGCCAGCAGC TGGACCAGGATCTGCTCGGCTGCCTCAAGTACTTCATCAACTTCTTCTTCTACAAATTCGGGCTGGAGATCTGCTTCCTGATGGCCGTGA ACGTGATCGGGCAGCGCATGAACTTTCTGGTGACCCTGCACGGTTGCTGGCTGGTGGCCATCCTCACCCGCAGGCACCGCCAGGCCATTG CCCGCCTCTGGCCCAACTACTGCCTCTTCCTGGCGCTGTTCCTGCTGTACCAGTACCTGCTGTGCCTGGGGATGCCCCCGGCCCTGTGCA TTGATTATCCCTGGCGCTGGAGCCGGGCCGTCCCCATGAACTCCGCACTCATCAAGTGGCTGTACCTGCCTGATTTCTTCCGGGCCCCCA ACTCCACCAACCTCATCAGCGACTTTCTCCTGCTGCTGTGCGCCTCCCAGCAGTGGCAGGTGTTCTCAGCTGAGCGCACAGAGGAGTGGC AGCGCATGGCTGGCGTCAACACCGACCGCCTGGAGCCGCTGCGGGGGGAGCCCAACCCCGTGCCCAACTTTATCCACTGCAGGTCCTACC TTGACATGCTGAAGGTGGCCGTCTTCCGATACCTGTTCTGGCTGGTGCTGGTGGTGGTGTTTGTCACGGGGGCCACCCGCATCAGCATCT TCGGGCTGGGCTACCTGCTGGCCTGCTTCTACCTGCTGCTCTTCGGCACGGCCCTGCTGCAGAGGGACACACGGGCCCGCCTCGTGCTGT GGGACTGCCTCATTCTGTACAACGTCACCGTCATCATCTCCAAGAACATGCTGTCGCTCCTGGCCTGCGTCTTCGTGGAGCAGATGCAGA CCGGCTTCTGCTGGGTCATCCAGCTCTTCAGCCTTGTATGCACCGTCAAGGGCTACTATGACCCCAAGGAGATGATGGACAGAGACCAGG ACTGCCTGCTGCCTGTGGAGGAGGCTGGCATCATCTGGGACAGCGTCTGCTTCTTCTTCCTGCTGCTGCAGCGCCGCGTCTTCCTTAGCC ATTACTACCTGCACGTCAGGGCCGACCTCCAGGCCACCGCCCTGCTAGCCTCCAGGGGCTTCGCCCTCTACAACGCTGCCAACCTCAAGA GCATTGACTTTCACCGCAGGATAGAGGAGAAGTCCCTGGCCCAGCTGAAAAGACAGATGGAGCGTATCCGTGCCAAGCAGGAGAAGCACA GGCAGGGCCGGGTGGACCGCAGTCGCCCCCAGGACACCCTGGGCCCCAAGGACCCCGGCCTGGAGCCAGGGCCCGACAGTCCAGGGGGCT CCTCCCCGCCACGGAGGCAGTGGTGGCGGCCCTGGCTGGACCACGCCACAGTCATCCACTCCGGGGACTACTTCCTGTTTGAGTCCGACA GTGAGGAAGAGGAGGAGGCTGTTCCTGAAGACCCGAGGCCGTCGGCACAGAGTGCCTTCCAGCTGGCGTACCAGGCATGGGTGACCAACG CCCAGGCGGTGCTGAGGCGGCGGCAGCAGGAGCAGGAGCAGGCAAGGCAGGAACAGGCAGGACAGCTACCCACAGGAGGTGGTCCCAGCC AGGAGGTGGAGCCAGCAGAGGGCCCCGAGGAGGCAGCGGCAGGCCGGAGCCATGTGGTGCAGAGGGTGCTGAGCACGGCGCAGTTCCTGT GGATGCTGGGGCAGGCGCTAGTGGATGAGCTGACACGCTGGCTGCAGGAGTTCACCCGGCACCACGGCACCATGAGCGACGTGCTGCGGG CAGAGCGCTACCTCCTCACACAGGAGCTCCTGCAGGGCGGCGAAGTGCACAGGGGCGTGCTGGATCAGCTGTACACAAGCCAGGCCGAGG CCACGCTGCCAGGCCCCACCGAGGCCCCCAATGCCCCAAGCACCGTGTCCAGTGGGCTGGGCGCGGAGGAGCCACTCAGCAGCATGACAG ACGACATGGGCAGCCCCCTGAGCACCGGCTACCACACGCGCAGTGGCAGTGAGGAGGCAGTCACCGACCCCGGGGAGCGTGAGGCTGGTG CCTCTCTGTACCAGGGACTGATGCGGACGGCCAGCGAGCTGCTCCTGGACAGGCGCCTGCGCATCCCAGAGCTGGAGGAGGCAGAGCTGT TTGCGGAGGGGCAGGGCCGGGCGCTGCGGCTGCTGCGGGCCGTGTACCAGTGTGTGGCCGCCCACTCGGAGCTGCTCTGCTACTTCATCA TCATCCTCAACCACATGGTCACGGCCTCCGCCGGCTCGCTGGTGCTGCCCGTGCTCGTCTTCCTGTGGGCCATGCTGTCGATCCCGAGGC CCAGCAAGCGCTTCTGGATGACGGCCATCGTCTTCACCGAGATCGCGGTGGTCGTCAAGTACCTGTTCCAGTTTGGGTTCTTCCCCTGGA ACAGCCACGTGGTGCTGCGGCGCTACGAGAACAAGCCCTACTTCCCGCCCCGCATCCTGGGCCTGGAGAAGACTGACGGCTACATCAAGT ACGACCTGGTGCAGCTCATGGCCCTTTTCTTCCACCGCTCCCAGCTGCTGTGCTATGGCCTCTGGGACCATGAGGAGGACTCACCATCCA AGGAGCATGACAAGAGCGGCGAGGAGGAGCAGGGAGCCGAGGAGGGGCCAGGGGTGCCTGCGGCCACCACCGAAGACCACATTCAGGTGG AAGCCAGGGTCGGACCCACGGACGGGACCCCAGAACCCCAAGTGGAGCTCAGGCCCCGTGATACGAGGCGCATCAGTCTACGTTTTAGAA GAAGGAAGAAGGAGGGCCCAGCACGGAAAGGAGCGGCAGCCATCGAAGCTGAGGACAGGGAGGAAGAAGAGGGGGAGGAAGAGAAAGAGG CCCCCACGGGGAGAGAGAAGAGGCCAAGCCGCTCTGGAGGAAGAGTAAGGGCGGCCGGGCGGCGGCTGCAGGGCTTCTGCCTGTCCCTGG CCCAGGGCACATATCGGCCGCTACGGCGCTTCTTCCACGACATCCTGCACACCAAGTACCGCGCAGCCACCGACGTCTATGCCCTCATGT TCCTGGCTGATGTTGTCGACTTCATCATCATCATTTTTGGCTTCTGGGCCTTTGGGAAGCACTCGGCGGCCACAGACATCACGTCCTCCC TATCAGACGACCAGGTACCCGAGGCTTTCCTGGTCATGCTGCTGATCCAGTTCAGTACCATGGTGGTTGACCGCGCCCTCTACCTGCGCA AGACCGTGCTGGGCAAGCTGGCCTTCCAGGTGGCGCTGGTGCTGGCCATCCACCTATGGATGTTCTTCATCCTGCCCGCCGTCACTGAGA GGATGTTCAACCAGAATGTGGTGGCCCAGCTCTGGTACTTCGTGAAGTGCATCTACTTCGCCCTGTCCGCCTACCAGATCCGCTGCGGCT ACCCCACCCGCATCCTCGGCAACTTCCTCACCAAGAAGTACAATCATCTCAACCTCTTCCTCTTCCAGGGGTTCCGGCTGGTGCCGTTCC TGGTGGAGCTGCGGGCAGTGATGGACTGGGTGTGGACGGACACCACGCTGTCCCTGTCCAGCTGGATGTGTGTGGAGGACATCTATGCCA ACATCTTCATCATCAAATGCAGCCGAGAGACAGAGAAGAAATACCCGCAGCCCAAAGGGCAGAAGAAGAAGAAGATCGTCAAGTACGGCA TGGGTGGCCTCATCATCCTCTTCCTCATCGCCATCATCTGGTTCCCACTGCTCTTCATGTCGCTGGTGCGCTCCGTGGTTGGGGTTGTCA ACCAGCCCATCGATGTCACCGTCACCCTGAAGCTGGGCGGCTATGAGCCGCTGTTCACCATGAGCGCCCAGCAGCCGTCCATCATCCCCT TCACGGCCCAGGCCTATGAGGAGCTGTCCCGGCAGTTTGACCCCCAGCCGCTGGCCATGCAGTTCATCAGCCAGTACAGCCCTGAGGACA TCGTCACGGCGCAGATTGAGGGCAGCTCCGGGGCGCTGTGGCGCATCAGTCCCCCCAGCCGTGCCCAGATGAAGCGGGAGCTCTACAACG GCACGGCCGACATCACCCTGCGCTTCACCTGGAACTTCCAGAGGGACCTGGCGAAGGGAGGCACTGTGGAGTATGCCAACGAGAAGCACA TGCTGGCCCTGGCCCCCAACAGCACTGCACGGCGGCAGCTGGCCAGCCTGCTCGAGGGCACCTCGGACCAGTCTGTGGTCATCCCTAATC TCTTCCCCAAGTACATCCGTGCCCCCAACGGGCCCGAAGCCAACCCTGTGAAGCAGCTGCAGCCCAATGAGGAGGCCGACTACCTCGGCG TGCGTATCCAGCTGCGGAGGGAGCAGGGTGCGGGGGCCACCGGCTTCCTCGAATGGTGGGTCATCGAGCTGCAGGAGTGCCGGACCGACT GCAACCTGCTGCCCATGGTCATTTTCAGTGACAAGGTCAGCCCACCGAGCCTCGGCTTCCTGGCTGGCTACGGCATCATGGGGCTGTACG TGTCCATCGTGCTGGTCATCGGCAAGTTCGTGCGCGGATTCTTCAGCGAGATCTCGCACTCCATTATGTTCGAGGAGCTGCCGTGCGTGG ACCGCATCCTCAAGCTCTGCCAGGACATCTTCCTGGTGCGGGAGACTCGGGAGCTGGAGCTGGAGGAGGAGTTGTACGCCAAGCTCATCT TCCTCTACCGCTCACCGGAGACCATGATCAAGTGGACTCGTGAGAAGGAGTAGGAGCTGCTGCTGGCGCCCGAGAGGGAAGGAGCCGGCC TGCTGGGCAGCGTGGCCACAAGGGGCGGCACTCCTCAGGCCGGGGGAGCCACTGCCCCGTCCAAGGCCGCCAGCTGTGATGCATCCTCCC GGCCTGCCTGAGCCCTGATGCTGCTGTCAGAGAAGGACACTGCGTCCCCACGGCCTGCGTGGCGCTGCCGTCCCCCACGTGTACTGTAGA GTTTTTTTTTTAATTAAAAAATGTTTTATTTATACAAATGGA >3004_3004_1_AGRN-PIEZO1_AGRN_chr1_970704_ENST00000379370_PIEZO1_chr16_88815887_ENST00000301015_length(amino acids)=2670AA_BP=168 MAGRSHPGPLRPLLPLLVVAACVLPGAGGTCPERALERREEEANVVLTGTVEEILNVDPVQHTYSCKVRVWRYLKGKDLVARESLLDGGN KVVISGFGDPLICDNQVSTGDTRIFFVNPAPPYLWPAHKNELMLNSSLMRITLRNLEEVEFCVEDKPGTHFTPVPPTPPDACLLRFSGLS LVYLLFLLLLPWFPGPTRCGLQGHTGRLLRALLGLSLLFLVAHLALQICLHIVPRLDQLLGPSCSRWETLSRHIGVTRLDLKDIPNAIRL VAPDLGILVVSSVCLGICGRLARNTRQSPHPRELDDDERDVDASPTAGLQEAATLAPTRRSRLAARFRVTAHWLLVAAGRVLAVTLLALA GIAHPSALSSVYLLLFLALCTWWACHFPISTRGFSRLCVAVGCFGAGHLICLYCYQMPLAQALLPPAGIWARVLGLKDFVGPTNCSSPHA LVLNTGLDWPVYASPGVLLLLCYATASLRKLRAYRPSGQRKEAAKGYEARELELAELDQWPQERESDQHVVPTAPDTEADNCIVHELTGQ SSVLRRPVRPKRAEPREASPLHSLGHLIMDQSYVCALIAMMVWSITYHSWLTFVLLLWACLIWTVRSRHQLAMLCSPCILLYGMTLCCLR YVWAMDLRPELPTTLGPVSLRQLGLEHTRYPCLDLGAMLLYTLTFWLLLRQFVKEKLLKWAESPAALTEVTVADTEPTRTQTLLQSLGEL VKGVYAKYWIYVCAGMFIVVSFAGRLVVYKIVYMFLFLLCLTLFQVYYSLWRKLLKAFWWLVVAYTMLVLIAVYTFQFQDFPAYWRNLTG FTDEQLGDLGLEQFSVSELFSSILVPGFFLLACILQLHYFHRPFMQLTDMEHVSLPGTRLPRWAHRQDAVSGTPLLREEQQEHQQQQQEE EEEEEDSRDEGLGVATPHQATQVPEGAAKWGLVAERLLELAAGFSDVLSRVQVFLRRLLELHVFKLVALYTVWVALKEVSVMNLLLVVLW AFALPYPRFRPMASCLSTVWTCVIIVCKMLYQLKVVNPQEYSSNCTEPFPNSTNLLPTEISQSLLYRGPVDPANWFGVRKGFPNLGYIQN HLQVLLLLVFEAIVYRRQEHYRRQHQLAPLPAQAVFASGTRQQLDQDLLGCLKYFINFFFYKFGLEICFLMAVNVIGQRMNFLVTLHGCW LVAILTRRHRQAIARLWPNYCLFLALFLLYQYLLCLGMPPALCIDYPWRWSRAVPMNSALIKWLYLPDFFRAPNSTNLISDFLLLLCASQ QWQVFSAERTEEWQRMAGVNTDRLEPLRGEPNPVPNFIHCRSYLDMLKVAVFRYLFWLVLVVVFVTGATRISIFGLGYLLACFYLLLFGT ALLQRDTRARLVLWDCLILYNVTVIISKNMLSLLACVFVEQMQTGFCWVIQLFSLVCTVKGYYDPKEMMDRDQDCLLPVEEAGIIWDSVC FFFLLLQRRVFLSHYYLHVRADLQATALLASRGFALYNAANLKSIDFHRRIEEKSLAQLKRQMERIRAKQEKHRQGRVDRSRPQDTLGPK DPGLEPGPDSPGGSSPPRRQWWRPWLDHATVIHSGDYFLFESDSEEEEEAVPEDPRPSAQSAFQLAYQAWVTNAQAVLRRRQQEQEQARQ EQAGQLPTGGGPSQEVEPAEGPEEAAAGRSHVVQRVLSTAQFLWMLGQALVDELTRWLQEFTRHHGTMSDVLRAERYLLTQELLQGGEVH RGVLDQLYTSQAEATLPGPTEAPNAPSTVSSGLGAEEPLSSMTDDMGSPLSTGYHTRSGSEEAVTDPGEREAGASLYQGLMRTASELLLD RRLRIPELEEAELFAEGQGRALRLLRAVYQCVAAHSELLCYFIIILNHMVTASAGSLVLPVLVFLWAMLSIPRPSKRFWMTAIVFTEIAV VVKYLFQFGFFPWNSHVVLRRYENKPYFPPRILGLEKTDGYIKYDLVQLMALFFHRSQLLCYGLWDHEEDSPSKEHDKSGEEEQGAEEGP GVPAATTEDHIQVEARVGPTDGTPEPQVELRPRDTRRISLRFRRRKKEGPARKGAAAIEAEDREEEEGEEEKEAPTGREKRPSRSGGRVR AAGRRLQGFCLSLAQGTYRPLRRFFHDILHTKYRAATDVYALMFLADVVDFIIIIFGFWAFGKHSAATDITSSLSDDQVPEAFLVMLLIQ FSTMVVDRALYLRKTVLGKLAFQVALVLAIHLWMFFILPAVTERMFNQNVVAQLWYFVKCIYFALSAYQIRCGYPTRILGNFLTKKYNHL NLFLFQGFRLVPFLVELRAVMDWVWTDTTLSLSSWMCVEDIYANIFIIKCSRETEKKYPQPKGQKKKKIVKYGMGGLIILFLIAIIWFPL LFMSLVRSVVGVVNQPIDVTVTLKLGGYEPLFTMSAQQPSIIPFTAQAYEELSRQFDPQPLAMQFISQYSPEDIVTAQIEGSSGALWRIS PPSRAQMKRELYNGTADITLRFTWNFQRDLAKGGTVEYANEKHMLALAPNSTARRQLASLLEGTSDQSVVIPNLFPKYIRAPNGPEANPV KQLQPNEEADYLGVRIQLRREQGAGATGFLEWWVIELQECRTDCNLLPMVIFSDKVSPPSLGFLAGYGIMGLYVSIVLVIGKFVRGFFSE ISHSIMFEELPCVDRILKLCQDIFLVRETRELELEEELYAKLIFLYRSPETMIKWTREKE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AGRN-PIEZO1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AGRN-PIEZO1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for AGRN-PIEZO1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | AGRN | C3808739 | MYASTHENIC SYNDROME, CONGENITAL, 8 | 3 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | AGRN | C0751883 | Congenital Myasthenic Syndromes, Postsynaptic | 2 | CTD_human;ORPHANET |
| Hgene | AGRN | C0751884 | Congenital Myasthenic Syndromes, Presynaptic | 2 | CTD_human;ORPHANET |
| Hgene | AGRN | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | AGRN | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | AGRN | C0751882 | Myasthenic Syndromes, Congenital | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | AGRN | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Tgene | C4551512 | DEHYDRATED HEREDITARY STOMATOCYTOSIS 1 WITH OR WITHOUT PSEUDOHYPERKALEMIA AND/OR PERINATAL EDEMA | 7 | GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C4225184 | LYMPHATIC MALFORMATION 6 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0272051 | Xerocytosis | 3 | GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C0272048 | stomatocytic anemia | 2 | GENOMICS_ENGLAND | |
| Tgene | C0677598 | Stomatocytosis Result | 2 | GENOMICS_ENGLAND |
