
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACLY-FAM117A (FusionGDB2 ID:HG47TG81558) |
Fusion Gene Summary for ACLY-FAM117A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACLY-FAM117A | Fusion gene ID: hg47tg81558 | Hgene | Tgene | Gene symbol | ACLY | FAM117A | Gene ID | 47 | 81558 |
| Gene name | ATP citrate lyase | family with sequence similarity 117 member A | |
| Synonyms | ACL|ATPCL|CLATP | - | |
| Cytomap | ('ACLY')('FAM117A') 17q21.2 | 17q21.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ATP-citrate synthaseATP-citrate (pro-S-)-lyasecitrate cleavage enzyme | protein FAM117AC/EBP-induced protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P53396 | Q9C073 | |
| Ensembl transtripts involved in fusion gene | ENST00000352035, ENST00000353196, ENST00000393896, ENST00000590151, ENST00000537919, ENST00000588779, | ||
| Fusion gene scores | * DoF score | 11 X 14 X 6=924 | 8 X 4 X 6=192 |
| # samples | 13 | 10 | |
| ** MAII score | log2(13/924*10)=-2.8293812283876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/192*10)=-0.941106310946431 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ACLY [Title/Abstract] AND FAM117A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACLY(40060982)-FAM117A(47810082), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ACLY-FAM117A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACLY-FAM117A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACLY-FAM117A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACLY-FAM117A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACLY-FAM117A seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACLY | GO:0006085 | acetyl-CoA biosynthetic process | 1371749 |
| Hgene | ACLY | GO:0006101 | citrate metabolic process | 1371749 |
| Hgene | ACLY | GO:0006107 | oxaloacetate metabolic process | 1371749 |
| Hgene | ACLY | GO:0008610 | lipid biosynthetic process | 23932781 |
 Fusion gene breakpoints across ACLY (5'-gene) Fusion gene breakpoints across ACLY (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
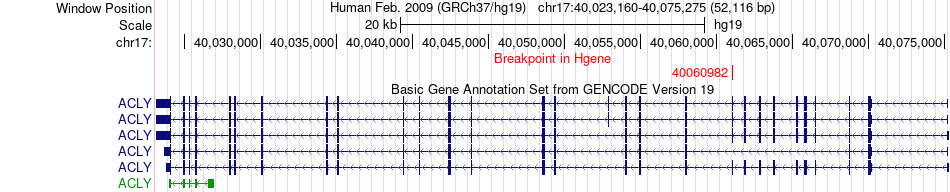 |
 Fusion gene breakpoints across FAM117A (3'-gene) Fusion gene breakpoints across FAM117A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
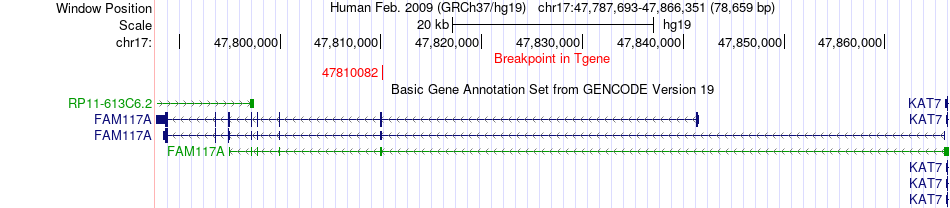 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCS | TCGA-N6-A4VF | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
Top |
Fusion Gene ORF analysis for ACLY-FAM117A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000352035 | ENST00000513602 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000352035 | ENST00000514018 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000353196 | ENST00000513602 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000353196 | ENST00000514018 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000393896 | ENST00000513602 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000393896 | ENST00000514018 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000590151 | ENST00000513602 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| 5CDS-5UTR | ENST00000590151 | ENST00000514018 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| Frame-shift | ENST00000352035 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| Frame-shift | ENST00000353196 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| In-frame | ENST00000393896 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| In-frame | ENST00000590151 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| intron-3CDS | ENST00000537919 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| intron-3CDS | ENST00000588779 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| intron-5UTR | ENST00000537919 | ENST00000513602 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| intron-5UTR | ENST00000537919 | ENST00000514018 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| intron-5UTR | ENST00000588779 | ENST00000513602 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
| intron-5UTR | ENST00000588779 | ENST00000514018 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000590151 | ACLY | chr17 | 40060982 | - | ENST00000240364 | FAM117A | chr17 | 47810082 | - | 3264 | 1175 | 29 | 1369 | 446 |
| ENST00000393896 | ACLY | chr17 | 40060982 | - | ENST00000240364 | FAM117A | chr17 | 47810082 | - | 3324 | 1235 | 8 | 1429 | 473 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000590151 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - | 0.005145579 | 0.99485445 |
| ENST00000393896 | ENST00000240364 | ACLY | chr17 | 40060982 | - | FAM117A | chr17 | 47810082 | - | 0.005963222 | 0.9940368 |
Top |
Fusion Genomic Features for ACLY-FAM117A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
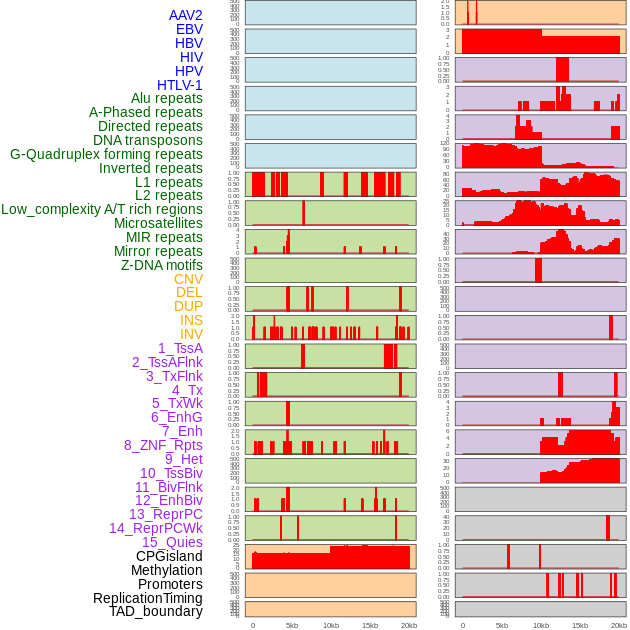 |
Top |
Fusion Protein Features for ACLY-FAM117A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:40060982/chr17:47810082) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ACLY | FAM117A |
| FUNCTION: Catalyzes the cleavage of citrate into oxaloacetate and acetyl-CoA, the latter serving as common substrate for de novo cholesterol and fatty acid synthesis. {ECO:0000269|PubMed:10653665, ECO:0000269|PubMed:1371749, ECO:0000269|PubMed:19286649, ECO:0000269|PubMed:23932781, ECO:0000269|PubMed:9116495}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000352035 | - | 10 | 29 | 4_265 | 355 | 1102.0 | Domain | Note=ATP-grasp |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000353196 | - | 10 | 28 | 4_265 | 355 | 1092.0 | Domain | Note=ATP-grasp |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000393896 | - | 10 | 28 | 4_265 | 355 | 1092.0 | Domain | Note=ATP-grasp |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000590151 | - | 10 | 29 | 4_265 | 355 | 1102.0 | Domain | Note=ATP-grasp |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000352035 | - | 10 | 29 | 109_111 | 355 | 1102.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000352035 | - | 10 | 29 | 66_67 | 355 | 1102.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000353196 | - | 10 | 28 | 109_111 | 355 | 1092.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000353196 | - | 10 | 28 | 66_67 | 355 | 1092.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000393896 | - | 10 | 28 | 109_111 | 355 | 1092.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000393896 | - | 10 | 28 | 66_67 | 355 | 1092.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000590151 | - | 10 | 29 | 109_111 | 355 | 1102.0 | Nucleotide binding | Note=ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000590151 | - | 10 | 29 | 66_67 | 355 | 1102.0 | Nucleotide binding | Note=ATP |
| Tgene | FAM117A | chr17:40060982 | chr17:47810082 | ENST00000240364 | 0 | 8 | 149_175 | 65 | 454.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | FAM117A | chr17:40060982 | chr17:47810082 | ENST00000240364 | 0 | 8 | 402_444 | 65 | 454.0 | Compositional bias | Note=Pro-rich |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000352035 | - | 10 | 29 | 701_721 | 355 | 1102.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000352035 | - | 10 | 29 | 752_778 | 355 | 1102.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000353196 | - | 10 | 28 | 701_721 | 355 | 1092.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000353196 | - | 10 | 28 | 752_778 | 355 | 1092.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000393896 | - | 10 | 28 | 701_721 | 355 | 1092.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000393896 | - | 10 | 28 | 752_778 | 355 | 1092.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000590151 | - | 10 | 29 | 701_721 | 355 | 1102.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000590151 | - | 10 | 29 | 752_778 | 355 | 1102.0 | Nucleotide binding | ATP |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000352035 | - | 10 | 29 | 779_789 | 355 | 1102.0 | Region | CoA-binding |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000353196 | - | 10 | 28 | 779_789 | 355 | 1092.0 | Region | CoA-binding |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000393896 | - | 10 | 28 | 779_789 | 355 | 1092.0 | Region | CoA-binding |
| Hgene | ACLY | chr17:40060982 | chr17:47810082 | ENST00000590151 | - | 10 | 29 | 779_789 | 355 | 1102.0 | Region | CoA-binding |
Top |
Fusion Gene Sequence for ACLY-FAM117A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1327_1327_1_ACLY-FAM117A_ACLY_chr17_40060982_ENST00000393896_FAM117A_chr17_47810082_ENST00000240364_length(transcript)=3324nt_BP=1235nt GCGAGCCGATGGGGGCGGGGAAAAGTCCGGCTGGGCCGGGACAAAAGCCGGATCCCGGGAAGCTACCGGCTGCTGGGGTGCTCCGGATTT TGCGGGGTTCGTCGGGCCTGTGGAAGAAGCGCCGCGCACGGACTTCGGCAGAGACAGGTAGAGCAGGTCTCTCTGCAGCCATGTCGGCCA AGGCAATTTCAGAGCAGACGGGCAAAGAACTCCTTTACAAGTTCATCTGTACCACCTCAGCCATCCAGAATCGGTTCAAGTATGCTCGGG TCACTCCTGACACAGACTGGGCCCGCTTGCTGCAGGACCACCCCTGGCTGCTCAGCCAGAACTTGGTAGTCAAGCCAGACCAGCTGATCA AACGTCGTGGAAAACTTGGTCTCGTTGGGGTCAACCTCACTCTGGATGGGGTCAAGTCCTGGCTGAAGCCACGGCTGGGACAGGAAGCCA CAGTTGGCAAGGCCACAGGCTTCCTCAAGAACTTTCTGATCGAGCCCTTCGTCCCCCACAGTCAGGCTGAGGAGTTCTATGTCTGCATCT ATGCCACCCGAGAAGGGGACTACGTCCTGTTCCACCACGAGGGGGGTGTGGACGTGGGTGATGTGGACGCCAAGGCCCAGAAGCTGCTTG TTGGCGTGGATGAGAAACTGAATCCTGAGGACATCAAAAAACACCTGTTGGTCCACGCCCCTGAAGACAAGAAAGAAATTCTGGCCAGTT TTATCTCCGGCCTCTTCAATTTCTACGAGGACTTGTACTTCACCTACCTCGAGATCAATCCCCTTGTAGTGACCAAAGATGGAGTCTATG TCCTTGACTTGGCGGCCAAGGTGGACGCCACTGCCGACTACATCTGCAAAGTGAAGTGGGGTGACATCGAGTTCCCTCCCCCCTTCGGGC GGGAGGCATATCCAGAGGAAGCCTACATTGCAGACCTCGATGCCAAAAGTGGGGCAAGCCTGAAGCTGACCTTGCTGAACCCCAAAGGGA GGATCTGGACCATGGTGGCCGGGGGTGGCGCCTCTGTCGTGTACAGCGATACCATCTGTGATCTAGGGGGTGTCAACGAGCTGGCAAACT ATGGGGAGTACTCAGGCGCCCCCAGCGAGCAGCAGACCTATGACTATGCCAAGACTATCCTCTCCCTCATGACCCGAGAGAAGCACCCAG ATGGCAAGATCCTCATCATTGGAGGCAGCATCGCAAACTTCACCAACGTGGCTGCCACGTTCAAGCCAGCGTCCCATGCTCGGTGGCCCC AGAAAAGTCAGTGTGTAGGCCTCAGCCACTTCAGGTCCGGCGTACATTCTCCCTGGACACCATCCTCAGCTCCTACCTTCTGGGCCAGTG GCCACGAGATGCTGATGGGGCCTTCACCTGCTGCACCAATGACAAGGCCACCCAGACGCCCCTGTCCTGGCAAGAGCTAGAAGGTGAGCG TGCCAGTTCCTGTGCACACAAGCGCTCAGCATCCTGGGGCAGCACAGACCACCGAAAAGAGATTTCCAAGTTGAAGCAACAACTGCAGAG GACGAAGCTGAGCCGCAGTGGGAAAGAGAAGGAGCGAGGTTCACCACTCCTAGGGGACCACGCAGTGCGGGGAGCACTGAGGGCGTCCCC TCCCAGCTTCCCCTCAGGGTCCCCTGTCTTGCGACTCAGCCCCTGCCTGCACAGGAGCCTGGAAGGGCTCAACCAAGAGCTGGAGGAGGT ATTTGTGAAGGAGCAGGGAGAAGAGGAGCTGCTGAGGATCCTTGATATCCCTGATGGGCACCGGGCCCCAGCTCCTCCCCAGAGTGGCAG CTGTGATCATCCCCTCCTCCTCCTGGAGCCTGGCAACCTTGCCAGCTCTCCTTCCATGTCCTTGGCATCTCCCCAGCCTTGTGGCCTGGC CAGTCATGAGGAACATCGGGGTGCCGCCGAGGAGCTGGCATCCACCCCCAACGACAAAGCCTCCTCTCCAGGACACCCAGCCTTTCTTGA AGATGGCAGCCCATCTCCAGTCCTTGCCTTTGCTGCCTCCCCTCGACCTAATCATAGCTACATCTTCAAACGGGAGCCCCCAGAAGGCTG TGAGAAAGTGCGTGTGTTTGAAGAAGCCACGTCTCCAGGTCCTGACCTGGCCTTCCTGACTTCCTGTCCTGACAAGAACAAAGTCCATTT CAACCCGACTGGCTCAGCCTTCTGCCCCGTCAACCTGATGAAGCCCCTCTTCCCCGGCATGGGCTTCATCTTCCGTAACTGCCCCTCAAA CCCGGGATCTCCCCTTCCCCCGGCCAGCCCCAGGCCACCACCTCGGAAGGATCCGGAAGCCTCCAAGGCCTCCCCACTGCCATTCGAGCC ATGGCAGCGCACCCCACCATCAGAAGAGCCTGTGCTTTTCCAGAGCTCCCTGATGGTCTGAGGGTCCCACCCCTGCCCCACTTTACCATA GAGACCAGTGCCTTGGTGGCAGGTCCCTCCCCAGGTCCCCTGAGATGGGGTATGGAGGGGCCCTTCCCTCTCGGCCTTCGAGCACTTTCT TTCACTTACTGTGTCAAAGCCCTGGGTCCTCTTTTTGATGGGCACCGGCCCCTCTGAACGTGATGGGACCTGCCTTCTCCACTAGTAGCT GGGCAGCTCACAATTCACACCTGTGTACCTGCCACATCCCTCACTTGGTGGAAAACACCCAGAAGGTCTTGAGTCCCCCACCCCTGGGTG TCAGTCCAAATGACTGTATAGGAGGCCCTTATTTTTGTCACAGAGCAAGCTGGCCATGAACGAAGGAGAGAAGACGCCACAGATTTCCTT CCCTCTCCTCCAGGAGACCATAAGATAGATCCCCCATCCTCTCAGCCCTATTCCCATGCCTCCCTCTCATTGGAGGAGCTGACCAAAGCA GCCCTAACGGGCCATAACACTTGACCAATTCAGCTGCTGGCAGAGGGAGGAAACAAGTGTTTTCCCAAGTGGCATTTTCATCTCGCTTTC ACCCTGACTAAAGATTGTCTTAAGTAGCAGCCCAGCCCGCCCAGCCCCAGGTGGGTAGTGGGGAGGAGAGCTGGCATTCCTCCAGGTGGC AAATGGCGACTCTATACTCTCCGCCCGCCCCAGGGCTGGATGGATTAGAAAAATGCCTATTTTTCTTGTATCGATGTAGAGACTCTATTT TCTCCCAAAGACACTATTTTTGCAGCTGTTTGAAGTTTGTATATTTTCCGTACTGCAGAGCTTACACAAAATTGAAGAATGTTAATGTTC GAGTTTTCTTATCTTGTGTTTAGAGGTTGTTTTTTGCAGATCTTGGTGTTAATAGACCAAATAAATAAATAAATATTCCCAGCA >1327_1327_1_ACLY-FAM117A_ACLY_chr17_40060982_ENST00000393896_FAM117A_chr17_47810082_ENST00000240364_length(amino acids)=473AA_BP=409 MGAGKSPAGPGQKPDPGKLPAAGVLRILRGSSGLWKKRRARTSAETGRAGLSAAMSAKAISEQTGKELLYKFICTTSAIQNRFKYARVTP DTDWARLLQDHPWLLSQNLVVKPDQLIKRRGKLGLVGVNLTLDGVKSWLKPRLGQEATVGKATGFLKNFLIEPFVPHSQAEEFYVCIYAT REGDYVLFHHEGGVDVGDVDAKAQKLLVGVDEKLNPEDIKKHLLVHAPEDKKEILASFISGLFNFYEDLYFTYLEINPLVVTKDGVYVLD LAAKVDATADYICKVKWGDIEFPPPFGREAYPEEAYIADLDAKSGASLKLTLLNPKGRIWTMVAGGGASVVYSDTICDLGGVNELANYGE YSGAPSEQQTYDYAKTILSLMTREKHPDGKILIIGGSIANFTNVAATFKPASHARWPQKSQCVGLSHFRSGVHSPWTPSSAPTFWASGHE MLMGPSPAAPMTRPPRRPCPGKS -------------------------------------------------------------- >1327_1327_2_ACLY-FAM117A_ACLY_chr17_40060982_ENST00000590151_FAM117A_chr17_47810082_ENST00000240364_length(transcript)=3264nt_BP=1175nt AGCTACCGGCTGCTGGGGTGCTCCGGATTTTGCGGGGTTCGTCGGGCCTGTGGAAGAAGCGCCGCGCACGGACTTCGGCAGAGACAGGTA GAGCAGGTCTCTCTGCAGCCATGTCGGCCAAGGCAATTTCAGAGCAGACGGGCAAAGAACTCCTTTACAAGTTCATCTGTACCACCTCAG CCATCCAGAATCGGTTCAAGTATGCTCGGGTCACTCCTGACACAGACTGGGCCCGCTTGCTGCAGGACCACCCCTGGCTGCTCAGCCAGA ACTTGGTAGTCAAGCCAGACCAGCTGATCAAACGTCGTGGAAAACTTGGTCTCGTTGGGGTCAACCTCACTCTGGATGGGGTCAAGTCCT GGCTGAAGCCACGGCTGGGACAGGAAGCCACAGTTGGCAAGGCCACAGGCTTCCTCAAGAACTTTCTGATCGAGCCCTTCGTCCCCCACA GTCAGGCTGAGGAGTTCTATGTCTGCATCTATGCCACCCGAGAAGGGGACTACGTCCTGTTCCACCACGAGGGGGGTGTGGACGTGGGTG ATGTGGACGCCAAGGCCCAGAAGCTGCTTGTTGGCGTGGATGAGAAACTGAATCCTGAGGACATCAAAAAACACCTGTTGGTCCACGCCC CTGAAGACAAGAAAGAAATTCTGGCCAGTTTTATCTCCGGCCTCTTCAATTTCTACGAGGACTTGTACTTCACCTACCTCGAGATCAATC CCCTTGTAGTGACCAAAGATGGAGTCTATGTCCTTGACTTGGCGGCCAAGGTGGACGCCACTGCCGACTACATCTGCAAAGTGAAGTGGG GTGACATCGAGTTCCCTCCCCCCTTCGGGCGGGAGGCATATCCAGAGGAAGCCTACATTGCAGACCTCGATGCCAAAAGTGGGGCAAGCC TGAAGCTGACCTTGCTGAACCCCAAAGGGAGGATCTGGACCATGGTGGCCGGGGGTGGCGCCTCTGTCGTGTACAGCGATACCATCTGTG ATCTAGGGGGTGTCAACGAGCTGGCAAACTATGGGGAGTACTCAGGCGCCCCCAGCGAGCAGCAGACCTATGACTATGCCAAGACTATCC TCTCCCTCATGACCCGAGAGAAGCACCCAGATGGCAAGATCCTCATCATTGGAGGCAGCATCGCAAACTTCACCAACGTGGCTGCCACGT TCAAGCCAGCGTCCCATGCTCGGTGGCCCCAGAAAAGTCAGTGTGTAGGCCTCAGCCACTTCAGGTCCGGCGTACATTCTCCCTGGACAC CATCCTCAGCTCCTACCTTCTGGGCCAGTGGCCACGAGATGCTGATGGGGCCTTCACCTGCTGCACCAATGACAAGGCCACCCAGACGCC CCTGTCCTGGCAAGAGCTAGAAGGTGAGCGTGCCAGTTCCTGTGCACACAAGCGCTCAGCATCCTGGGGCAGCACAGACCACCGAAAAGA GATTTCCAAGTTGAAGCAACAACTGCAGAGGACGAAGCTGAGCCGCAGTGGGAAAGAGAAGGAGCGAGGTTCACCACTCCTAGGGGACCA CGCAGTGCGGGGAGCACTGAGGGCGTCCCCTCCCAGCTTCCCCTCAGGGTCCCCTGTCTTGCGACTCAGCCCCTGCCTGCACAGGAGCCT GGAAGGGCTCAACCAAGAGCTGGAGGAGGTATTTGTGAAGGAGCAGGGAGAAGAGGAGCTGCTGAGGATCCTTGATATCCCTGATGGGCA CCGGGCCCCAGCTCCTCCCCAGAGTGGCAGCTGTGATCATCCCCTCCTCCTCCTGGAGCCTGGCAACCTTGCCAGCTCTCCTTCCATGTC CTTGGCATCTCCCCAGCCTTGTGGCCTGGCCAGTCATGAGGAACATCGGGGTGCCGCCGAGGAGCTGGCATCCACCCCCAACGACAAAGC CTCCTCTCCAGGACACCCAGCCTTTCTTGAAGATGGCAGCCCATCTCCAGTCCTTGCCTTTGCTGCCTCCCCTCGACCTAATCATAGCTA CATCTTCAAACGGGAGCCCCCAGAAGGCTGTGAGAAAGTGCGTGTGTTTGAAGAAGCCACGTCTCCAGGTCCTGACCTGGCCTTCCTGAC TTCCTGTCCTGACAAGAACAAAGTCCATTTCAACCCGACTGGCTCAGCCTTCTGCCCCGTCAACCTGATGAAGCCCCTCTTCCCCGGCAT GGGCTTCATCTTCCGTAACTGCCCCTCAAACCCGGGATCTCCCCTTCCCCCGGCCAGCCCCAGGCCACCACCTCGGAAGGATCCGGAAGC CTCCAAGGCCTCCCCACTGCCATTCGAGCCATGGCAGCGCACCCCACCATCAGAAGAGCCTGTGCTTTTCCAGAGCTCCCTGATGGTCTG AGGGTCCCACCCCTGCCCCACTTTACCATAGAGACCAGTGCCTTGGTGGCAGGTCCCTCCCCAGGTCCCCTGAGATGGGGTATGGAGGGG CCCTTCCCTCTCGGCCTTCGAGCACTTTCTTTCACTTACTGTGTCAAAGCCCTGGGTCCTCTTTTTGATGGGCACCGGCCCCTCTGAACG TGATGGGACCTGCCTTCTCCACTAGTAGCTGGGCAGCTCACAATTCACACCTGTGTACCTGCCACATCCCTCACTTGGTGGAAAACACCC AGAAGGTCTTGAGTCCCCCACCCCTGGGTGTCAGTCCAAATGACTGTATAGGAGGCCCTTATTTTTGTCACAGAGCAAGCTGGCCATGAA CGAAGGAGAGAAGACGCCACAGATTTCCTTCCCTCTCCTCCAGGAGACCATAAGATAGATCCCCCATCCTCTCAGCCCTATTCCCATGCC TCCCTCTCATTGGAGGAGCTGACCAAAGCAGCCCTAACGGGCCATAACACTTGACCAATTCAGCTGCTGGCAGAGGGAGGAAACAAGTGT TTTCCCAAGTGGCATTTTCATCTCGCTTTCACCCTGACTAAAGATTGTCTTAAGTAGCAGCCCAGCCCGCCCAGCCCCAGGTGGGTAGTG GGGAGGAGAGCTGGCATTCCTCCAGGTGGCAAATGGCGACTCTATACTCTCCGCCCGCCCCAGGGCTGGATGGATTAGAAAAATGCCTAT TTTTCTTGTATCGATGTAGAGACTCTATTTTCTCCCAAAGACACTATTTTTGCAGCTGTTTGAAGTTTGTATATTTTCCGTACTGCAGAG CTTACACAAAATTGAAGAATGTTAATGTTCGAGTTTTCTTATCTTGTGTTTAGAGGTTGTTTTTTGCAGATCTTGGTGTTAATAGACCAA ATAAATAAATAAATATTCCCAGCA >1327_1327_2_ACLY-FAM117A_ACLY_chr17_40060982_ENST00000590151_FAM117A_chr17_47810082_ENST00000240364_length(amino acids)=446AA_BP=382 MRGSSGLWKKRRARTSAETGRAGLSAAMSAKAISEQTGKELLYKFICTTSAIQNRFKYARVTPDTDWARLLQDHPWLLSQNLVVKPDQLI KRRGKLGLVGVNLTLDGVKSWLKPRLGQEATVGKATGFLKNFLIEPFVPHSQAEEFYVCIYATREGDYVLFHHEGGVDVGDVDAKAQKLL VGVDEKLNPEDIKKHLLVHAPEDKKEILASFISGLFNFYEDLYFTYLEINPLVVTKDGVYVLDLAAKVDATADYICKVKWGDIEFPPPFG REAYPEEAYIADLDAKSGASLKLTLLNPKGRIWTMVAGGGASVVYSDTICDLGGVNELANYGEYSGAPSEQQTYDYAKTILSLMTREKHP DGKILIIGGSIANFTNVAATFKPASHARWPQKSQCVGLSHFRSGVHSPWTPSSAPTFWASGHEMLMGPSPAAPMTRPPRRPCPGKS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACLY-FAM117A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACLY-FAM117A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Hgene | ACLY | P53396 | DB11936 | Bempedoic acid | Inhibitor | Small molecule | Approved|Investigational |
Top |
Related Diseases for ACLY-FAM117A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ACLY | C0018801 | Heart failure | 1 | CTD_human |
| Hgene | ACLY | C0018802 | Congestive heart failure | 1 | CTD_human |
| Hgene | ACLY | C0023212 | Left-Sided Heart Failure | 1 | CTD_human |
| Hgene | ACLY | C0028754 | Obesity | 1 | CTD_human |
| Hgene | ACLY | C0235527 | Heart Failure, Right-Sided | 1 | CTD_human |
| Hgene | ACLY | C1959583 | Myocardial Failure | 1 | CTD_human |
| Hgene | ACLY | C1961112 | Heart Decompensation | 1 | CTD_human |
| Hgene | ACLY | C2239176 | Liver carcinoma | 1 | CTD_human |
