
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ATP2A2-ARPC3 (FusionGDB2 ID:HG488TG10094) |
Fusion Gene Summary for ATP2A2-ARPC3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ATP2A2-ARPC3 | Fusion gene ID: hg488tg10094 | Hgene | Tgene | Gene symbol | ATP2A2 | ARPC3 | Gene ID | 488 | 10094 |
| Gene name | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 | actin related protein 2/3 complex subunit 3 | |
| Synonyms | ATP2B|DAR|DD|SERCA2 | ARC21|p21-Arc | |
| Cytomap | ('ATP2A2')('ARPC3') 12q24.11 | 12q24.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | sarcoplasmic/endoplasmic reticulum calcium ATPase 2ATPase Ca++ transporting cardiac muscle slow twitch 2ATPase, Ca++ dependent, slow-twitch, cardiac muscle-2SR Ca(2+)-ATPase 2calcium pump 2calcium-transporting ATPase sarcoplasmic reticulum type, slow | actin-related protein 2/3 complex subunit 3ARP2/3 protein complex subunit p21actin related protein 2/3 complex, subunit 3, 21kDaarp2/3 complex 21 kDa subunit | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000550248, ENST00000308664, ENST00000395494, ENST00000539276, ENST00000552636, | ||
| Fusion gene scores | * DoF score | 17 X 22 X 11=4114 | 6 X 4 X 4=96 |
| # samples | 27 | 7 | |
| ** MAII score | log2(27/4114*10)=-3.9295104814741 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/96*10)=-0.45567948377619 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ATP2A2 [Title/Abstract] AND ARPC3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ATP2A2(110734542)-ARPC3(110878193), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ATP2A2-ARPC3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATP2A2-ARPC3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ATP2A2 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 16402920 |
| Hgene | ATP2A2 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration | 16402920 |
| Hgene | ATP2A2 | GO:0070588 | calcium ion transmembrane transport | 16402920 |
| Hgene | ATP2A2 | GO:1903515 | calcium ion transport from cytosol to endoplasmic reticulum | 16402920 |
| Tgene | ARPC3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 11741539 |
 Fusion gene breakpoints across ATP2A2 (5'-gene) Fusion gene breakpoints across ATP2A2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
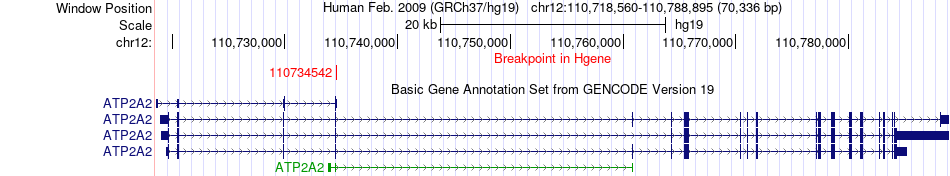 |
 Fusion gene breakpoints across ARPC3 (3'-gene) Fusion gene breakpoints across ARPC3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
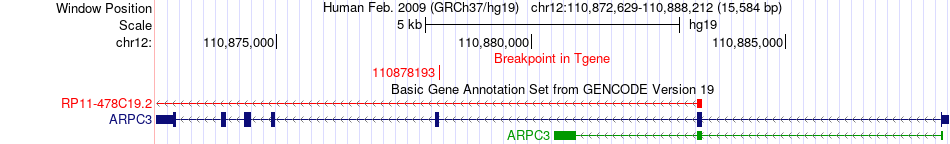 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8366-01A | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
Top |
Fusion Gene ORF analysis for ATP2A2-ARPC3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000550248 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| 3UTR-intron | ENST00000550248 | ENST00000471641 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000308664 | ENST00000471641 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000395494 | ENST00000471641 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000539276 | ENST00000471641 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000308664 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000395494 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000539276 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| intron-3CDS | ENST00000552636 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
| intron-intron | ENST00000552636 | ENST00000471641 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000308664 | ATP2A2 | chr12 | 110734542 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1898 | 1137 | 329 | 1567 | 412 |
| ENST00000395494 | ATP2A2 | chr12 | 110734542 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1787 | 1026 | 218 | 1456 | 412 |
| ENST00000539276 | ATP2A2 | chr12 | 110734542 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1333 | 572 | 109 | 1002 | 297 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000308664 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - | 0.002040749 | 0.99795926 |
| ENST00000395494 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - | 0.002038574 | 0.9979614 |
| ENST00000539276 | ENST00000228825 | ATP2A2 | chr12 | 110734542 | + | ARPC3 | chr12 | 110878193 | - | 0.000989583 | 0.99901044 |
Top |
Fusion Genomic Features for ATP2A2-ARPC3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
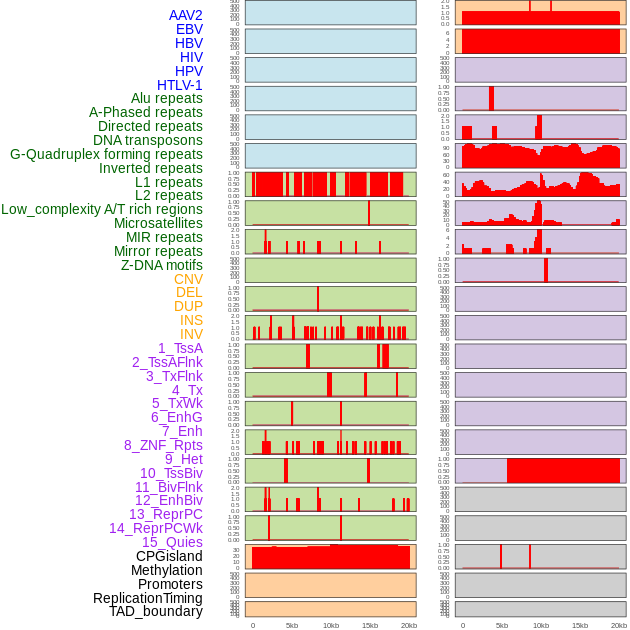 |
Top |
Fusion Protein Features for ATP2A2-ARPC3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:110734542/chr12:110878193) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 1_48 | 154 | 998.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 70_89 | 154 | 998.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 1_48 | 154 | 1016.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 70_89 | 154 | 1016.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 1_48 | 154 | 1043.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 70_89 | 154 | 1043.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 49_69 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 90_110 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 49_69 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 90_110 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 49_69 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 90_110 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 111_253 | 154 | 998.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 274_295 | 154 | 998.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 314_756 | 154 | 998.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 777_786 | 154 | 998.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 808_827 | 154 | 998.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 851_896 | 154 | 998.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 917_929 | 154 | 998.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 949_963 | 154 | 998.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 985_1042 | 154 | 998.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 111_253 | 154 | 1016.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 274_295 | 154 | 1016.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 314_756 | 154 | 1016.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 777_786 | 154 | 1016.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 808_827 | 154 | 1016.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 851_896 | 154 | 1016.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 917_929 | 154 | 1016.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 949_963 | 154 | 1016.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 985_1042 | 154 | 1016.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 111_253 | 154 | 1043.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 274_295 | 154 | 1043.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 314_756 | 154 | 1043.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 777_786 | 154 | 1043.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 808_827 | 154 | 1043.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 851_896 | 154 | 1043.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 917_929 | 154 | 1043.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 949_963 | 154 | 1043.0 | Topological domain | Lumenal |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 985_1042 | 154 | 1043.0 | Topological domain | Cytoplasmic |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 254_273 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 296_313 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 757_776 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 828_850 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 897_916 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D8 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 930_948 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D9 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 964_984 | 154 | 998.0 | Transmembrane | Helical%3B Name%3D10 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 254_273 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 296_313 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 757_776 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 828_850 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 897_916 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D8 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 930_948 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D9 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 964_984 | 154 | 1016.0 | Transmembrane | Helical%3B Name%3D10 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 254_273 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 296_313 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 757_776 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 828_850 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D7 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 897_916 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D8 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 930_948 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D9 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 964_984 | 154 | 1043.0 | Transmembrane | Helical%3B Name%3D10 |
Top |
Fusion Gene Sequence for ATP2A2-ARPC3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >7825_7825_1_ATP2A2-ARPC3_ATP2A2_chr12_110734542_ENST00000308664_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1898nt_BP=1137nt GCGCGGGAGGAGGGAGCCGGGAGGAGGGGGCGGGGCCGCGCCGCCCGCGCCGCGCTGGGCGCTCTCGGCCAATGAGCGGCGTCCACATGC CGCGGCGGCGGCGAAAGGGGAGGCAGCGGCCGATAAATGCTATTAGAGCAGCCGCCGCGGAGCCGTCCCCGACGCCACCTCCTTTTCCTT CGCCGCAGTTTCCTCCGCCGCTGTCGGGCGTGCGGCGCTGAGGGACCCGGGCGAGCGCGCCGCGCACCGCCCCGCCGGCTCGCCTCCCTC GCCGCGTTCCGCCCTCAGTGGTCTGCCGGGCGCCCCCTCCTCCGGCCCGGGCGGGGCCTCTGATCGCCTCAAGAGAGCGGGGAGGGGGCT CGGGGGCCGCGGCCTGCCCTCCCGGCGGGCGGCTGAGGGCGAGGGAGGCCCTCCCTTCTGGCGAGGGGAGGGAGGGTGGGTCAGGAGCCC CCAACCCGCCCTGCGGAGCTCGGGGCCGCGCGAGGGGCGGTTGTCTGGGGGAGGGGGCGCGGGGTGATTCAGCGCCCGGCGAGGCGGAAG CGGCCGCAAGAGGAGGAGGGGAGAGCCCGTCCGCGCCTGGGCTCCCGGGGTGGCACGAGCCCGCGGCCGGAGTGCGAGGCGGAGGCGAGG AGGCCGCGGGGACGGGAGGCGAGGCCGGCCGGGCCCCCGAAGCCATGGAGAACGCGCACACCAAGACGGTGGAGGAGGTGCTGGGCCACT TCGGCGTCAACGAGAGTACGGGGCTGAGCCTGGAACAGGTCAAGAAGCTTAAGGAGAGATGGGGCTCCAACGAGTTACCGGCTGAAGAAG GAAAAACCTTGCTGGAACTTGTGATTGAGCAGTTTGAAGACTTGCTAGTTAGGATTTTATTACTGGCAGCATGTATATCTTTTGTTTTGG CTTGGTTTGAAGAAGGTGAAGAAACAATTACAGCCTTTGTAGAACCTTTTGTAATTTTACTCATATTAGTAGCCAATGCAATTGTGGGTG TATGGCAGGAAAGAAATGCTGAAAATGCCATCGAAGCCCTTAAGGAATATGAGCCTGAAATGGGCAAAGTGTATCGACAGGACAGAAAGA GTGTGCAGCGGATTAAAGCTAAAGACATAGTTCCTGGTGATATTGTAGAAATTGCTGCAAAAGATACAGATATTGTGGATGAAGCCATCT ATTACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTACATTTCTG AATGTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCATTCCTG GAGAGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAACAGCTAA GGCAAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGAAGAGAC AGTTCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTTCCAGCA AGATGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGATCAAGAA TTTGGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAGTAATAA TAAAGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAATTCACATT CCATAATG >7825_7825_1_ATP2A2-ARPC3_ATP2A2_chr12_110734542_ENST00000308664_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=412AA_BP=269 MIASRERGGGSGAAACPPGGRLRAREALPSGEGREGGSGAPNPPCGARGRARGGCLGEGARGDSAPGEAEAAARGGGESPSAPGLPGWHE PAAGVRGGGEEAAGTGGEAGRAPEAMENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDLLVRIL LLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNAENAIEALKEYEPEMGKVYRQDRKSVQRIKAKDIVPGDIVEIAA KDTDIVDEAIYYFKANVFFKNYEIKNEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKPANKQED EVMRAYLQQLRQETGLRLCEKVFDPQNDKPSKWWTCFVKRQFMNKSLSGPGQ -------------------------------------------------------------- >7825_7825_2_ATP2A2-ARPC3_ATP2A2_chr12_110734542_ENST00000395494_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1787nt_BP=1026nt GGCAGCGGCCGATAAATGCTATTAGAGCAGCCGCCGCGGAGCCGTCCCCGACGCCACCTCCTTTTCCTTCGCCGCAGTTTCCTCCGCCGC TGTCGGGCGTGCGGCGCTGAGGGACCCGGGCGAGCGCGCCGCGCACCGCCCCGCCGGCTCGCCTCCCTCGCCGCGTTCCGCCCTCAGTGG TCTGCCGGGCGCCCCCTCCTCCGGCCCGGGCGGGGCCTCTGATCGCCTCAAGAGAGCGGGGAGGGGGCTCGGGGGCCGCGGCCTGCCCTC CCGGCGGGCGGCTGAGGGCGAGGGAGGCCCTCCCTTCTGGCGAGGGGAGGGAGGGTGGGTCAGGAGCCCCCAACCCGCCCTGCGGAGCTC GGGGCCGCGCGAGGGGCGGTTGTCTGGGGGAGGGGGCGCGGGGTGATTCAGCGCCCGGCGAGGCGGAAGCGGCCGCAAGAGGAGGAGGGG AGAGCCCGTCCGCGCCTGGGCTCCCGGGGTGGCACGAGCCCGCGGCCGGAGTGCGAGGCGGAGGCGAGGAGGCCGCGGGGACGGGAGGCG AGGCCGGCCGGGCCCCCGAAGCCATGGAGAACGCGCACACCAAGACGGTGGAGGAGGTGCTGGGCCACTTCGGCGTCAACGAGAGTACGG GGCTGAGCCTGGAACAGGTCAAGAAGCTTAAGGAGAGATGGGGCTCCAACGAGTTACCGGCTGAAGAAGGAAAAACCTTGCTGGAACTTG TGATTGAGCAGTTTGAAGACTTGCTAGTTAGGATTTTATTACTGGCAGCATGTATATCTTTTGTTTTGGCTTGGTTTGAAGAAGGTGAAG AAACAATTACAGCCTTTGTAGAACCTTTTGTAATTTTACTCATATTAGTAGCCAATGCAATTGTGGGTGTATGGCAGGAAAGAAATGCTG AAAATGCCATCGAAGCCCTTAAGGAATATGAGCCTGAAATGGGCAAAGTGTATCGACAGGACAGAAAGAGTGTGCAGCGGATTAAAGCTA AAGACATAGTTCCTGGTGATATTGTAGAAATTGCTGCAAAAGATACAGATATTGTGGATGAAGCCATCTATTACTTCAAGGCCAATGTCT TCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTACATTTCTGAATGTCTGAAGAAACTGCAAA AGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCATTCCTGGAGAGCCTGGTTTTCCACTTA ACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAACAGCTAAGGCAAGAGACTGGACTGAGAC TTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGAAGAGACAGTTCATGAACAAGAGTCTTT CAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTTCCAGCAAGATGTACACAATCTTTTGCC TTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGATCAAGAATTTGGGTGGGAGAAAAGAAAG TGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAGTAATAATAAAGAAATTTCTAACATTCC ATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAATTCACATTCCATAATG >7825_7825_2_ATP2A2-ARPC3_ATP2A2_chr12_110734542_ENST00000395494_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=412AA_BP=269 MIASRERGGGSGAAACPPGGRLRAREALPSGEGREGGSGAPNPPCGARGRARGGCLGEGARGDSAPGEAEAAARGGGESPSAPGLPGWHE PAAGVRGGGEEAAGTGGEAGRAPEAMENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDLLVRIL LLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNAENAIEALKEYEPEMGKVYRQDRKSVQRIKAKDIVPGDIVEIAA KDTDIVDEAIYYFKANVFFKNYEIKNEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKPANKQED EVMRAYLQQLRQETGLRLCEKVFDPQNDKPSKWWTCFVKRQFMNKSLSGPGQ -------------------------------------------------------------- >7825_7825_3_ATP2A2-ARPC3_ATP2A2_chr12_110734542_ENST00000539276_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1333nt_BP=572nt CCCGTCCGCGCCTGGGCTCCCGGGGTGGCACGAGCCCGCGGCCGGAGTGCGAGGCGGAGGCGAGGAGGCCGCGGGGACGGGAGGCGAGGC CGGCCGGGCCCCCGAAGCCATGGAGAACGCGCACACCAAGACGGTGGAGGAGGTGCTGGGCCACTTCGGCGTCAACGAGAGTACGGGGCT GAGCCTGGAACAGGTCAAGAAGCTTAAGGAGAGATGGGGCTCCAACGAGTTACCGGCTGAAGAAGGAAAAACCTTGCTGGAACTTGTGAT TGAGCAGTTTGAAGACTTGCTAGTTAGGATTTTATTACTGGCAGCATGTATATCTTTTGTTTTGGCTTGGTTTGAAGAAGGTGAAGAAAC AATTACAGCCTTTGTAGAACCTTTTGTAATTTTACTCATATTAGTAGCCAATGCAATTGTGGGTGTATGGCAGGAAAGAAATGCTGAAAA TGCCATCGAAGCCCTTAAGGAATATGAGCCTGAAATGGGCAAAGTGTATCGACAGGACAGAAAGAGTGTGCAGCGGATTAAAGCTAAAGA CATAGTTCCTGGTGATATTGTAGAAATTGCTGCAAAAGATACAGATATTGTGGATGAAGCCATCTATTACTTCAAGGCCAATGTCTTCTT CAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTACATTTCTGAATGTCTGAAGAAACTGCAAAAGTG CAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCATTCCTGGAGAGCCTGGTTTTCCACTTAACGC AATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAACAGCTAAGGCAAGAGACTGGACTGAGACTTTG TGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGAAGAGACAGTTCATGAACAAGAGTCTTTCAGG ACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTTCCAGCAAGATGTACACAATCTTTTGCCTTTA TTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGATCAAGAATTTGGGTGGGAGAAAAGAAAGTGGG TTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAGTAATAATAAAGAAATTTCTAACATTCCATGT CAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAATTCACATTCCATAATG >7825_7825_3_ATP2A2-ARPC3_ATP2A2_chr12_110734542_ENST00000539276_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=297AA_BP=154 MENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDLLVRILLLAACISFVLAWFEEGEETITAFVE PFVILLILVANAIVGVWQERNAENAIEALKEYEPEMGKVYRQDRKSVQRIKAKDIVPGDIVEIAAKDTDIVDEAIYYFKANVFFKNYEIK NEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKPANKQEDEVMRAYLQQLRQETGLRLCEKVFDP QNDKPSKWWTCFVKRQFMNKSLSGPGQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ATP2A2-ARPC3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 575_594 | 154.33333333333334 | 998.0 | HAX1 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 575_594 | 154.33333333333334 | 1016.0 | HAX1 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 575_594 | 154.33333333333334 | 1043.0 | HAX1 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 787_807 | 154.33333333333334 | 998.0 | PLN |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 931_942 | 154.33333333333334 | 998.0 | PLN |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 787_807 | 154.33333333333334 | 1016.0 | PLN |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 931_942 | 154.33333333333334 | 1016.0 | PLN |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 787_807 | 154.33333333333334 | 1043.0 | PLN |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 931_942 | 154.33333333333334 | 1043.0 | PLN |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000308664 | + | 5 | 21 | 788_1042 | 154.33333333333334 | 998.0 | TMEM64 and PDIA3 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000395494 | + | 5 | 19 | 788_1042 | 154.33333333333334 | 1016.0 | TMEM64 and PDIA3 |
| Hgene | ATP2A2 | chr12:110734542 | chr12:110878193 | ENST00000539276 | + | 5 | 20 | 788_1042 | 154.33333333333334 | 1043.0 | TMEM64 and PDIA3 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ATP2A2-ARPC3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ATP2A2-ARPC3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ATP2A2 | C0022595 | Keratosis Follicularis | 12 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ATP2A2 | C0265971 | Acrokeratosis Verruciformis of Hopf | 3 | CTD_human;ORPHANET;UNIPROT |
| Hgene | ATP2A2 | C0011570 | Mental Depression | 2 | PSYGENET |
| Hgene | ATP2A2 | C0011581 | Depressive disorder | 2 | PSYGENET |
| Hgene | ATP2A2 | C0473575 | Acantholytic Dyskeratotic Epidermal Nevus | 2 | CTD_human |
| Hgene | ATP2A2 | C0525045 | Mood Disorders | 2 | PSYGENET |
| Hgene | ATP2A2 | C0002152 | Alloxan Diabetes | 1 | CTD_human |
| Hgene | ATP2A2 | C0011853 | Diabetes Mellitus, Experimental | 1 | CTD_human |
| Hgene | ATP2A2 | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Hgene | ATP2A2 | C0018799 | Heart Diseases | 1 | CTD_human |
| Hgene | ATP2A2 | C0018800 | Cardiomegaly | 1 | CTD_human |
| Hgene | ATP2A2 | C0018801 | Heart failure | 1 | CTD_human |
| Hgene | ATP2A2 | C0018802 | Congestive heart failure | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | ATP2A2 | C0023212 | Left-Sided Heart Failure | 1 | CTD_human |
| Hgene | ATP2A2 | C0027055 | Myocardial Reperfusion Injury | 1 | CTD_human |
| Hgene | ATP2A2 | C0036341 | Schizophrenia | 1 | PSYGENET |
| Hgene | ATP2A2 | C0038220 | Status Epilepticus | 1 | CTD_human |
| Hgene | ATP2A2 | C0038433 | Streptozotocin Diabetes | 1 | CTD_human |
| Hgene | ATP2A2 | C0206145 | Stunned Myocardium | 1 | CTD_human |
| Hgene | ATP2A2 | C0206146 | Myocardial Stunning | 1 | CTD_human |
| Hgene | ATP2A2 | C0235527 | Heart Failure, Right-Sided | 1 | CTD_human |
| Hgene | ATP2A2 | C0242698 | Ventricular Dysfunction, Left | 1 | CTD_human |
| Hgene | ATP2A2 | C0270823 | Petit mal status | 1 | CTD_human |
| Hgene | ATP2A2 | C0311335 | Grand Mal Status Epilepticus | 1 | CTD_human |
| Hgene | ATP2A2 | C0376416 | Hibernation, Myocardial | 1 | CTD_human |
| Hgene | ATP2A2 | C0393734 | Complex Partial Status Epilepticus | 1 | CTD_human |
| Hgene | ATP2A2 | C0751522 | Status Epilepticus, Subclinical | 1 | CTD_human |
| Hgene | ATP2A2 | C0751523 | Non-Convulsive Status Epilepticus | 1 | CTD_human |
| Hgene | ATP2A2 | C0751524 | Simple Partial Status Epilepticus | 1 | CTD_human |
| Hgene | ATP2A2 | C0853897 | Diabetic Cardiomyopathies | 1 | CTD_human |
| Hgene | ATP2A2 | C1383860 | Cardiac Hypertrophy | 1 | CTD_human |
| Hgene | ATP2A2 | C1959583 | Myocardial Failure | 1 | CTD_human |
| Hgene | ATP2A2 | C1961112 | Heart Decompensation | 1 | CTD_human |
