
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACO2-ARHGAP8 (FusionGDB2 ID:HG50TG23779) |
Fusion Gene Summary for ACO2-ARHGAP8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACO2-ARHGAP8 | Fusion gene ID: hg50tg23779 | Hgene | Tgene | Gene symbol | ACO2 | ARHGAP8 | Gene ID | 50 | 23779 |
| Gene name | aconitase 2 | Rho GTPase activating protein 8 | |
| Synonyms | ACONM|HEL-S-284|ICRD|OCA8|OPA9 | BPGAP1|PP610 | |
| Cytomap | ('ACO2')('ARHGAP8') 22q13.2 | 22q13.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | aconitate hydratase, mitochondrialaconitase 2, mitochondrialcitrate hydro-lyaseepididymis secretory sperm binding protein Li 284mitochondrial aconitase | rho GTPase-activating protein 8BCH domain-containing Cdc42GAP-like proteinBNIP-2 and Cdc42GAP homology domain-containing, proline-rich and Cdc42GAP-like protein subtype-1rho-type GTPase-activating protein 8 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q99798 | P85298 | |
| Ensembl transtripts involved in fusion gene | ENST00000216254, ENST00000396512, ENST00000466237, | ||
| Fusion gene scores | * DoF score | 17 X 8 X 10=1360 | 6 X 9 X 5=270 |
| # samples | 22 | 7 | |
| ** MAII score | log2(22/1360*10)=-2.62803122261304 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/270*10)=-1.94753258010586 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ACO2 [Title/Abstract] AND ARHGAP8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACO2(41895866)-ARHGAP8(45110471), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ACO2-ARHGAP8 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACO2-ARHGAP8 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACO2-ARHGAP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACO2-ARHGAP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACO2-ARHGAP8 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ACO2-ARHGAP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACO2 | GO:0006099 | tricarboxylic acid cycle | 9630632 |
| Hgene | ACO2 | GO:0006101 | citrate metabolic process | 9630632 |
| Tgene | ARHGAP8 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 20179103 |
 Fusion gene breakpoints across ACO2 (5'-gene) Fusion gene breakpoints across ACO2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
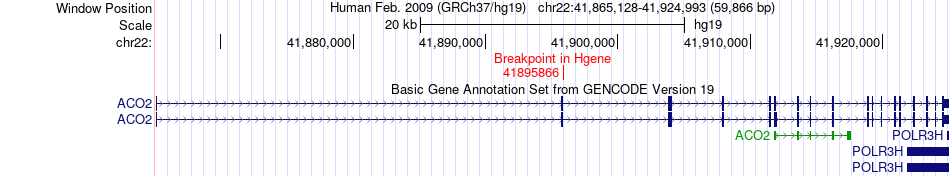 |
 Fusion gene breakpoints across ARHGAP8 (3'-gene) Fusion gene breakpoints across ARHGAP8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
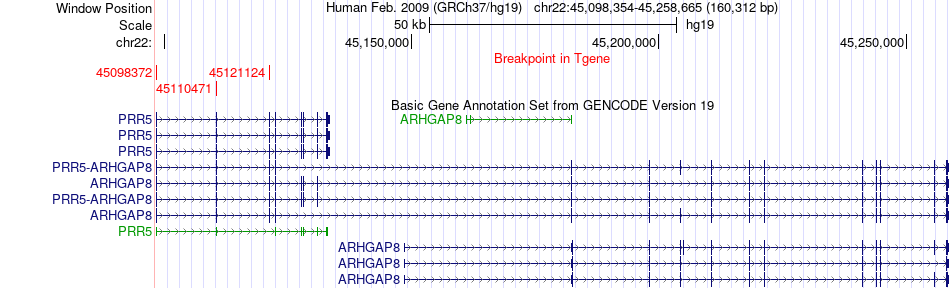 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM-01A | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM-01A | ACO2 | chr22 | 41895866 | - | ARHGAP8 | chr22 | 45110471 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM-01A | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| ChimerDB4 | UCEC | TCGA-AJ-A3EM-01A | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
Top |
Fusion Gene ORF analysis for ACO2-ARHGAP8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000216254 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000216254 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000216254 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000216254 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000216254 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000216254 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000216254 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000216254 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000216254 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000216254 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000216254 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000216254 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000216254 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000396512 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000396512 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000396512 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000396512 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000396512 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000396512 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000396512 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000396512 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000396512 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000396512 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| 5CDS-intron | ENST00000396512 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| 5CDS-intron | ENST00000396512 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| 5CDS-intron | ENST00000396512 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| Frame-shift | ENST00000216254 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| Frame-shift | ENST00000396512 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| In-frame | ENST00000216254 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| In-frame | ENST00000216254 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| In-frame | ENST00000216254 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| In-frame | ENST00000216254 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| In-frame | ENST00000396512 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| In-frame | ENST00000396512 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| In-frame | ENST00000396512 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| In-frame | ENST00000396512 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| intron-3CDS | ENST00000466237 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| intron-3CDS | ENST00000466237 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| intron-3CDS | ENST00000466237 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| intron-3CDS | ENST00000466237 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| intron-3CDS | ENST00000466237 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| intron-intron | ENST00000466237 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| intron-intron | ENST00000466237 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| intron-intron | ENST00000466237 | ENST00000336963 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| intron-intron | ENST00000466237 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| intron-intron | ENST00000466237 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| intron-intron | ENST00000466237 | ENST00000356099 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| intron-intron | ENST00000466237 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| intron-intron | ENST00000466237 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| intron-intron | ENST00000466237 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| intron-intron | ENST00000466237 | ENST00000389774 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
| intron-intron | ENST00000466237 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45098372 | + |
| intron-intron | ENST00000466237 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + |
| intron-intron | ENST00000466237 | ENST00000469872 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000216254 | ACO2 | chr22 | 41895866 | + | ENST00000517296 | ARHGAP8 | chr22 | 45110471 | + | 2104 | 195 | 7 | 1992 | 661 |
| ENST00000216254 | ACO2 | chr22 | 41895866 | + | ENST00000389773 | ARHGAP8 | chr22 | 45110471 | + | 1945 | 195 | 7 | 1755 | 582 |
| ENST00000396512 | ACO2 | chr22 | 41895866 | + | ENST00000517296 | ARHGAP8 | chr22 | 45110471 | + | 2099 | 190 | 2 | 1987 | 661 |
| ENST00000396512 | ACO2 | chr22 | 41895866 | + | ENST00000389773 | ARHGAP8 | chr22 | 45110471 | + | 1940 | 190 | 2 | 1750 | 582 |
| ENST00000216254 | ACO2 | chr22 | 41895866 | + | ENST00000517296 | ARHGAP8 | chr22 | 45121124 | + | 2023 | 195 | 7 | 1911 | 634 |
| ENST00000216254 | ACO2 | chr22 | 41895866 | + | ENST00000389773 | ARHGAP8 | chr22 | 45121124 | + | 1864 | 195 | 7 | 1674 | 555 |
| ENST00000396512 | ACO2 | chr22 | 41895866 | + | ENST00000517296 | ARHGAP8 | chr22 | 45121124 | + | 2018 | 190 | 2 | 1906 | 634 |
| ENST00000396512 | ACO2 | chr22 | 41895866 | + | ENST00000389773 | ARHGAP8 | chr22 | 45121124 | + | 1859 | 190 | 2 | 1669 | 555 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000216254 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + | 0.017317306 | 0.9826827 |
| ENST00000216254 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + | 0.017884467 | 0.9821155 |
| ENST00000396512 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + | 0.017112203 | 0.98288774 |
| ENST00000396512 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110471 | + | 0.017710026 | 0.98228997 |
| ENST00000216254 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + | 0.024145449 | 0.9758546 |
| ENST00000216254 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + | 0.010431885 | 0.98956805 |
| ENST00000396512 | ENST00000517296 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + | 0.023871291 | 0.9761287 |
| ENST00000396512 | ENST00000389773 | ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121124 | + | 0.010197055 | 0.98980296 |
Top |
Fusion Genomic Features for ACO2-ARHGAP8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110470 | + | 1.64E-11 | 1 |
| ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121123 | + | 5.16E-08 | 1 |
| ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45110470 | + | 1.64E-11 | 1 |
| ACO2 | chr22 | 41895866 | + | ARHGAP8 | chr22 | 45121123 | + | 5.16E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
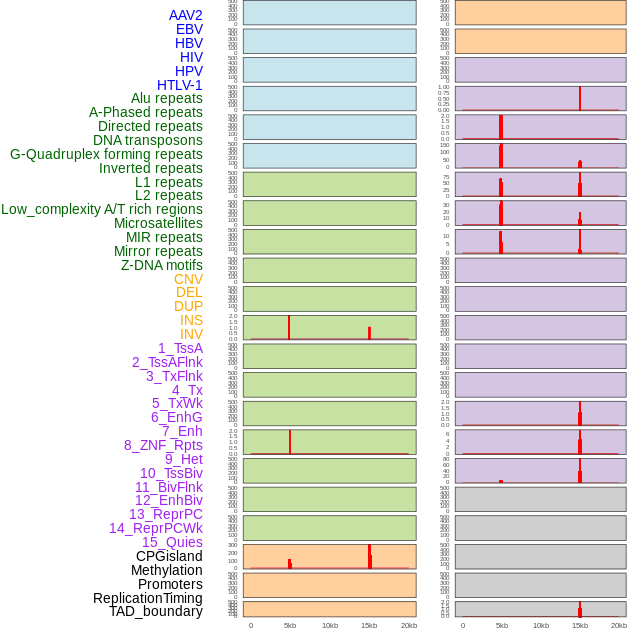 |
Top |
Fusion Protein Features for ACO2-ARHGAP8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:41895866/chr22:45110471) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ACO2 | ARHGAP8 |
| FUNCTION: Catalyzes the isomerization of citrate to isocitrate via cis-aconitate. {ECO:0000250|UniProtKB:P16276}. | FUNCTION: GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45110471 | ENST00000356099 | 0 | 12 | 13_199 | 0 | 434.0 | Domain | CRAL-TRIO | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45110471 | ENST00000356099 | 0 | 12 | 226_412 | 0 | 434.0 | Domain | Rho-GAP | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45110471 | ENST00000389774 | 0 | 13 | 13_199 | 0 | 465.0 | Domain | CRAL-TRIO | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45110471 | ENST00000389774 | 0 | 13 | 226_412 | 0 | 465.0 | Domain | Rho-GAP | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45121124 | ENST00000356099 | 0 | 12 | 13_199 | 0 | 434.0 | Domain | CRAL-TRIO | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45121124 | ENST00000356099 | 0 | 12 | 226_412 | 0 | 434.0 | Domain | Rho-GAP | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45121124 | ENST00000389774 | 0 | 13 | 13_199 | 0 | 465.0 | Domain | CRAL-TRIO | |
| Tgene | ARHGAP8 | chr22:41895866 | chr22:45121124 | ENST00000389774 | 0 | 13 | 226_412 | 0 | 465.0 | Domain | Rho-GAP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACO2 | chr22:41895866 | chr22:45110471 | ENST00000216254 | + | 2 | 18 | 192_194 | 57 | 781.0 | Region | Substrate binding |
| Hgene | ACO2 | chr22:41895866 | chr22:45110471 | ENST00000216254 | + | 2 | 18 | 670_671 | 57 | 781.0 | Region | Substrate binding |
| Hgene | ACO2 | chr22:41895866 | chr22:45121124 | ENST00000216254 | + | 2 | 18 | 192_194 | 57 | 781.0 | Region | Substrate binding |
| Hgene | ACO2 | chr22:41895866 | chr22:45121124 | ENST00000216254 | + | 2 | 18 | 670_671 | 57 | 781.0 | Region | Substrate binding |
Top |
Fusion Gene Sequence for ACO2-ARHGAP8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1341_1341_1_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45110471_ENST00000389773_length(transcript)=1945nt_BP=195nt CTCATCTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGC CTCAGTCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAA CATTGTTCGCAAACGCATCCACAACGGGGTGATCGCCGTCTTCCAGCGCAAGGGGCTGCCCGACCAGGAGCTCTTCAGCCTCAACGAGGG CGTCCGGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGTGATCCTTCG GGACAAGATTCGCTTCTATGAGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGAGGCCCTCGGTGGT GCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGGTGGCAGGGGATGA CCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGCTGGAGTATTTGAA GTACACACTGGACCAATACGTTGAGAACGATTATACCATCGTCTATTTCCACTACGGGCTGAACAGCCGGAACAAGCCTTCCCTGGGCTG GCTCCAGAGCGCATACAAGGAGTTCGATAGGAAGTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCAAGGT CCTGTGGAACATCTTGAAGCCCCTCATCAGTCACAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAACACCT TAAATACGACCAGCTGGTCATCCCTCCCGAAGTTTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTCCCAC CAAGACACCACCGCCGCGGCCCCCGCTGCCCACACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAACTCAT CCCCCCTGTGCTGAGGTTCACAGTGACGTACCTGAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGCAGAC CGTCCGCGAGATCCAGAGGCTCTACAACCAAGGGAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCCTGAA GACCTTCCTGCGAGAGCTGCCCCAGCCGCTTCTGACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCCTGCG TGTCACTGGCTGCCGCCAGATCTTACGGAGCCTCCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGGTGTC CCGGGAGAGCATCTTCAACAAAATGAACAGCTCTAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCTCCTC CCTGAGTGCCCTTGTGCCCCTGAACATGTTCACTGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTGGGGA GCACGGCCTGGCACCATGGGAACAGGGGAGCAGGGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCAAGCC TACCCTACCTCCGAGTCCCCTGATGGCAGCCAGAAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACACCTGT CTGTGCACTTGTATGTTTTGTAAACTTGGCATCTGTAAAAATAACCAGCCATTAGATGAATTCAGAACCTTCTAATGAAAACTCCATGCC TCTGGTCCTTGGACTCTTGTCCATGGTTCCTGAGCTGTGGACCGGGATAGAATAA >1341_1341_1_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45110471_ENST00000389773_length(amino acids)=582AA_BP=63 MSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRIHNGVIAVFQRKGLPDQELFSLNEGVR QLLKTELGSFFTEYLQNQLLTKGMVILRDKIRFYEELQRDKAAAAAVLGAVRKRPSVVPMAGQDPALSTSHPFYDVARHGILQVAGDDRF GRRVVTFSCCRMPPSHELDHQRLLEYLKYTLDQYVENDYTIVYFHYGLNSRNKPSLGWLQSAYKEFDRKYKKNLKALYVVHPTSFIKVLW NILKPLISHKFGKKVIYFNYLSELHEHLKYDQLVIPPEVLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPP VLRFTVTYLREKGLRTEGLFRRSASVQTVREIQRLYNQGKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVT GCRQILRSLPEHNYVVLRYLMGFLHAVSRESIFNKMNSSNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHG LAPWEQGSRAAPLQEAVPRTQATGLTKPTLPPSPLMAARRRL -------------------------------------------------------------- >1341_1341_2_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45110471_ENST00000517296_length(transcript)=2104nt_BP=195nt CTCATCTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGC CTCAGTCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAA CATTGTTCGCAAACGCATCCACAACGGGGTGATCGCCGTCTTCCAGCGCAAGGGGCTGCCCGACCAGGAGCTCTTCAGCCTCAACGAGGG CGTCCGGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGTGATCCTTCG GGACAAGATTCGCTTCTATGAGGGACAGAAGCTGCTGGACTCACTGGCAGAGACCTGGGACTTCTTCTTCAGTGACGTGCTGCCCATGCT GCAGGCCATCTTCTACCCGGTGCAGGGCAAGGAGCCATCGGTGCGCCAGCTGGCCCTGCTGCACTTCCGGAATGCCATCACCCTCAGTGT GAAGCTAGAGGATGCGCTGGCCCGGGCCCATGCCCGTGTGCCCCCTGCCATCGTGCAGATGCTGCTGGTGCTGCAGGGGGTACATGAGTC CAGGGGCGTGACTGAGGACTACCTGCGCCTGGAGACGCTGGTCCAGAAGGTGGTGTCGCCATACCTGGGCACCTACGGCCTCCACTCCAG CGAGGGGCCCTTCACCCATTCCTGCATCCTGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGAGGCC CTCGGTGGTGCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGGTGGC AGGGGATGACCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGCTGGA GTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCAAGGTCCTGTGGAACATCTTGAAGCCCCTCATCAGTCA CAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAACACCTTAAATACGACCAGCTGGTCATCCCTCCCGAAGT TTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTCCCACCAAGACACCACCGCCGCGGCCCCCGCTGCCCAC ACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAACTCATCCCCCCTGTGCTGAGGTTCACAGTGACGTACCT GAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGCAGACCGTCCGCGAGATCCAGAGGCTCTACAACCAAGG GAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCCTGAAGACCTTCCTGCGAGAGCTGCCCCAGCCGCTTCT GACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCCTGCGTGTCACTGGCTGCCGCCAGATCTTACGGAGCCT CCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGGTGTCCCGGGAGAGCATCTTCAACAAAATGAACAGCTC TAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCTCCTCCCTGAGTGCCCTTGTGCCCCTGAACATGTTCAC TGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTGGGGAGCACGGCCTGGCACCATGGGAACAGGGGAGCAG GGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCAAGCCTACCCTACCTCCGAGTCCCCTGATGGCAGCCAG AAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACACCTGTCTGTGCACTTGTATGTTTTGTAAACTTGGCATC TGTAAAAATAACCAGCCATTAGATGAATTCAGAA >1341_1341_2_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45110471_ENST00000517296_length(amino acids)=661AA_BP=63 MSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRIHNGVIAVFQRKGLPDQELFSLNEGVR QLLKTELGSFFTEYLQNQLLTKGMVILRDKIRFYEGQKLLDSLAETWDFFFSDVLPMLQAIFYPVQGKEPSVRQLALLHFRNAITLSVKL EDALARAHARVPPAIVQMLLVLQGVHESRGVTEDYLRLETLVQKVVSPYLGTYGLHSSEGPFTHSCILELQRDKAAAAAVLGAVRKRPSV VPMAGQDPALSTSHPFYDVARHGILQVAGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYKKNLKALYVVHPTSFIKVLWNILKPLISHKF GKKVIYFNYLSELHEHLKYDQLVIPPEVLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPPVLRFTVTYLRE KGLRTEGLFRRSASVQTVREIQRLYNQGKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVTGCRQILRSLPE HNYVVLRYLMGFLHAVSRESIFNKMNSSNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHGLAPWEQGSRAA PLQEAVPRTQATGLTKPTLPPSPLMAARRRL -------------------------------------------------------------- >1341_1341_3_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45121124_ENST00000389773_length(transcript)=1864nt_BP=195nt CTCATCTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGC CTCAGTCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAA CATTGTTCGCAAACGGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGT GATCCTTCGGGACAAGATTCGCTTCTATGAGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGAGGCC CTCGGTGGTGCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGGTGGC AGGGGATGACCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGCTGGA GTATTTGAAGTACACACTGGACCAATACGTTGAGAACGATTATACCATCGTCTATTTCCACTACGGGCTGAACAGCCGGAACAAGCCTTC CCTGGGCTGGCTCCAGAGCGCATACAAGGAGTTCGATAGGAAGTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTT CATCAAGGTCCTGTGGAACATCTTGAAGCCCCTCATCAGTCACAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCA CGAACACCTTAAATACGACCAGCTGGTCATCCCTCCCGAAGTTTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCC GCCTCCCACCAAGACACCACCGCCGCGGCCCCCGCTGCCCACACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGG CGAACTCATCCCCCCTGTGCTGAGGTTCACAGTGACGTACCTGAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAG CGTGCAGACCGTCCGCGAGATCCAGAGGCTCTACAACCAAGGGAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGT GATCCTGAAGACCTTCCTGCGAGAGCTGCCCCAGCCGCTTCTGACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAG CAGCCTGCGTGTCACTGGCTGCCGCCAGATCTTACGGAGCCTCCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCA TGCGGTGTCCCGGGAGAGCATCTTCAACAAAATGAACAGCTCTAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGG GGTCTCCTCCCTGAGTGCCCTTGTGCCCCTGAACATGTTCACTGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGC ACCTGGGGAGCACGGCCTGGCACCATGGGAACAGGGGAGCAGGGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCT CACCAAGCCTACCCTACCTCCGAGTCCCCTGATGGCAGCCAGAAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCC CACACCTGTCTGTGCACTTGTATGTTTTGTAAACTTGGCATCTGTAAAAATAACCAGCCATTAGATGAATTCAGAACCTTCTAATGAAAA CTCCATGCCTCTGGTCCTTGGACTCTTGTCCATGGTTCCTGAGCTGTGGACCGGGATAGAATAA >1341_1341_3_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45121124_ENST00000389773_length(amino acids)=555AA_BP=62 MSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRQLLKTELGSFFTEYLQNQLLTKGMVIL RDKIRFYEELQRDKAAAAAVLGAVRKRPSVVPMAGQDPALSTSHPFYDVARHGILQVAGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYL KYTLDQYVENDYTIVYFHYGLNSRNKPSLGWLQSAYKEFDRKYKKNLKALYVVHPTSFIKVLWNILKPLISHKFGKKVIYFNYLSELHEH LKYDQLVIPPEVLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPPVLRFTVTYLREKGLRTEGLFRRSASVQ TVREIQRLYNQGKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVTGCRQILRSLPEHNYVVLRYLMGFLHAV SRESIFNKMNSSNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHGLAPWEQGSRAAPLQEAVPRTQATGLTK PTLPPSPLMAARRRL -------------------------------------------------------------- >1341_1341_4_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45121124_ENST00000517296_length(transcript)=2023nt_BP=195nt CTCATCTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGC CTCAGTCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAA CATTGTTCGCAAACGGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGT GATCCTTCGGGACAAGATTCGCTTCTATGAGGGACAGAAGCTGCTGGACTCACTGGCAGAGACCTGGGACTTCTTCTTCAGTGACGTGCT GCCCATGCTGCAGGCCATCTTCTACCCGGTGCAGGGCAAGGAGCCATCGGTGCGCCAGCTGGCCCTGCTGCACTTCCGGAATGCCATCAC CCTCAGTGTGAAGCTAGAGGATGCGCTGGCCCGGGCCCATGCCCGTGTGCCCCCTGCCATCGTGCAGATGCTGCTGGTGCTGCAGGGGGT ACATGAGTCCAGGGGCGTGACTGAGGACTACCTGCGCCTGGAGACGCTGGTCCAGAAGGTGGTGTCGCCATACCTGGGCACCTACGGCCT CCACTCCAGCGAGGGGCCCTTCACCCATTCCTGCATCCTGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAG GAAGAGGCCCTCGGTGGTGCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCT GCAGGTGGCAGGGGATGACCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCG GCTGCTGGAGTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCAAGGTCCTGTGGAACATCTTGAAGCCCCT CATCAGTCACAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAACACCTTAAATACGACCAGCTGGTCATCCC TCCCGAAGTTTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTCCCACCAAGACACCACCGCCGCGGCCCCC GCTGCCCACACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAACTCATCCCCCCTGTGCTGAGGTTCACAGT GACGTACCTGAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGCAGACCGTCCGCGAGATCCAGAGGCTCTA CAACCAAGGGAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCCTGAAGACCTTCCTGCGAGAGCTGCCCCA GCCGCTTCTGACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCCTGCGTGTCACTGGCTGCCGCCAGATCTT ACGGAGCCTCCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGGTGTCCCGGGAGAGCATCTTCAACAAAAT GAACAGCTCTAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCTCCTCCCTGAGTGCCCTTGTGCCCCTGAA CATGTTCACTGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTGGGGAGCACGGCCTGGCACCATGGGAACA GGGGAGCAGGGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCAAGCCTACCCTACCTCCGAGTCCCCTGAT GGCAGCCAGAAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACACCTGTCTGTGCACTTGTATGTTTTGTAAA CTTGGCATCTGTAAAAATAACCAGCCATTAGATGAATTCAGAA >1341_1341_4_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000216254_ARHGAP8_chr22_45121124_ENST00000517296_length(amino acids)=634AA_BP=62 MSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRQLLKTELGSFFTEYLQNQLLTKGMVIL RDKIRFYEGQKLLDSLAETWDFFFSDVLPMLQAIFYPVQGKEPSVRQLALLHFRNAITLSVKLEDALARAHARVPPAIVQMLLVLQGVHE SRGVTEDYLRLETLVQKVVSPYLGTYGLHSSEGPFTHSCILELQRDKAAAAAVLGAVRKRPSVVPMAGQDPALSTSHPFYDVARHGILQV AGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYKKNLKALYVVHPTSFIKVLWNILKPLISHKFGKKVIYFNYLSELHEHLKYDQLVIPPE VLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPPVLRFTVTYLREKGLRTEGLFRRSASVQTVREIQRLYNQ GKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVTGCRQILRSLPEHNYVVLRYLMGFLHAVSRESIFNKMNS SNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHGLAPWEQGSRAAPLQEAVPRTQATGLTKPTLPPSPLMAA RRRL -------------------------------------------------------------- >1341_1341_5_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45110471_ENST00000389773_length(transcript)=1940nt_BP=190nt CTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGCCTCAG TCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAACATTG TTCGCAAACGCATCCACAACGGGGTGATCGCCGTCTTCCAGCGCAAGGGGCTGCCCGACCAGGAGCTCTTCAGCCTCAACGAGGGCGTCC GGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGTGATCCTTCGGGACA AGATTCGCTTCTATGAGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGAGGCCCTCGGTGGTGCCCA TGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGGTGGCAGGGGATGACCGCT TTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGCTGGAGTATTTGAAGTACA CACTGGACCAATACGTTGAGAACGATTATACCATCGTCTATTTCCACTACGGGCTGAACAGCCGGAACAAGCCTTCCCTGGGCTGGCTCC AGAGCGCATACAAGGAGTTCGATAGGAAGTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCAAGGTCCTGT GGAACATCTTGAAGCCCCTCATCAGTCACAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAACACCTTAAAT ACGACCAGCTGGTCATCCCTCCCGAAGTTTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTCCCACCAAGA CACCACCGCCGCGGCCCCCGCTGCCCACACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAACTCATCCCCC CTGTGCTGAGGTTCACAGTGACGTACCTGAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGCAGACCGTCC GCGAGATCCAGAGGCTCTACAACCAAGGGAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCCTGAAGACCT TCCTGCGAGAGCTGCCCCAGCCGCTTCTGACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCCTGCGTGTCA CTGGCTGCCGCCAGATCTTACGGAGCCTCCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGGTGTCCCGGG AGAGCATCTTCAACAAAATGAACAGCTCTAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCTCCTCCCTGA GTGCCCTTGTGCCCCTGAACATGTTCACTGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTGGGGAGCACG GCCTGGCACCATGGGAACAGGGGAGCAGGGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCAAGCCTACCC TACCTCCGAGTCCCCTGATGGCAGCCAGAAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACACCTGTCTGTG CACTTGTATGTTTTGTAAACTTGGCATCTGTAAAAATAACCAGCCATTAGATGAATTCAGAACCTTCTAATGAAAACTCCATGCCTCTGG TCCTTGGACTCTTGTCCATGGTTCCTGAGCTGTGGACCGGGATAGAATAA >1341_1341_5_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45110471_ENST00000389773_length(amino acids)=582AA_BP=63 LSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRIHNGVIAVFQRKGLPDQELFSLNEGVR QLLKTELGSFFTEYLQNQLLTKGMVILRDKIRFYEELQRDKAAAAAVLGAVRKRPSVVPMAGQDPALSTSHPFYDVARHGILQVAGDDRF GRRVVTFSCCRMPPSHELDHQRLLEYLKYTLDQYVENDYTIVYFHYGLNSRNKPSLGWLQSAYKEFDRKYKKNLKALYVVHPTSFIKVLW NILKPLISHKFGKKVIYFNYLSELHEHLKYDQLVIPPEVLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPP VLRFTVTYLREKGLRTEGLFRRSASVQTVREIQRLYNQGKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVT GCRQILRSLPEHNYVVLRYLMGFLHAVSRESIFNKMNSSNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHG LAPWEQGSRAAPLQEAVPRTQATGLTKPTLPPSPLMAARRRL -------------------------------------------------------------- >1341_1341_6_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45110471_ENST00000517296_length(transcript)=2099nt_BP=190nt CTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGCCTCAG TCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAACATTG TTCGCAAACGCATCCACAACGGGGTGATCGCCGTCTTCCAGCGCAAGGGGCTGCCCGACCAGGAGCTCTTCAGCCTCAACGAGGGCGTCC GGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGTGATCCTTCGGGACA AGATTCGCTTCTATGAGGGACAGAAGCTGCTGGACTCACTGGCAGAGACCTGGGACTTCTTCTTCAGTGACGTGCTGCCCATGCTGCAGG CCATCTTCTACCCGGTGCAGGGCAAGGAGCCATCGGTGCGCCAGCTGGCCCTGCTGCACTTCCGGAATGCCATCACCCTCAGTGTGAAGC TAGAGGATGCGCTGGCCCGGGCCCATGCCCGTGTGCCCCCTGCCATCGTGCAGATGCTGCTGGTGCTGCAGGGGGTACATGAGTCCAGGG GCGTGACTGAGGACTACCTGCGCCTGGAGACGCTGGTCCAGAAGGTGGTGTCGCCATACCTGGGCACCTACGGCCTCCACTCCAGCGAGG GGCCCTTCACCCATTCCTGCATCCTGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGAGGCCCTCGG TGGTGCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGGTGGCAGGGG ATGACCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGCTGGAGTACA AGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCAAGGTCCTGTGGAACATCTTGAAGCCCCTCATCAGTCACAAGT TTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAACACCTTAAATACGACCAGCTGGTCATCCCTCCCGAAGTTTTGC GGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTCCCACCAAGACACCACCGCCGCGGCCCCCGCTGCCCACACAGC AGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAACTCATCCCCCCTGTGCTGAGGTTCACAGTGACGTACCTGAGAG AGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGCAGACCGTCCGCGAGATCCAGAGGCTCTACAACCAAGGGAAGC CCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCCTGAAGACCTTCCTGCGAGAGCTGCCCCAGCCGCTTCTGACCT TCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCCTGCGTGTCACTGGCTGCCGCCAGATCTTACGGAGCCTCCCAG AGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGGTGTCCCGGGAGAGCATCTTCAACAAAATGAACAGCTCTAACC TGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCTCCTCCCTGAGTGCCCTTGTGCCCCTGAACATGTTCACTGAAC TGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTGGGGAGCACGGCCTGGCACCATGGGAACAGGGGAGCAGGGCAG CCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCAAGCCTACCCTACCTCCGAGTCCCCTGATGGCAGCCAGAAGAC GTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACACCTGTCTGTGCACTTGTATGTTTTGTAAACTTGGCATCTGTAA AAATAACCAGCCATTAGATGAATTCAGAA >1341_1341_6_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45110471_ENST00000517296_length(amino acids)=661AA_BP=63 LSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRIHNGVIAVFQRKGLPDQELFSLNEGVR QLLKTELGSFFTEYLQNQLLTKGMVILRDKIRFYEGQKLLDSLAETWDFFFSDVLPMLQAIFYPVQGKEPSVRQLALLHFRNAITLSVKL EDALARAHARVPPAIVQMLLVLQGVHESRGVTEDYLRLETLVQKVVSPYLGTYGLHSSEGPFTHSCILELQRDKAAAAAVLGAVRKRPSV VPMAGQDPALSTSHPFYDVARHGILQVAGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYKKNLKALYVVHPTSFIKVLWNILKPLISHKF GKKVIYFNYLSELHEHLKYDQLVIPPEVLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPPVLRFTVTYLRE KGLRTEGLFRRSASVQTVREIQRLYNQGKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVTGCRQILRSLPE HNYVVLRYLMGFLHAVSRESIFNKMNSSNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHGLAPWEQGSRAA PLQEAVPRTQATGLTKPTLPPSPLMAARRRL -------------------------------------------------------------- >1341_1341_7_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45121124_ENST00000389773_length(transcript)=1859nt_BP=190nt CTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGCCTCAG TCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAACATTG TTCGCAAACGGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGTGATCC TTCGGGACAAGATTCGCTTCTATGAGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGAGGCCCTCGG TGGTGCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGGTGGCAGGGG ATGACCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGCTGGAGTATT TGAAGTACACACTGGACCAATACGTTGAGAACGATTATACCATCGTCTATTTCCACTACGGGCTGAACAGCCGGAACAAGCCTTCCCTGG GCTGGCTCCAGAGCGCATACAAGGAGTTCGATAGGAAGTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCA AGGTCCTGTGGAACATCTTGAAGCCCCTCATCAGTCACAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAAC ACCTTAAATACGACCAGCTGGTCATCCCTCCCGAAGTTTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTC CCACCAAGACACCACCGCCGCGGCCCCCGCTGCCCACACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAAC TCATCCCCCCTGTGCTGAGGTTCACAGTGACGTACCTGAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGC AGACCGTCCGCGAGATCCAGAGGCTCTACAACCAAGGGAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCC TGAAGACCTTCCTGCGAGAGCTGCCCCAGCCGCTTCTGACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCC TGCGTGTCACTGGCTGCCGCCAGATCTTACGGAGCCTCCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGG TGTCCCGGGAGAGCATCTTCAACAAAATGAACAGCTCTAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCT CCTCCCTGAGTGCCCTTGTGCCCCTGAACATGTTCACTGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTG GGGAGCACGGCCTGGCACCATGGGAACAGGGGAGCAGGGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCA AGCCTACCCTACCTCCGAGTCCCCTGATGGCAGCCAGAAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACAC CTGTCTGTGCACTTGTATGTTTTGTAAACTTGGCATCTGTAAAAATAACCAGCCATTAGATGAATTCAGAACCTTCTAATGAAAACTCCA TGCCTCTGGTCCTTGGACTCTTGTCCATGGTTCCTGAGCTGTGGACCGGGATAGAATAA >1341_1341_7_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45121124_ENST00000389773_length(amino acids)=555AA_BP=62 LSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRQLLKTELGSFFTEYLQNQLLTKGMVIL RDKIRFYEELQRDKAAAAAVLGAVRKRPSVVPMAGQDPALSTSHPFYDVARHGILQVAGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYL KYTLDQYVENDYTIVYFHYGLNSRNKPSLGWLQSAYKEFDRKYKKNLKALYVVHPTSFIKVLWNILKPLISHKFGKKVIYFNYLSELHEH LKYDQLVIPPEVLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPPVLRFTVTYLREKGLRTEGLFRRSASVQ TVREIQRLYNQGKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVTGCRQILRSLPEHNYVVLRYLMGFLHAV SRESIFNKMNSSNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHGLAPWEQGSRAAPLQEAVPRTQATGLTK PTLPPSPLMAARRRL -------------------------------------------------------------- >1341_1341_8_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45121124_ENST00000517296_length(transcript)=2018nt_BP=190nt CTTTGTCAGTGCACAAAATGGCGCCCTACAGCCTACTGGTGACTCGGCTGCAGAAAGCTCTGGGTGTGCGGCAGTACCATGTGGCCTCAG TCCTGTGCCAACGGGCCAAGGTGGCGATGAGCCACTTTGAGCCCAACGAGTACATCCATTATGACCTGCTAGAGAAGAACATTAACATTG TTCGCAAACGGCAGCTGTTGAAGACAGAGCTGGGGTCCTTCTTCACGGAGTACCTGCAGAACCAGCTGCTGACAAAAGGCATGGTGATCC TTCGGGACAAGATTCGCTTCTATGAGGGACAGAAGCTGCTGGACTCACTGGCAGAGACCTGGGACTTCTTCTTCAGTGACGTGCTGCCCA TGCTGCAGGCCATCTTCTACCCGGTGCAGGGCAAGGAGCCATCGGTGCGCCAGCTGGCCCTGCTGCACTTCCGGAATGCCATCACCCTCA GTGTGAAGCTAGAGGATGCGCTGGCCCGGGCCCATGCCCGTGTGCCCCCTGCCATCGTGCAGATGCTGCTGGTGCTGCAGGGGGTACATG AGTCCAGGGGCGTGACTGAGGACTACCTGCGCCTGGAGACGCTGGTCCAGAAGGTGGTGTCGCCATACCTGGGCACCTACGGCCTCCACT CCAGCGAGGGGCCCTTCACCCATTCCTGCATCCTGGAGCTGCAGAGAGACAAGGCGGCGGCGGCTGCTGTGCTGGGTGCAGTGAGGAAGA GGCCCTCGGTGGTGCCCATGGCTGGCCAGGATCCTGCGCTGAGCACGAGTCACCCGTTCTACGACGTGGCCAGACATGGCATTCTGCAGG TGGCAGGGGATGACCGCTTTGGAAGACGTGTTGTCACGTTCAGCTGCTGCCGGATGCCACCCTCCCACGAGCTGGACCACCAGCGGCTGC TGGAGTACAAGAAGAACTTGAAGGCCCTCTACGTGGTGCACCCCACCAGCTTCATCAAGGTCCTGTGGAACATCTTGAAGCCCCTCATCA GTCACAAGTTTGGGAAGAAAGTCATCTATTTCAACTACCTGAGTGAGCTCCACGAACACCTTAAATACGACCAGCTGGTCATCCCTCCCG AAGTTTTGCGGTACGATGAGAAGCTCCAGAGCCTGCACGAGGGCCGGACGCCGCCTCCCACCAAGACACCACCGCCGCGGCCCCCGCTGC CCACACAGCAGTTTGGCGTCAGTCTGCAATACCTCAAAGACAAAAATCAAGGCGAACTCATCCCCCCTGTGCTGAGGTTCACAGTGACGT ACCTGAGAGAGAAAGGCCTGCGCACCGAGGGCCTGTTCCGGAGATCCGCCAGCGTGCAGACCGTCCGCGAGATCCAGAGGCTCTACAACC AAGGGAAGCCCGTGAACTTTGACGACTACGGGGACATTCACATCCCTGCCGTGATCCTGAAGACCTTCCTGCGAGAGCTGCCCCAGCCGC TTCTGACCTTCCAGGCCTACGAGCAGATTCTCGGGATCACCTGTGTGGAGAGCAGCCTGCGTGTCACTGGCTGCCGCCAGATCTTACGGA GCCTCCCAGAGCACAACTACGTCGTCCTCCGCTACCTCATGGGCTTCCTGCATGCGGTGTCCCGGGAGAGCATCTTCAACAAAATGAACA GCTCTAACCTGGCCTGTGTCTTCGGGCTGAATTTGATCTGGCCATCCCAGGGGGTCTCCTCCCTGAGTGCCCTTGTGCCCCTGAACATGT TCACTGAACTGCTGATCGAGTACTATGAAAAGATCTTCAGCACCCCGGAGGCACCTGGGGAGCACGGCCTGGCACCATGGGAACAGGGGA GCAGGGCAGCCCCTTTGCAGGAGGCTGTGCCACGGACACAAGCCACGGGCCTCACCAAGCCTACCCTACCTCCGAGTCCCCTGATGGCAG CCAGAAGACGTCTCTAGTGTTGCGAACACTCTGTATATTTCGAGCTACCTCCCACACCTGTCTGTGCACTTGTATGTTTTGTAAACTTGG CATCTGTAAAAATAACCAGCCATTAGATGAATTCAGAA >1341_1341_8_ACO2-ARHGAP8_ACO2_chr22_41895866_ENST00000396512_ARHGAP8_chr22_45121124_ENST00000517296_length(amino acids)=634AA_BP=62 LSVHKMAPYSLLVTRLQKALGVRQYHVASVLCQRAKVAMSHFEPNEYIHYDLLEKNINIVRKRQLLKTELGSFFTEYLQNQLLTKGMVIL RDKIRFYEGQKLLDSLAETWDFFFSDVLPMLQAIFYPVQGKEPSVRQLALLHFRNAITLSVKLEDALARAHARVPPAIVQMLLVLQGVHE SRGVTEDYLRLETLVQKVVSPYLGTYGLHSSEGPFTHSCILELQRDKAAAAAVLGAVRKRPSVVPMAGQDPALSTSHPFYDVARHGILQV AGDDRFGRRVVTFSCCRMPPSHELDHQRLLEYKKNLKALYVVHPTSFIKVLWNILKPLISHKFGKKVIYFNYLSELHEHLKYDQLVIPPE VLRYDEKLQSLHEGRTPPPTKTPPPRPPLPTQQFGVSLQYLKDKNQGELIPPVLRFTVTYLREKGLRTEGLFRRSASVQTVREIQRLYNQ GKPVNFDDYGDIHIPAVILKTFLRELPQPLLTFQAYEQILGITCVESSLRVTGCRQILRSLPEHNYVVLRYLMGFLHAVSRESIFNKMNS SNLACVFGLNLIWPSQGVSSLSALVPLNMFTELLIEYYEKIFSTPEAPGEHGLAPWEQGSRAAPLQEAVPRTQATGLTKPTLPPSPLMAA RRRL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACO2-ARHGAP8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACO2-ARHGAP8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ACO2-ARHGAP8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ACO2 | C3281192 | INFANTILE CEREBELLAR-RETINAL DEGENERATION | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ACO2 | C4225384 | OPTIC ATROPHY 9 | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | ACO2 | C0035222 | Respiratory Distress Syndrome, Adult | 1 | CTD_human |
| Hgene | ACO2 | C2239176 | Liver carcinoma | 1 | CTD_human |
