
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:COMMD10-AP3S1 (FusionGDB2 ID:HG51397TG1176) |
Fusion Gene Summary for COMMD10-AP3S1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: COMMD10-AP3S1 | Fusion gene ID: hg51397tg1176 | Hgene | Tgene | Gene symbol | COMMD10 | AP3S1 | Gene ID | 51397 | 1176 |
| Gene name | COMM domain containing 10 | adaptor related protein complex 3 subunit sigma 1 | |
| Synonyms | PTD002 | CLAPS3|Sigma3A | |
| Cytomap | ('COMMD10')('AP3S1') 5q23.1 | 5q22.3-q23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | COMM domain-containing protein 10 | AP-3 complex subunit sigma-1adapter-related protein complex 3 subunit sigma-1adaptor related protein complex 3 sigma 1 subunitclathrin adaptor complex AP3, sigma-3A subunitclathrin-associated/assembly/adapter protein, small 3clathrin-associated/assem | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q9Y6G5 | Q92572 | |
| Ensembl transtripts involved in fusion gene | ENST00000274458, ENST00000515539, ENST00000503424, | ENST00000503424, ENST00000274458, ENST00000515539, | |
| Fusion gene scores | * DoF score | 11 X 9 X 8=792 | 6 X 8 X 7=336 |
| # samples | 15 | 10 | |
| ** MAII score | log2(15/792*10)=-2.40053792958373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/336*10)=-1.74846123300404 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: COMMD10 [Title/Abstract] AND AP3S1 [Title/Abstract] AND fusion [Title/Abstract] Common fusion transcripts identified in colorectal cancer cell lines by high-throughput RNA sequencing (pmid: 24151535) | ||
| Most frequent breakpoint | COMMD10(115428397)-AP3S1(115230784), # samples:6 | ||
| Anticipated loss of major functional domain due to fusion event. | AP3S1-COMMD10 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AP3S1-COMMD10 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. COMMD10-AP3S1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. COMMD10-AP3S1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across COMMD10 (5'-gene) Fusion gene breakpoints across COMMD10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
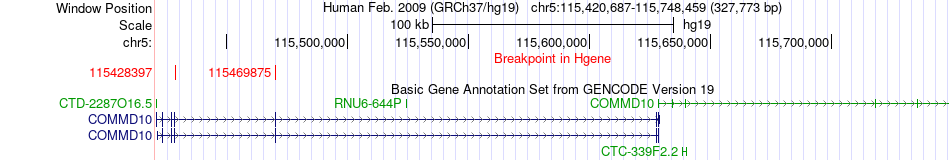 |
 Fusion gene breakpoints across AP3S1 (3'-gene) Fusion gene breakpoints across AP3S1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
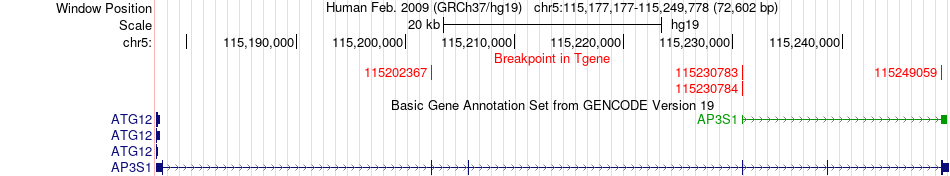 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-2F-A9KT-01A | COMMD10 | chr5 | 115428397 | - | AP3S1 | chr5 | 115230784 | + |
| ChimerDB4 | BLCA | TCGA-GV-A3QI-01A | COMMD10 | chr5 | 115428397 | - | AP3S1 | chr5 | 115230784 | + |
| ChimerDB4 | LGG | TCGA-DB-5273-01A | COMMD10 | chr5 | 115428397 | - | AP3S1 | chr5 | 115230784 | + |
| ChimerDB4 | LGG | TCGA-DH-A7US-01A | COMMD10 | chr5 | 115428397 | - | AP3S1 | chr5 | 115230784 | + |
| ChimerDB4 | LGG | TCGA-DH-A7US | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| ChimerDB4 | LUAD | TCGA-55-A490 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
| ChimerDB4 | Non-Cancer | ERR315443 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| ChimerDB4 | STAD | TCGA-CG-5721-01A | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| ChimerDB4 | UCEC | TCGA-D1-A0ZR-01A | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| ChimerKB4 | . | . | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
Top |
Fusion Gene ORF analysis for COMMD10-AP3S1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000274458 | ENST00000505423 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| 5CDS-3UTR | ENST00000515539 | ENST00000505423 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| 5CDS-intron | ENST00000274458 | ENST00000505423 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
| 5CDS-intron | ENST00000515539 | ENST00000505423 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
| In-frame | ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| In-frame | ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
| In-frame | ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| In-frame | ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| In-frame | ENST00000274458 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| In-frame | ENST00000274458 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| In-frame | ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| In-frame | ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
| In-frame | ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| In-frame | ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| In-frame | ENST00000515539 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| In-frame | ENST00000515539 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| intron-3CDS | ENST00000503424 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| intron-3CDS | ENST00000503424 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
| intron-3CDS | ENST00000503424 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| intron-3CDS | ENST00000503424 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| intron-3CDS | ENST00000503424 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + |
| intron-3CDS | ENST00000503424 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + |
| intron-3UTR | ENST00000503424 | ENST00000505423 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + |
| intron-intron | ENST00000503424 | ENST00000505423 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000274458 | COMMD10 | chr5 | 115428397 | + | ENST00000316788 | AP3S1 | chr5 | 115230783 | + | 1361 | 461 | 14 | 769 | 251 |
| ENST00000274458 | COMMD10 | chr5 | 115428397 | + | ENST00000505423 | AP3S1 | chr5 | 115230783 | + | 1072 | 461 | 14 | 661 | 215 |
| ENST00000515539 | COMMD10 | chr5 | 115428397 | + | ENST00000316788 | AP3S1 | chr5 | 115230783 | + | 1391 | 491 | 134 | 799 | 221 |
| ENST00000515539 | COMMD10 | chr5 | 115428397 | + | ENST00000505423 | AP3S1 | chr5 | 115230783 | + | 1102 | 491 | 134 | 691 | 185 |
| ENST00000274458 | COMMD10 | chr5 | 115428397 | + | ENST00000316788 | AP3S1 | chr5 | 115230784 | + | 1361 | 461 | 14 | 769 | 251 |
| ENST00000274458 | COMMD10 | chr5 | 115428397 | + | ENST00000505423 | AP3S1 | chr5 | 115230784 | + | 1072 | 461 | 14 | 661 | 215 |
| ENST00000515539 | COMMD10 | chr5 | 115428397 | + | ENST00000316788 | AP3S1 | chr5 | 115230784 | + | 1391 | 491 | 134 | 799 | 221 |
| ENST00000515539 | COMMD10 | chr5 | 115428397 | + | ENST00000505423 | AP3S1 | chr5 | 115230784 | + | 1102 | 491 | 134 | 691 | 185 |
| ENST00000274458 | COMMD10 | chr5 | 115428397 | + | ENST00000316788 | AP3S1 | chr5 | 115230783 | + | 1361 | 461 | 14 | 769 | 251 |
| ENST00000274458 | COMMD10 | chr5 | 115428397 | + | ENST00000505423 | AP3S1 | chr5 | 115230783 | + | 1072 | 461 | 14 | 661 | 215 |
| ENST00000515539 | COMMD10 | chr5 | 115428397 | + | ENST00000316788 | AP3S1 | chr5 | 115230783 | + | 1391 | 491 | 134 | 799 | 221 |
| ENST00000515539 | COMMD10 | chr5 | 115428397 | + | ENST00000505423 | AP3S1 | chr5 | 115230783 | + | 1102 | 491 | 134 | 691 | 185 |
| ENST00000274458 | COMMD10 | chr5 | 115469875 | + | ENST00000316788 | AP3S1 | chr5 | 115202367 | + | 1676 | 572 | 14 | 1084 | 356 |
| ENST00000515539 | COMMD10 | chr5 | 115469875 | + | ENST00000316788 | AP3S1 | chr5 | 115202367 | + | 1706 | 602 | 134 | 1114 | 326 |
| ENST00000274458 | COMMD10 | chr5 | 115469875 | + | ENST00000316788 | AP3S1 | chr5 | 115249059 | + | 1292 | 572 | 14 | 700 | 228 |
| ENST00000515539 | COMMD10 | chr5 | 115469875 | + | ENST00000316788 | AP3S1 | chr5 | 115249059 | + | 1322 | 602 | 134 | 730 | 198 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + | 0.00074004 | 0.99926 |
| ENST00000274458 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + | 0.001622059 | 0.9983779 |
| ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + | 0.00087729 | 0.9991227 |
| ENST00000515539 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230784 | + | 0.002249681 | 0.99775034 |
| ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + | 0.00074004 | 0.99926 |
| ENST00000274458 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + | 0.001622059 | 0.9983779 |
| ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + | 0.00087729 | 0.9991227 |
| ENST00000515539 | ENST00000505423 | COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + | 0.002249681 | 0.99775034 |
| ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + | 0.00045288 | 0.99954706 |
| ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202367 | + | 0.000424362 | 0.9995757 |
| ENST00000274458 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + | 0.00125994 | 0.99874014 |
| ENST00000515539 | ENST00000316788 | COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249059 | + | 0.00128717 | 0.9987129 |
Top |
Fusion Genomic Features for COMMD10-AP3S1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249058 | + | 0.000118543 | 0.9998814 |
| COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + | 8.28E-05 | 0.99991715 |
| COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202366 | + | 4.10E-05 | 0.999959 |
| COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115249058 | + | 0.000118543 | 0.9998814 |
| COMMD10 | chr5 | 115428397 | + | AP3S1 | chr5 | 115230783 | + | 8.28E-05 | 0.99991715 |
| COMMD10 | chr5 | 115469875 | + | AP3S1 | chr5 | 115202366 | + | 4.10E-05 | 0.999959 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
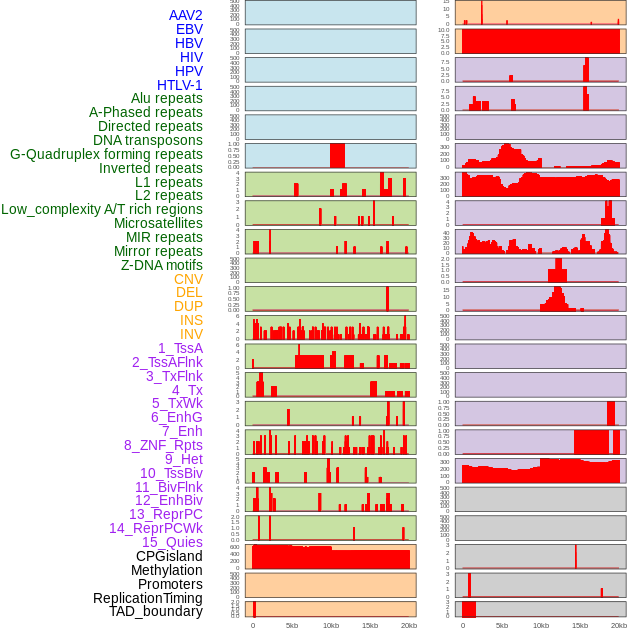 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
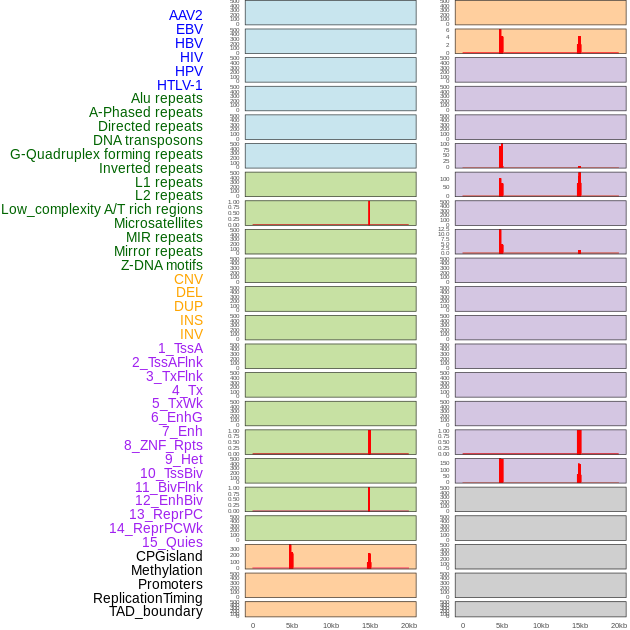 |
Top |
Fusion Protein Features for COMMD10-AP3S1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:115428397/chr5:115230784) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| COMMD10 | AP3S1 |
| FUNCTION: May modulate activity of cullin-RING E3 ubiquitin ligase (CRL) complexes (PubMed:21778237). May down-regulate activation of NF-kappa-B (PubMed:15799966). {ECO:0000269|PubMed:15799966, ECO:0000305|PubMed:21778237}. | FUNCTION: Part of the AP-3 complex, an adaptor-related complex which is not clathrin-associated. The complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and may be directly involved in trafficking to lysosomes. In concert with the BLOC-1 complex, AP-3 is required to target cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | COMMD10 | chr5:115428397 | chr5:115230783 | ENST00000274458 | + | 4 | 7 | 133_202 | 133 | 203.0 | Domain | COMM |
| Hgene | COMMD10 | chr5:115428397 | chr5:115230784 | ENST00000274458 | + | 4 | 7 | 133_202 | 133 | 203.0 | Domain | COMM |
| Hgene | COMMD10 | chr5:115469875 | chr5:115202367 | ENST00000274458 | + | 5 | 7 | 133_202 | 170 | 203.0 | Domain | COMM |
| Hgene | COMMD10 | chr5:115469875 | chr5:115249059 | ENST00000274458 | + | 5 | 7 | 133_202 | 170 | 203.0 | Domain | COMM |
Top |
Fusion Gene Sequence for COMMD10-AP3S1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >18503_18503_1_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230783_ENST00000316788_length(transcript)=1361nt_BP=461nt GCGGCCGCCATTGGCTGGGTTCGGCGCAGCTAACAGACGGCGGCAGTGCGAGAAAGCCGAAGATGGCGGTCCCCGCGGCGCTGATCCTAC GGGAGAGCCCCAGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGATTTCCACGGTTGCTCACTCGGATTCTTCAAA AACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGGCATTTTCTCTAGAGAAACAAGATCTTCACC TAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAGCAGCTTTGCAGCAGCAATTAGAGAACATTC ATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAACAGTTGAAAAGTTCCGGCAGAGAATTCTGG CTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGGATTTGATTTTCCATGTAGACAAGGTTCACA ATATTCTTGCAGAAATGGTGATGGGGGGAATGGTATTGGAGACAAATATGAATGAGATTGTTACACAAATTGATGCACAAAATAAGCTGG AAAAATCTGAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTA ACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAA GAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTA TAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTT TTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATA TTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACA GTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAACATTATGACTTTTTTCTCTTAGTTTAAATAAACTCCAAGGTAACTGGACTTC TAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAATAATTTTTTTTACAACCCTCTGACTCAACAAAGTAAATAAAAGTATATTTT ATCACTATTAA >18503_18503_1_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230783_ENST00000316788_length(amino acids)=251AA_BP=149 MGSAQLTDGGSARKPKMAVPAALILRESPSMKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLET ISFILEQAVYHNVKPAALQQQLENIHLRQDKAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKVHNILAE MVMGGMVLETNMNEIVTQIDAQNKLEKSEAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_2_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230783_ENST00000505423_length(transcript)=1072nt_BP=461nt GCGGCCGCCATTGGCTGGGTTCGGCGCAGCTAACAGACGGCGGCAGTGCGAGAAAGCCGAAGATGGCGGTCCCCGCGGCGCTGATCCTAC GGGAGAGCCCCAGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGATTTCCACGGTTGCTCACTCGGATTCTTCAAA AACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGGCATTTTCTCTAGAGAAACAAGATCTTCACC TAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAGCAGCTTTGCAGCAGCAATTAGAGAACATTC ATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAACAGTTGAAAAGTTCCGGCAGAGAATTCTGG CTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGGATTTGATTTTCCATGTAGACAAGGCTGGCT TAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTA TAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACC ATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATC AGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACT GCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACA ACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTT >18503_18503_2_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230783_ENST00000505423_length(amino acids)=215AA_BP=149 MGSAQLTDGGSARKPKMAVPAALILRESPSMKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLET ISFILEQAVYHNVKPAALQQQLENIHLRQDKAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKAGLAGAP ARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_3_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230784_ENST00000316788_length(transcript)=1361nt_BP=461nt GCGGCCGCCATTGGCTGGGTTCGGCGCAGCTAACAGACGGCGGCAGTGCGAGAAAGCCGAAGATGGCGGTCCCCGCGGCGCTGATCCTAC GGGAGAGCCCCAGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGATTTCCACGGTTGCTCACTCGGATTCTTCAAA AACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGGCATTTTCTCTAGAGAAACAAGATCTTCACC TAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAGCAGCTTTGCAGCAGCAATTAGAGAACATTC ATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAACAGTTGAAAAGTTCCGGCAGAGAATTCTGG CTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGGATTTGATTTTCCATGTAGACAAGGTTCACA ATATTCTTGCAGAAATGGTGATGGGGGGAATGGTATTGGAGACAAATATGAATGAGATTGTTACACAAATTGATGCACAAAATAAGCTGG AAAAATCTGAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTA ACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAA GAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTA TAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTT TTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATA TTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACA GTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAACATTATGACTTTTTTCTCTTAGTTTAAATAAACTCCAAGGTAACTGGACTTC TAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAATAATTTTTTTTACAACCCTCTGACTCAACAAAGTAAATAAAAGTATATTTT ATCACTATTAA >18503_18503_3_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230784_ENST00000316788_length(amino acids)=251AA_BP=149 MGSAQLTDGGSARKPKMAVPAALILRESPSMKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLET ISFILEQAVYHNVKPAALQQQLENIHLRQDKAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKVHNILAE MVMGGMVLETNMNEIVTQIDAQNKLEKSEAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_4_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230784_ENST00000505423_length(transcript)=1072nt_BP=461nt GCGGCCGCCATTGGCTGGGTTCGGCGCAGCTAACAGACGGCGGCAGTGCGAGAAAGCCGAAGATGGCGGTCCCCGCGGCGCTGATCCTAC GGGAGAGCCCCAGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGATTTCCACGGTTGCTCACTCGGATTCTTCAAA AACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGGCATTTTCTCTAGAGAAACAAGATCTTCACC TAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAGCAGCTTTGCAGCAGCAATTAGAGAACATTC ATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAACAGTTGAAAAGTTCCGGCAGAGAATTCTGG CTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGGATTTGATTTTCCATGTAGACAAGGCTGGCT TAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTA TAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACC ATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATC AGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACT GCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACA ACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTT >18503_18503_4_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000274458_AP3S1_chr5_115230784_ENST00000505423_length(amino acids)=215AA_BP=149 MGSAQLTDGGSARKPKMAVPAALILRESPSMKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLET ISFILEQAVYHNVKPAALQQQLENIHLRQDKAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKAGLAGAP ARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_5_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230783_ENST00000316788_length(transcript)=1391nt_BP=491nt GACACTCTGGTGGGAAAGGCTCACGCTGCTGCTGGAGGGTCTTTAATAGCCAACATGTGTTAATACCCCTTTTAAATCAGTATCGAATAG GGGACCGAATTTTTGGTTGGTAGAAGAATCTAGCCTGGCTTTGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGAT TTCCACGGTTGCTCACTCGGATTCTTCAAAAACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGG CATTTTCTCTAGAGAAACAAGATCTTCACCTAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAG CAGCTTTGCAGCAGCAATTAGAGAACATTCATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAA CAGTTGAAAAGTTCCGGCAGAGAATTCTGGCTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGG ATTTGATTTTCCATGTAGACAAGGTTCACAATATTCTTGCAGAAATGGTGATGGGGGGAATGGTATTGGAGACAAATATGAATGAGATTG TTACACAAATTGATGCACAAAATAAGCTGGAAAAATCTGAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATA TGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAA AGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTT TTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTA AAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCT ACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCAT TCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAACATTATGACTTTTTTCTCTTAG TTTAAATAAACTCCAAGGTAACTGGACTTCTAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAATAATTTTTTTTACAACCCTCT GACTCAACAAAGTAAATAAAAGTATATTTTATCACTATTAA >18503_18503_5_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230783_ENST00000316788_length(amino acids)=221AA_BP=119 MKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLETISFILEQAVYHNVKPAALQQQLENIHLRQD KAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKVHNILAEMVMGGMVLETNMNEIVTQIDAQNKLEKSEA GLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_6_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230783_ENST00000505423_length(transcript)=1102nt_BP=491nt GACACTCTGGTGGGAAAGGCTCACGCTGCTGCTGGAGGGTCTTTAATAGCCAACATGTGTTAATACCCCTTTTAAATCAGTATCGAATAG GGGACCGAATTTTTGGTTGGTAGAAGAATCTAGCCTGGCTTTGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGAT TTCCACGGTTGCTCACTCGGATTCTTCAAAAACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGG CATTTTCTCTAGAGAAACAAGATCTTCACCTAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAG CAGCTTTGCAGCAGCAATTAGAGAACATTCATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAA CAGTTGAAAAGTTCCGGCAGAGAATTCTGGCTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGG ATTTGATTTTCCATGTAGACAAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCC CAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAA ATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGT CAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCC TATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAA GTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCT ATATGTCACACAGTTGTTATTT >18503_18503_6_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230783_ENST00000505423_length(amino acids)=185AA_BP=119 MKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLETISFILEQAVYHNVKPAALQQQLENIHLRQD KAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPN LPSFK -------------------------------------------------------------- >18503_18503_7_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230784_ENST00000316788_length(transcript)=1391nt_BP=491nt GACACTCTGGTGGGAAAGGCTCACGCTGCTGCTGGAGGGTCTTTAATAGCCAACATGTGTTAATACCCCTTTTAAATCAGTATCGAATAG GGGACCGAATTTTTGGTTGGTAGAAGAATCTAGCCTGGCTTTGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGAT TTCCACGGTTGCTCACTCGGATTCTTCAAAAACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGG CATTTTCTCTAGAGAAACAAGATCTTCACCTAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAG CAGCTTTGCAGCAGCAATTAGAGAACATTCATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAA CAGTTGAAAAGTTCCGGCAGAGAATTCTGGCTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGG ATTTGATTTTCCATGTAGACAAGGTTCACAATATTCTTGCAGAAATGGTGATGGGGGGAATGGTATTGGAGACAAATATGAATGAGATTG TTACACAAATTGATGCACAAAATAAGCTGGAAAAATCTGAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATA TGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAA AGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTT TTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTA AAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCT ACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCAT TCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAACATTATGACTTTTTTCTCTTAG TTTAAATAAACTCCAAGGTAACTGGACTTCTAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAATAATTTTTTTTACAACCCTCT GACTCAACAAAGTAAATAAAAGTATATTTTATCACTATTAA >18503_18503_7_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230784_ENST00000316788_length(amino acids)=221AA_BP=119 MKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLETISFILEQAVYHNVKPAALQQQLENIHLRQD KAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKVHNILAEMVMGGMVLETNMNEIVTQIDAQNKLEKSEA GLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_8_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230784_ENST00000505423_length(transcript)=1102nt_BP=491nt GACACTCTGGTGGGAAAGGCTCACGCTGCTGCTGGAGGGTCTTTAATAGCCAACATGTGTTAATACCCCTTTTAAATCAGTATCGAATAG GGGACCGAATTTTTGGTTGGTAGAAGAATCTAGCCTGGCTTTGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGAT TTCCACGGTTGCTCACTCGGATTCTTCAAAAACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGG CATTTTCTCTAGAGAAACAAGATCTTCACCTAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAG CAGCTTTGCAGCAGCAATTAGAGAACATTCATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAA CAGTTGAAAAGTTCCGGCAGAGAATTCTGGCTCCCTGTAAGGTATTTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGG ATTTGATTTTCCATGTAGACAAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCC CAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAA ATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGT CAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCC TATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAA GTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCT ATATGTCACACAGTTGTTATTT >18503_18503_8_COMMD10-AP3S1_COMMD10_chr5_115428397_ENST00000515539_AP3S1_chr5_115230784_ENST00000505423_length(amino acids)=185AA_BP=119 MKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLETISFILEQAVYHNVKPAALQQQLENIHLRQD KAEAFVNTWSSMGQETVEKFRQRILAPCKVFVETLDKCFENVCELDLIFHVDKAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPN LPSFK -------------------------------------------------------------- >18503_18503_9_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000274458_AP3S1_chr5_115202367_ENST00000316788_length(transcript)=1676nt_BP=572nt GCGGCCGCCATTGGCTGGGTTCGGCGCAGCTAACAGACGGCGGCAGTGCGAGAAAGCCGAAGATGGCGGTCCCCGCGGCGCTGATCCTAC GGGAGAGCCCCAGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGATTTCCACGGTTGCTCACTCGGATTCTTCAAA AACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGGCATTTTCTCTAGAGAAACAAGATCTTCACC TAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAGCAGCTTTGCAGCAGCAATTAGAGAACATTC ATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAACAGTTGAAAAGTTCCGGCAGAGAATTCTGG CTCCCTGTAAGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCAAGCTGTGT TACAACTCGGAGTGAACAATGAAGATTCAAAGAGTGAAGATACACAACAGCAAATCATCAGGGAGACTTTCCATTTGGTATCTAAGAGAG ATGAAAATGTTTGTAATTTCCTAGAAGGAGGATTATTAATTGGAGGATCTGACAACAAACTGATTTATAGACATTATGCAACGTTATATT TTGTCTTCTGTGTGGATTCTTCAGAAAGTGAACTTGGCATTTTAGATCTAATTCAAGTATTTGTGGAAACATTAGACAAATGTTTTGAAA ATGTCTGTGAGCTGGATTTGATTTTCCATGTAGACAAGGTTCACAATATTCTTGCAGAAATGGTGATGGGGGGAATGGTATTGGAGACAA ATATGAATGAGATTGTTACACAAATTGATGCACAAAATAAGCTGGAAAAATCTGAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTAT CAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTA AATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGA GGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCT TGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGC CTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGA TCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAACATTATG ACTTTTTTCTCTTAGTTTAAATAAACTCCAAGGTAACTGGACTTCTAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAATAATTT TTTTTACAACCCTCTGACTCAACAAAGTAAATAAAAGTATATTTTATCACTATTAA >18503_18503_9_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000274458_AP3S1_chr5_115202367_ENST00000316788_length(amino acids)=356AA_BP=186 MGSAQLTDGGSARKPKMAVPAALILRESPSMKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLET ISFILEQAVYHNVKPAALQQQLENIHLRQDKAEAFVNTWSSMGQETVEKFRQRILAPCKLETVGWQLNLQMAHSAQAKLKSPQAVLQLGV NNEDSKSEDTQQQIIRETFHLVSKRDENVCNFLEGGLLIGGSDNKLIYRHYATLYFVFCVDSSESELGILDLIQVFVETLDKCFENVCEL DLIFHVDKVHNILAEMVMGGMVLETNMNEIVTQIDAQNKLEKSEAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_10_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000274458_AP3S1_chr5_115249059_ENST00000316788_length(transcript)=1292nt_BP=572nt GCGGCCGCCATTGGCTGGGTTCGGCGCAGCTAACAGACGGCGGCAGTGCGAGAAAGCCGAAGATGGCGGTCCCCGCGGCGCTGATCCTAC GGGAGAGCCCCAGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGATTTCCACGGTTGCTCACTCGGATTCTTCAAA AACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGGCATTTTCTCTAGAGAAACAAGATCTTCACC TAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAGCAGCTTTGCAGCAGCAATTAGAGAACATTC ATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAACAGTTGAAAAGTTCCGGCAGAGAATTCTGG CTCCCTGTAAGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTCAAGCAAAACTAAAATCTCCTCAAGCTGTGT TACAACTCGGAGTGAACAATGAAGATTCAAAGGCTGGCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTC CTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTC CCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTG GATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTG AGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTG TTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATT CCTTCTGCTATATGTCACACAGTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAACATTATGACTTTTTTCTCTTAGTTTAAATAA ACTCCAAGGTAACTGGACTTCTAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAATAATTTTTTTTACAACCCTCTGACTCAACA AAGTAAATAAAAGTATATTTTATCACTATTAA >18503_18503_10_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000274458_AP3S1_chr5_115249059_ENST00000316788_length(amino acids)=228AA_BP=186 MGSAQLTDGGSARKPKMAVPAALILRESPSMKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLET ISFILEQAVYHNVKPAALQQQLENIHLRQDKAEAFVNTWSSMGQETVEKFRQRILAPCKLETVGWQLNLQMAHSAQAKLKSPQAVLQLGV NNEDSKAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_11_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000515539_AP3S1_chr5_115202367_ENST00000316788_length(transcript)=1706nt_BP=602nt GACACTCTGGTGGGAAAGGCTCACGCTGCTGCTGGAGGGTCTTTAATAGCCAACATGTGTTAATACCCCTTTTAAATCAGTATCGAATAG GGGACCGAATTTTTGGTTGGTAGAAGAATCTAGCCTGGCTTTGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGAT TTCCACGGTTGCTCACTCGGATTCTTCAAAAACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGG CATTTTCTCTAGAGAAACAAGATCTTCACCTAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAG CAGCTTTGCAGCAGCAATTAGAGAACATTCATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAA CAGTTGAAAAGTTCCGGCAGAGAATTCTGGCTCCCTGTAAGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTC AAGCAAAACTAAAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGAGTGAAGATACACAACAGCAAATCATCA GGGAGACTTTCCATTTGGTATCTAAGAGAGATGAAAATGTTTGTAATTTCCTAGAAGGAGGATTATTAATTGGAGGATCTGACAACAAAC TGATTTATAGACATTATGCAACGTTATATTTTGTCTTCTGTGTGGATTCTTCAGAAAGTGAACTTGGCATTTTAGATCTAATTCAAGTAT TTGTGGAAACATTAGACAAATGTTTTGAAAATGTCTGTGAGCTGGATTTGATTTTCCATGTAGACAAGGTTCACAATATTCTTGCAGAAA TGGTGATGGGGGGAATGGTATTGGAGACAAATATGAATGAGATTGTTACACAAATTGATGCACAAAATAAGCTGGAAAAATCTGAGGCTG GCTTAGCAGGAGCTCCAGCCCGTGCTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCA GTATAAAAGTGCCAAACCTGCCCTCTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTT ACCATGCAGTTGTTTACCAAAAATAGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAA ATCAGTATGAAAGTTCAATGTTGCTGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAA ACTGCATTCCTGTGCCCACCTACGGCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGT ACAACCTAAAATGTTGGTGTTTTGTATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTTGGAGA ACCAAGTATGTATTGCATGAAAACATTATGACTTTTTTCTCTTAGTTTAAATAAACTCCAAGGTAACTGGACTTCTAAAGCACCTTTCTG TTTGCCTGATATCTACTTTAGCAATAATTTTTTTTACAACCCTCTGACTCAACAAAGTAAATAAAAGTATATTTTATCACTATTAA >18503_18503_11_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000515539_AP3S1_chr5_115202367_ENST00000316788_length(amino acids)=326AA_BP=156 MKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLETISFILEQAVYHNVKPAALQQQLENIHLRQD KAEAFVNTWSSMGQETVEKFRQRILAPCKLETVGWQLNLQMAHSAQAKLKSPQAVLQLGVNNEDSKSEDTQQQIIRETFHLVSKRDENVC NFLEGGLLIGGSDNKLIYRHYATLYFVFCVDSSESELGILDLIQVFVETLDKCFENVCELDLIFHVDKVHNILAEMVMGGMVLETNMNEI VTQIDAQNKLEKSEAGLAGAPARAVSAVKNMNLPEIPRNINIGDISIKVPNLPSFK -------------------------------------------------------------- >18503_18503_12_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000515539_AP3S1_chr5_115249059_ENST00000316788_length(transcript)=1322nt_BP=602nt GACACTCTGGTGGGAAAGGCTCACGCTGCTGCTGGAGGGTCTTTAATAGCCAACATGTGTTAATACCCCTTTTAAATCAGTATCGAATAG GGGACCGAATTTTTGGTTGGTAGAAGAATCTAGCCTGGCTTTGCATGAAGAAAGCAGTGTCACTGATAAATGCAATAGATACAGGAAGAT TTCCACGGTTGCTCACTCGGATTCTTCAAAAACTTCACCTGAAGGCTGAGAGCAGTTTCAGTGAAGAAGAGGAAGAAAAACTTCAAGCGG CATTTTCTCTAGAGAAACAAGATCTTCACCTAGTTCTTGAAACAATATCATTTATTTTAGAACAGGCAGTGTATCACAATGTGAAGCCAG CAGCTTTGCAGCAGCAATTAGAGAACATTCATCTTAGACAAGACAAAGCTGAAGCATTTGTCAATACGTGGTCTTCTATGGGTCAAGAAA CAGTTGAAAAGTTCCGGCAGAGAATTCTGGCTCCCTGTAAGCTAGAGACCGTTGGATGGCAGCTTAACCTTCAGATGGCTCACTCTGCTC AAGCAAAACTAAAATCTCCTCAAGCTGTGTTACAACTCGGAGTGAACAATGAAGATTCAAAGGCTGGCTTAGCAGGAGCTCCAGCCCGTG CTGTATCAGCTGTAAAGAATATGAATCTTCCTGAGATCCCAAGAAATATTAACATTGGTGACATCAGTATAAAAGTGCCAAACCTGCCCT CTTTTAAATAAAAATGTAAAAAGGCCACTCCCAGGTAAAATCCAGGGGGAAGAGTCATCTAAGTTTACCATGCAGTTGTTTACCAAAAAT AGAGGAGGAGAGTCTTAACTTTTGCTCTTGGATTTAAGTCAAGGTACTGTATAGAAGTTGTGTAAAATCAGTATGAAAGTTCAATGTTGC TGTTCTTGCTCAGTGATTTTAAAGAAATTGAGTAGTTCCTATGTGATTTTTTTTTTTCTTTTCTAAACTGCATTCCTGTGCCCACCTACG GCATGCCTCTATGTATTGGCTACTACAGTGTTTTAAAAAGTGTTTCAGATATTTCTCTAATTATGTACAACCTAAAATGTTGGTGTTTTG TATGGATCACAAGTGCAGCATTCCTTAATTCCTTCTGCTATATGTCACACAGTTGTTATTTGGAGAACCAAGTATGTATTGCATGAAAAC ATTATGACTTTTTTCTCTTAGTTTAAATAAACTCCAAGGTAACTGGACTTCTAAAGCACCTTTCTGTTTGCCTGATATCTACTTTAGCAA TAATTTTTTTTACAACCCTCTGACTCAACAAAGTAAATAAAAGTATATTTTATCACTATTAA >18503_18503_12_COMMD10-AP3S1_COMMD10_chr5_115469875_ENST00000515539_AP3S1_chr5_115249059_ENST00000316788_length(amino acids)=198AA_BP=156 MKKAVSLINAIDTGRFPRLLTRILQKLHLKAESSFSEEEEEKLQAAFSLEKQDLHLVLETISFILEQAVYHNVKPAALQQQLENIHLRQD KAEAFVNTWSSMGQETVEKFRQRILAPCKLETVGWQLNLQMAHSAQAKLKSPQAVLQLGVNNEDSKAGLAGAPARAVSAVKNMNLPEIPR NINIGDISIKVPNLPSFK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for COMMD10-AP3S1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for COMMD10-AP3S1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for COMMD10-AP3S1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
